 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Nanoparticles (NPs) have been emerging as potential players in modern medicine with clinical applications ranging from therapeutic purposes to antimicrobial agents. However, before applications in medical agents, some in vitro studies should be done to explore their biological responses.
Aim
In this study, protein binding, anticancer and antibacterial activates of zero valent iron (ZVFe) were explored.
Materials and methods
ZVFe nanoparticles were synthesized and fully characterized by X-ray diffraction, field-emission scanning electron microscope, and dynamic light scattering analyses. Afterward, the interaction of ZVFe NPs with human serum albumin (HSA) was examined using a range of techniques including intrinsic fluorescence, circular dichroism, and UV–visible spectroscopic methods. Molecular docking study was run to determine the kind of interaction between ZVFe NPs and HSA. The anticancer influence of ZVFe NPs on SH-SY5Y was examined by MTT and flow cytometry analysis, whereas human white blood cells were used as the control cell. Also, the antibacterial effect of ZVFe NPs was examined on Pseudomonas aeruginosa (ATCC 27853), Escherichia coli (ATCC 25922), and Staphylococcus aureus (ATCC 25923).
Results
X-ray diffraction, transmission electron microscope, and dynamic light scattering analyses verified the synthesis of ZVFe NPs in a nanosized diameter. Fluorescence spectroscopy analysis showed that ZVFe NPs spontaneously formed a complex with HSA through hydrogen bonds and van der Waals interactions. Also, circular dichroism spectroscopy study revealed that ZVFe NPs did not change the secondary structure of HSA. Moreover, UV–visible data presented that melting temperature (Tm) of HSA in the absence and presence of ZVFe NPs was almost identical. Molecular dynamic study also showed that ZVFe NP came into contact with polar residues on the surface of HSA molecule. Cellular assays showed that ZVFe NPs can induce cell mortality in a dose-dependent manner against SH-SY5Y cells, whereas these NPs did not trigger significant cell mortality against normal white bloods in the concentration range studied (1–100 µg/mL). Antibacterial assays showed a noteworthy inhibition on both bacterial strains.
Conclusion
In conclusion, it was revealed that ZVFe NPs did not induce a substantial influence on the structure of protein and cytotoxicity against normal cell, whereas they derived significant anticancer and antibacterial effects.
Introduction
In the past few years, there has been a rapid interest in the application of nanoparticles (NPs) in the biochemical sensing,Citation1 diagnosis,Citation2 and therapy systems.Citation3 NPs are expected to play an important role in the formation of several technological,Citation4 biological,Citation5 and biomedical innovations.Citation6 To date, a large number of NPs have been designed and synthesized, especially those made from noble metals NPs.Citation7,Citation8 Through the potential features of pharmacodynamics and pharmacokinetics, metal NPs can be offered as an excellent candidate in the medicinal chemistry.Citation9,Citation10 Hence, synthesis and development of unique anticancerCitation11,Citation12 and antimicrobial drugs derivativesCitation12 might be productive in the design of biologically active agents. Zero valent iron (ZVFe) NPs that show unique optical, electronic, and chemical features provide efficient uses in chemistry, biology, and environmental sciences because of suitable surface bioconjugation with molecular probes and significant optical or magnetic properties.Citation13 Especially in modern biological researches, ZVFe NPs with the correct substrate could be implemented to take advantage of the degradation potentials of microbial communities.Citation14 Also, it has been well documented that ZVFe NPs could increase head and neck cancer cells mortalityCitation15,Citation16 and ovarian cancer cellsCitation16 through mitochondria-mediated apoptosis or autophagy.
As nanobiomedicine has been developing a growing interest in the application of NPs as anticancer or antimicrobial drugs, determining their adverse effects regarding their impacts on the structural changes of some carrier proteins like human serum albumin (HSA) and cytotoxicity against normal cells become crucially important.Citation17
NPs may become lodged in tissues, and the high durability and reactivity of some NPs raise concerns of their outcomes in the healthy systems.Citation18,Citation19 Indeed, at this time, not enough data exist to evaluate the biomedical exposure for most engineered NPs. Some NPs that accumulate in animal tissues may have the ability to easily pass through the cell membranes or cross the blood–brain barrier and attack the cells. This may be a potential feature for implementations such as targeted drug delivery and other disorders treatments, but could lead to unintended influences in some cases. Thus, the influence of NPs targeted to human is inevitable as they can come into contact with body through biomedical applications. Furthermore, the small size of NPs is similar to that of most biomacromolecules, and this results in the interaction of NPs with proteins. Therefore, it is vital to explore in advance the detailed interaction mechanism between NPs and biological systems such as cell and proteins before using them as medical agents. For example, Maji et alCitation20 showed that plant extract-mediated silver NPs provide potential protein binding and antibacterial and anticancer activities. Karthika et alCitation21 reported the fabrication of silver and gold NPs by plant extract and investigated their DNA/protein interactions, anticancer and antimicrobial activities. Khan and Al-ThabaitiCitation22 revealed that green-fabricated ZVFe NPs have a strong albumin binding and significant antimicrobial activities.
Therefore, we aimed to fabricate ZVFe NPs with chemical method and investigate their interaction with HSA by biophysical and molecular docking studies. Afterwards, their anticancer and antibacterial effects against human neuroblastoma cell line (SH-SY5Y) and three bacterial strains were investigated, respectively.
HSA is known as a soluble protein in the blood circulatory system showing a number of physiological roles including transporters for a variety of organic and inorganic compounds and controlling the osmotic pressure and pH of blood.Citation23 HSA is composed of 585 residues with dominant α-helix conformation, three structural domains (I–III), and numerous efficient drug binding sites.Citation24 HSA has been widely used as one of the most extensively studied proteins, not only because of its biomedical significance, low cost, availability, and wide approval in the pharmaceutical industry, but also due to its ligand-binding properties.Citation25,Citation26
The white blood cells (WBCs) and SH-SY5Y cellsCitation27,Citation28 are highly sensitive and commonly used for assessing the NP toxicity and their anticancer effects, respectively.
Also, the antibacterial effect of ZVFe NPs was investigated on Pseudomonas aeruginosa, Escherichia coli, and Staphylococcus aureus.
Experimental
Synthesis of ZVFe NPs
The method of synthesis was modified according to Yuvakkumar et al.Citation29 For the synthesis of ZVFe NPs, an aqueous solution of 0.14 M FeSO4⋅7H2O was dissolved in a 4/1 (v/v) ethanol/water mixture and stirred well with an electric rod for 15 minutes. Then, sodium borohydride solution (100 mL of 0.2 M) was added dropwise into the mixture with gentle stirring for another 30 minutes. The product was separated by centrifugation at 5,000 rpm for 15 minutes and washed three times with ethanol to remove excess borohydrate. Prepared NPs were finally dried under vacuum overnight.
X-ray diffraction analysis
Powder X-ray diffraction (XRD) analysis of ZVFe NP was carried out using Bruker D8 Advance Diffractometer (Bruker AXS, Karlsruhe, Germany) with Cu Kα radiation (λ=1.54 Å) at 45 kV and 40 mA. ZVFe NP was placed in a glass holder and scanned over a 2θ range of 20°–80°. Scan rate was 0.5 min−1.
Field-emission transmission electron microscope study
Field-emission transmission electron microscope (FETEM) analysis of ZVFe NP was carried out using a Philips JSM-6360LA instrument (Philips, Eindhoven, Netherlands). Samples were prepared by sonication for 30 minutes using a sonicator (Misonix S3000).
Dynamic light scattering study
Hydrodynamic diameter, polydispersity index (PDI), and zeta potential of ZVFe with a concentration of 50 µM was measured by using a dynamic light scattering (DLS; Malvern, He/Ne 633 nm laser) at room temperature. For each sample, three independent measurements were done with five scans for each run.
Fluorescence spectroscopy
Fluorescence emission spectra were recorded at three different temperatures of 298, 310, and 315 K on a Cary Eclipse VAR-IAN fluorescence spectrophotometer (Hitachi, Japan). All measurements were carried out at an excitation wavelength of 280 nm and spectral bandwidths of 5 nm. Static fluorescence spectra of HSA and ZVFe NP mixtures were determined in 20 mM phosphate buffer, pH 7.5 at a fixed protein concentration of 2 µM, and varying concentrations (1–15 µM) of ZVFe NP. The spectrum of the buffer and ZVFe NP samples were subtracted from those of protein–NP mixture.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded from 190 to 260 nm at 0.2 nm with a scan of rate of 2 points per second at 298 K, with three scans averaged for each CD spectrum. The data were expressed as ellipticity (mdeg), which was obtained in mdeg directly from the instrument. The far CD records of HSA (3 µM) was measured in the absence and presence of varying concentrations of ZVFe NP (3–30 µM), using a spectropolarimeter (model 215; Aviv, Lakewood, NJ, USA). Each record of HSA was subtracted from that of ZVFe NP sample as control.
Thermal melting measurement
The absorbance of HSA as a function of temperature was measured at 295 nm using a UV–visible (UV–vis) spectrophotometer equipped with a thermal bath. The absorbance of HSA (3 µM) in the absence and presence of ZVFe NP (3 M) was recorded as a function of temperature over the range of 30°C–90°C at a heating rate of 1°C min−1. The melting temperature (Tm) was calculated from the midpoint of the melting curves.
Molecular docking study
The ZVFe NP was clustered in our previous study.Citation30 Briefly, geometry of ZVFe was optimized by the CASTEP module in Materials Studio 5.0 developed by Accelrys Software Inc. Local density approximation functional and ultrasoft pseudo potentials were used to model electron–ion interactions. HEX 6.3 software was used to run molecular docking study. Visualization of the ligand site was performed by using CHIMERA (www.cgl.ucsf.edu/chimera) and PyMOL (http://pymol.sourceforge.net/) tools.
Cell culture
SH-SY5Y cell line was purchased from Pasture Institute (Tehran, Iran) and was cultured in plastic tissue culture flasks in Eagle’s minimum essential medium and Ham’s F12 (1:1) supplemented with 10% FBS and 100 U/mL penicillin–streptomycin–neomycin mixture. The cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. WBCs were separated and cultured based on the method reported in our previous paper.Citation31 Blood samples were obtained after written informed consent was confirmed in accordance with the Declaration of Helsinki, according to protocol approved by the Ethical Committee of Pharmaceutical Sciences Branch, Islamic Azad University of Tehran, Tehran, Iran.
3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide assay
MTT test has been carried out to evaluate cells viability. Following the 24 hours exposure to varying concentrations (1–100 µg/mL) of ZVFe and replacement of the medium, the cells (5×103 per well) were incubated in 37°C for 4 hours with MTT (Sigma-Aldrich, St Louis, MO, USA) solution (0.5 mg/mL). Then, supernatants were gently removed and formazan salt was extracted in DMSO. Measurement of the optical density was performed at 570 nm using a microplate reader (Expert 96, Asys Hitch, Ec Austria). DMSO and NP were used as blank references.
Flow cytometry analysis
Apoptosis induction by ZVFe NPs was assessed by flow cytometric analysis of negative control and ZVFe NPs (IC50 concentration) incubated cells according to the manufacturer’s protocol (BD Biosciences, San Jose, CA, USA). Briefly, SH-SY5Y cells (3×105 per well) were treated with IC50 concentration of ZVFe NPs for 24 hours, collected, washed, and resuspended in 500 µL of 1× Annexin-binding buffer, incubated at room temperature with Annexin V-FITC and PI stain, and analyzed by flow cytometry (BD Biosciences).
Antibacterial assay
The antibacterial potential of synthesized ZVFe NP was analyzed initially by well-diffusion method using following Gram-negative and Gram-positive strains of bacteria.
Totally, three bacterial strains, including P. aeruginosa (ATCC 27853), E. coli (ATCC 25922) as Gram-negative and S. aureus (ATCC 25923) as Gram-positive strains, were used for analyzing the antibacterial potential of ZVFe NP. Bacteria were subcultured from pure cultures of different strains of bacteria on Mueller–Hinton Broth for overnight at 37°C. The turbidity of bacterial culture was adjusted to 0.5 McFerland (1.5×108 CFU/mL) standard. Each bacterial strain was swabbed uniformly onto separate Muller–Hinton agar plates using sterile cotton swabs under sterile condition. Wells with 8 mm diameters were prepared by punching a sterile cork borer onto agar plates and removing the agar to form a well. One hundred microliters of 250 µg/mL (w/v) concentration of ZVFe NP were then applied to wells. After incubation for 24 hours, at 35°C–37°C, the zones of inhibition around the wells were measured in millimeter using a caliper. The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of ZVFe NP were determined against bacterial strains by the microdilution method.Citation31
Statistical analysis
All experiments were carried out in triplicate and data were shown as mean ± SD of three independent experiments. The significance of difference between control- and sample-treated groups was calculated by one-way ANOVA followed by Dunnett’s t-test using SPSS version 20 (SPSS Inc., Chicago, IL, USA). P-value <0.05 was nominated as the level of significance.
Results and discussion
ZVFe NP characterization
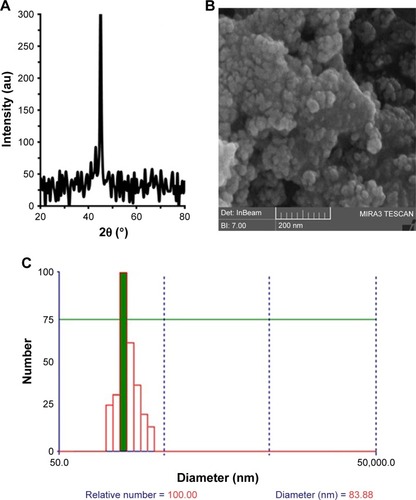
The oxidation state and phase of ZVFe NP were analyzed by employing XRD technique (). It was shown that the Fe NP was in its zero valent state and no appearance of oxide/hydroxide ligand was revealed on the surfaces of the ZVFe NPs. The peak appearing at 2θ value of ~45° is in good agreement with the value for Fe0 reported.Citation32
Figure 1 The XRD pattern of ZVFe NP (A); FESEM image of ZVFe NPs (B); DLS histogram of ZVFe NPs (C).
Abbreviations: DLS, dynamic light scattering; FESEM, field-emission scanning electron microscope; XRD, X-ray diffraction; ZVFe NPs, zero valent iron nanoparticles.
Particle size was calculated with the XRD by using Scherer equation:Citation32
FESEM is widely employed as a potential technique to detect the size distribution and morphology of particles.Citation33 The diameter and morphology of ZVFe NPs are shown in . demonstrated that the diameter of ZVFe NPs was about 30 nm and NPs exhibited homogeneous and nearly spherical shape. By using DLS, we monitored the hydrodynamic radius of ZVFe NPs at room temperature. DLS exhibits a hydrodynamic radius value of 83.88 nm with a PDI value of 0.21 (). Zeta potential value was also obtained from the DLS study. It was determined that the zeta potential value of ZVFe NPs was around −31.91±6.24 mV. These data also confirm the presence of monodispersed ZVFe NPs in the solution and high colloidal stability of the fabricated NPs.
Thermodynamic parameters
Fluorescence spectroscopy is a sensitive and simple tool in the protein conformational changes study. Quenching studies utilizing intrinsic Trp fluorescence of proteins can yield important data regarding the microenvironment of the aromatic fluorophore.Citation34–Citation36 The quenching mechanism can be classified as dynamic quenching (collisional system) or static quenching (complex formation) between fluorophores and quenchers.Citation37 Dynamic and static quenching can be distinguished by their different relations on temperature and viscosity.Citation38 The probability of fluorescence quenching with ligands depends on the rate of collision of the quencher and the ground-state fluorophore (static quenching) or excited fluorophore (dynamic quenching).Citation39
As shown in , the fluorescence intensity of HSA decreases as the ZVFe NP concentration increases at 298 K (), 310 K (), and 315 K (). To determine the mode of quenching and NP-induced conformational changes, further analysis was done.
Figure 2 Fluorescence quenching of HSA upon interaction with ZVFe NP at three different temperatures: 298 K (A), 310 K (B), and 315 K (C).
Note: The fluorescence spectra of HSA (2 µM) in the presence of varying concentrations (0 [dark blue], 1 [brown], 2 [green], 4 [bright blue], 6 [purple], 8 [gray], 10 [bright brown], and 15 [red] µM) of ZVFe NPs.
Abbreviations: HSA, human serum albumin; ZVFe NPs, zero valent iron nanoparticles.
![Figure 2 Fluorescence quenching of HSA upon interaction with ZVFe NP at three different temperatures: 298 K (A), 310 K (B), and 315 K (C).Note: The fluorescence spectra of HSA (2 µM) in the presence of varying concentrations (0 [dark blue], 1 [brown], 2 [green], 4 [bright blue], 6 [purple], 8 [gray], 10 [bright brown], and 15 [red] µM) of ZVFe NPs.Abbreviations: HSA, human serum albumin; ZVFe NPs, zero valent iron nanoparticles.](/cms/asset/e960dcd0-65f6-4089-9851-d24d5747c274/dijn_a_12190680_f0002_c.jpg)
Mode of interaction
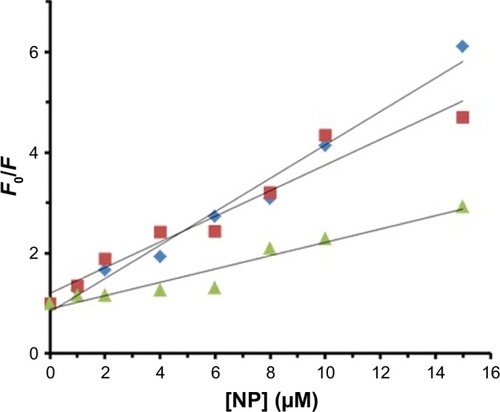
Static quenching is invariably related to the temperature, whereas collision quenching is directly related to the temperature. Therefore, fluorescence experiments were performed at three different temperatures (298, 310, and 315 K) to reveal the mode of fluorescence quenching by using the Stern– Volmer equation (EquationEquation 1(1) ):Citation40,Citation41
Table 1 KSV parameters at three different temperatures for the interaction of HSA with ZVFe NPs
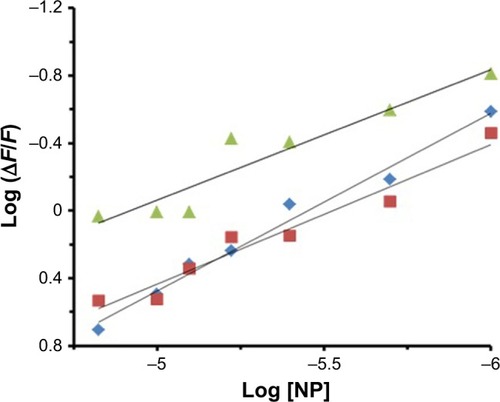
Binding parameters
Several equations can be used for calculating the binding constant of the interaction between a protein and a ligand. One of the most routinely used equations is the Hill equation, since it can determine the correct stoichiometry for the binding sites of protein and ligands.Citation42 To determine the binding parameters between ZVFe NP and HSA, EquationEquation 2(2) was used:Citation42
Table 2 Binding parameters at three different temperatures for the interaction of HSA with ZVFe NPs
Figure 4 Hill plot for the binding of HSA with ZVFe NPs at 298 K (■), 310 K (♦), and 315 K (▲).
Abbreviations: HSA, human serum albumin; ZVFe NPs, zero valent iron nanoparticles.
Thermodynamic parameters
In general, intermolecular interacting forces between a ligand and a protein include hydrogen bonding, van der Waals interaction, electrostatic forces, and hydrophobic network.Citation43 The thermodynamic parameters such as enthalpy change (ΔH°), standard entropy change (ΔS°), and standard Gibbs free energy change (ΔG°) of interaction were determined using van’t Hoff equation (EquationEquation 3(3) ):Citation43
Table 3 Thermodynamic parameters at three different temperatures for the interaction of HSA with ZVFe NPs
Figure 5 Van’t Hoff plot for the binding of HSA with ZVFe NPs.
Abbreviations: HSA, human serum albumin; ZVFe NPs, zero valent iron nanoparticles.
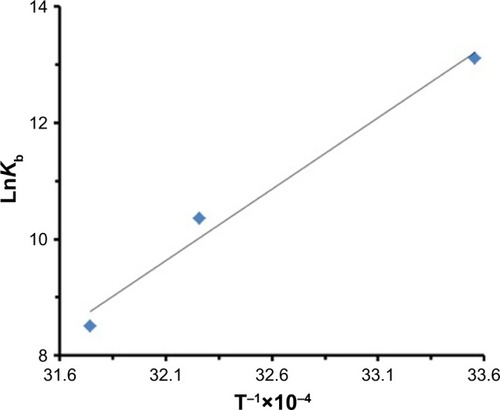
The sign and magnitude of thermodynamic parameters are associated with the involving forces during protein– ligand interaction. ΔH°>0 and ΔS°>0 exhibit hydrophobic interactions, ΔH°<0 and ΔS°<0 show hydrogen bonds and van der Waals interactions, and ΔH°<0 and ΔS°>0 represent electrostatic interactions.Citation40 Negative values of ΔH° (−53.21 kJ/mol) and ΔS° (−42.44 kJ/mol) showed that the NP-induced conformational changes of protein is induced by an enthalpy-driven manner.Citation40 Likewise, ΔH°<0 and ΔS°<0 are characteristics of hydrogen bonds and van der Waals interactions in aqueous solution. Due to the large surface-to-volume-ratio of NPs, water molecules are substantially adsorbed on their surface and can mediate the establishment of hydrogen bonds between NP and protein.
Water is an extremely good acceptor and donator of hydrogen bonds. So, if a protein is partially unfolded in solution due to its interaction with NP, geometrically perfect hydrogen bonds can be formed with a very polarized molecule, water, on the surface of NP.
Secondary structural studies
Due to the influence of NP on the spectra of HSA, it is crucial to determine the structural changes of HSA in the presence of ZVFe NP. CD is a sensitive technique to monitor the secondary structural changes of protein.Citation45 Far UV–CD (190–260 nm) spectra of HSA and HSA–ZVFe NP are shown in . In HSA spectrum, there are two bands in the UV region, at 208 and 222 nm, which are characteristic of the α-helical conformation of HSA. Both transitions at 208 and 222 nm are known as a broad n→π* transition for the peptide bond of the α-helix.Citation46 shows that the spectra obtained for HSA in the presence of various concentrations of ZVFe NPs were also similar to the spectrum of HSA in the absence of NPs. Therefore, it can be concluded that the relative α-helix structure of HSA remains unchanged in the presence of ZVFe NP.
UV–vis spectroscopy study
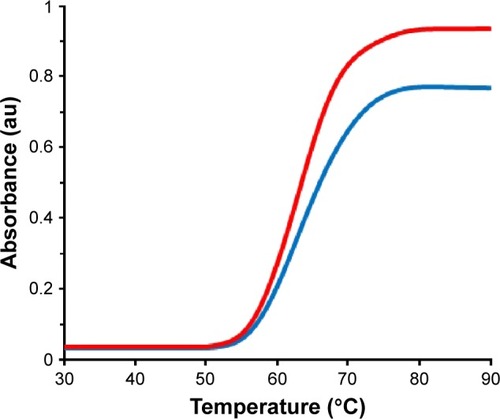
UV–vis absorption measurements are commonly used to investigate structural changes of proteins and complex formation.Citation47 Thermal behaviors of HSA in the absence and presence of ZVFe NP can provide detailed information about the interaction strength of NPs with proteins. It is well known that, when the temperature in the solution goes up, the folded state of protein shifts toward the denatured structure and generates a hyperchromic influence on the absorption spectra of protein residues. In order to reveal this transition process, the Tm point, which is defined as the temperature, where half of the total native protein are denatured, is usually introduced.
The melting curves of HSA at 295 nm in the absence and presence of ZVFe NPs are presented in . Here, the thermal denaturation experiment revealed Tm values of around 64°C and 65°C for HSA in the presence and absence of the ZVFe NP, respectively. These almost identical values of Tm for HSA in the absence and presence of ZVFe NP are in good agreement with CD data. Therefore, it may be concluded that HSA structure remains almost unchanged after interaction with ZVFe NPs.
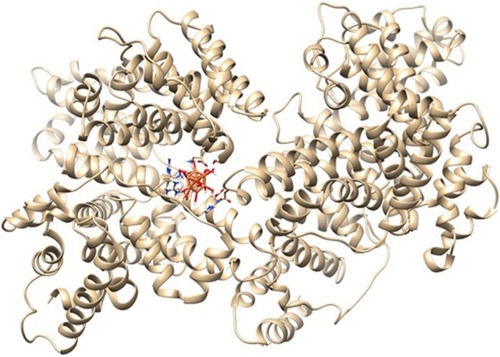
Molecular docking study
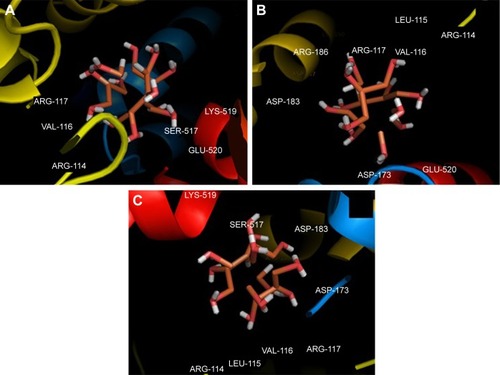
Molecular docking study was carried out to support the experimental data indicating that hydrogen bonds are the main forces in the interaction of HSA with ZVFe NPs. The X-ray crystallographic 3D structure of HSA at 2.5 A resolution (Protein Data Bank [PDB] ID: 1AO6) was downloaded from the online protein data bank Research Collaboratory for Structural Bioinformatics (RCSB) PDB (http://www.pdb.org).Citation48 The molecular docking was performed with designed ZVFe cluster. The resulting E-value was found to be −280.64. The docked complex is shown in . The ligand with surrounding active site residues within 3.5 Å is given in . The nearest interacting residues are SER-517, ASP-173, ASP-183, ARG-114, LEU-115, VAL-116, ARG-117, LYS-519, GLU-520, LYS-519, and ARG-186. These residues possess hydroxyl, carboxyl, and amine groups, which provide a potential medium to form hydrogen bonds with water molecules on ZVFe NP surface. Indeed, it may be indicated that the type of interaction between NPs and macromolecules depends on a number of factors, such as concentration, pH, temperature, and type of NPs/proteins. Actually, the type of interaction between NPs and proteins can be weakened or strengthened by changing these factors. It seems ZVFe NPs form hydrogen bonds and van der Waals interaction with HSA, which are known as relatively weak forces. Therefore, these interactions do not induce substantial conformational changes on the HSA structure.
MTT assay
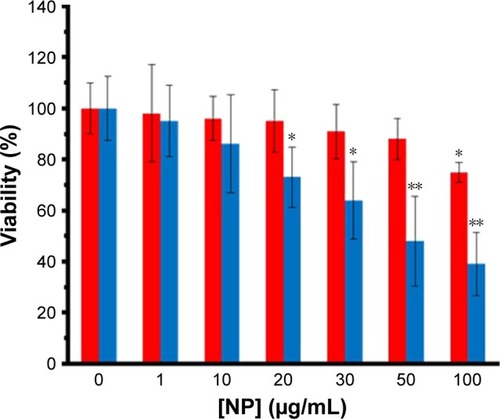
Different cells were employed to reveal the in vitro cytotoxic effect of ZVFe NPs. SH-SY5Y and WBCs were incubated with different concentrations (1, 10, 20, 30, 50, 100 µg/mL) of ZVFe NPs for 24 hours. Cell viability was then assessed by MTT assay. It was observed that ZVFe NPs induced mortality of cancer cells () and growth inhibitions were 95.37%±14.02%, 86.13%±19.27%, 73.31%±11.87% (P<0.05), 64.19%±15.16% (P<0.05), 48.21%±17.62% (P<0.01), and 39.11%±12.51% (P<0.01) in SH-SY5Y cells after treatment with 1, 10, 20, 30, 50, and 100 µg/mL of ZVFe NPs, respectively. However, it was revealed that the WBCs viability was 98%±19.05%, 96%±8.68%, 95%±12.16%, 91%±10.66%, 88%±8.09%, and 75%±3.97% (P<0.05), after treatment with 1, 10, 20, 30, 50, and 100 µg/mL of ZVFe NPs, respectively. As presented in , some cytotoxic activity was already revealed at 20 µg/mL dose of ZVFe NPs on SH-SY5Y cells, whereas maximal effect was obtained at a concentration of 100 µg/mL in WBCs. Therefore, it was concluded that ZVFe NPs with concentrations of <100 µg/mL did not induce cytotoxicity on normal cells, so they could be used as potential candidates to act as nontoxic drug carriers or anticancer agents. The IC50 concentration of ZVFe NPs against SH-SY5Y cells and WBCs were calculated to be 47.89±81 µg/mL and >100 µg/mL, respectively. In fact, the nano state of ZVFe intensified the difference between cytotoxicity outcome in cancer and normal cells. Based on these observations, the present paper focused on SH-SY5Y cells for subsequent assays.
Figure 10 The viability percent of SH-SY5Y (blue bars) cells and WBCs (red bars) in the presence of varying concentrations of ZVFe NPs after 24 hours.
Note: *P<0.05 and **P<0.01 represent the significant differences between ZVFe NPs-treated groups and control.
Abbreviations: WBCs, white blood cells; ZVFe NPs, zero valent iron nanoparticles.
Quantification of apoptosis
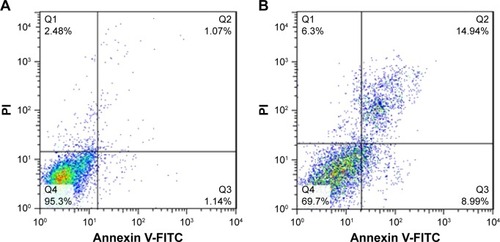
Quantification of apoptosis induction by ZVFe NPs on SH-SY5Y cells (IC50 concentration) was carried out by using flow cytometry. displays the ZVFe NPs-triggered apoptosis in SH-SY5Y cells after 24 hours incubation at IC50 concentration. The flow cytometry analysis results indicated that the rate of early apoptosis (Q3), late apoptosis (Q2), and necrosis (Q1) in control cells were 1.14%, 1.07%, and 2.48%, respectively (). However, treatment of the SH-SY5Y cells with IC50 concentration of ZVFe NPs for 24 hours enhanced the rate of early apoptosis (8.99%, P<0.001), late apoptosis (14.94%, P<0.001), and necrosis (6.3%, P<0. 01; ).
Antibacterial assay
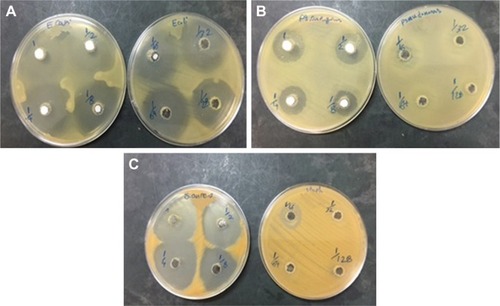
The antibacterial activity of ZVFe NP against one Gram-positive and two Gram-negative strains of bacteria are illustrated in . Also, the inhibition zone diameter of ZVFe NP against E. coli, P. aeruginosa, and S. aureus is displayed in . The results showed a noteworthy inhibitory effect of ZVFe NPs on both bacterial strains. shows the MIC and MBC of ZVFe NP against three strains of bacteria. It was observed that ZVFe NP induced strong antibacterial effect against E. coli, P. aeruginosa, and S. aureus with an MIC of 1.96, 31.25, and 15.75 µg/mL, respectively. The MBC observed in the present study was 1.96 µg/mL for E. coli, 31.25 µg/mL for S. aureus, and 62.5 µg/mL for P. aeruginosa.
Table 4 MIC and MBC of ZVFe NP against three strains of bacteria
Discussion
In pharmaceutical industry, it is critical to determine and specify the pharmacokinetics and pharmacological influence of drugs like NPs.Citation49,Citation50 It is invariably assumed that the probable influence of drugs (NPs) heavily depends on their binding affinity to carrier proteins such as albumin.Citation51 The interaction of NPs with proteins also affects the rate at which the NPs are delivered to targeted sites of reaction.Citation52 Hence, analyzing the interaction of NPs with albumin is essential not only because the unbound NPs may affect a number of overcritical pharmacokinetic constants including the constant distribution volume, but also because it provides a crucial effect to express the administration regimen dose.Citation53 Hence, it is important to examine that albumin can eventually influence the NP’s absorption, tissue distribution and pharmacodynamics, metabolism, and excretion features. In addition, the application of NPs, which can enter the body through the different routes and can interact with the albumin, could alter the conformation of albumin.
Uncontrolled side effects such as pH, temperature, and other denaturing conditions can influence the binding of an NP to albumin.Citation54 Indeed, conformational changes of protein in the presence of NPs can weaken or strengthen the binding affinity of proteins to NPs. Furthermore, a comprehensive characterization of albumin in the presence of NPs and thermodynamic parameters is also necessary not only for understanding its pivotal physiological features, but also most recently to develop nanosystems as targeted delivery vehicles for several therapeutic and diagnostic NPs. Hence, the data about NP–protein interaction may provide potential and central details in the field of nanomedicine.
We showed that ZVFe NP binds to HSA through hydrogen bonds and does not play a significant role in the structural changes of HSA. The reason that hydrogen bonds do not induce a net destabilizing/stabilizing impact on protein conformation lies in the fact that in order to establish protein–NPs hydrogen bonds, another set of hydrogen bonds on the surface of protein should be disrupted.Citation55 Interaction of protein with NP causes destabilization of protein conformation and exposure of the hydrophobic core. Contacting the dipoles that form the hydrogen bonds with these hydrophobic patches is energetically unfavorable. So, the overall effect of hydrogen bonds in NP-induced conformational alterations of proteins is that of inducing structural destabilization, not denaturation.
Biophysical study of zinc oxide interaction with albumin demonstrated no structural perturbation in the secondary structure of protein; however, some minor tertiary changes were reported.Citation56 Gold NPs were indicated to cause conformational changes in the structure of albumin in a dose-dependent manner,Citation57 whereas no major conformational change was reported for albumin when adsorbed onto the carbon C60 fullerene NP.Citation58 The interaction of magnetic iron oxide NPs with albumin was also investigated by spectroscopic methods.Citation59 The results indicated that the interaction was spontaneous and the electrostatic interactions played key roles in the interaction process. In addition, secondary structural alteration of albumin in the presence of MNPs was shown. It seems that the chemical composition of NPs may play an important role in inducing conformational changes of proteins upon interaction.
One of the crucial challenges in the development of anticancer drugs is increasing the therapeutic effectiveness of anticancer NPs and minimizing their side effects on normal cells. In our MTT assay, ZVFe NPs demonstrated a broad range of cytotoxicity with most mortality effect on SH-SY5Y cells but not on normal WBCs. Although, the mechanism of cancer-specific toxicity of ZVFe NPs is still unknown, this selective killing effect of ZVFe NPs in cancer cells is clinically important. Recently, selectivity in killing a wide spectrum of cancer cells by NPs including zinc oxide NPs,Citation60 iron oxide NPs,Citation61 and copper oxide NPsCitation62 has been reported.
However, oxidative stress induced by ZVFe NPs in human bronchial epithelial cells has been reported.Citation63 Therefore, these conflicting results may suggest that further studies should be done to explore the mechanisms behind the cytotoxic effects of NPs.
The effect of ZVFe NPs on bacterial activity was also investigated. The synthesized ZVFe NPs showed a noteworthy inhibitory effect on tested pathogenic bacteria. Based on these findings, bactericidal action of ZVFe NPs was affective. The different sensitivities of Gram-positive and Gram-negative bacteria against ZVFe NPs could be explained in accordance with the morphological differences between these microorganisms, due to different polarities of their cell wall. Other studies have demonstrated that the size of NPs can also affect their bactericidal activities. For example, Lee et alCitation64 reported that the inactivation of E. coli by ZVFe NPs could be induced by infiltration of the NPs through E. coli membranes. A number of other investigations on zinc oxideCitation65,Citation66 and magnesium oxide NPsCitation67 have also shown that antibacterial activity of NPs depend on their dimension.
In this study, the size of ZVFe NPs was about 30 nm, which enabled the NPs to cross the cell membrane and induce some adverse effects on the viability of bacteria. It may be suggested that ZVFe NPs could interact with intracellular oxygen, causing oxidative stress induction and finally driving cell membrane leakage.Citation64 Herein, the concentration of ZVFe NPs was also a crucial factor to induce bactericidal activity against pathogenic bacteria. This behavior was reported by other studies when they explored the antimicrobial impacts of silver,Citation68 zinc oxide,Citation65 and iron oxideCitation69 NPs on S. aureus and E. coli. Also, it should be noted that the fabrication route of NPs also influences the bactericidal activity. Indeed, different synthetic methods can form different NPs with various physicochemical and antibacterial properties.Citation70
Conclusion
The results of this study indicate that ZVFe NPs potentially bind to HSA through hydrogen bonds and van der Waals interactions and induce marginal structural changes on the HSA structure. Cytotoxicity assay revealed the selective killing of cancer cells by ZVFe NPs. Antibacterial examination showed that ZVFe NPs provide a potential antibacterial effect against E. coli, P. aeruginosa, and S. aureus.
Therefore, it may be concluded that ZVFe NP may be used in medicine as a potential anticancer and/or antibacterial drug. However, it should be noted that some in vivo studies are required to more explore the anticancer and antibacterial activities of ZVFe NPs.
Acknowledgments
The research has been supported by Tehran University of Medical Sciences and Health Services grant no. 96033036163, Tehran, Iran.
Disclosure
The authors report no conflicts of interest in this work.
References
- BurnsASenguptaPZedaykoTBairdBWiesnerUCore/Shell fluorescent silica nanoparticles for chemical sensing: towards single-particle laboratoriesSmall20062672372617193111
- KahnNLavieOPazMSegevYHaickHDynamic nanoparticle-based flexible sensors: diagnosis of ovarian carcinoma from exhaled breathNano Lett201515107023702826352191
- ChenPCMwakwariSCOyelereAKGold nanoparticles: from nano-medicine to nanosensingNanotechnol Sci Appl200814524198460
- GehrkeIGeiserASomborn-SchulzAInnovations in nanotechnology for water treatmentNanotechnol Sci Appl20158125609931
- BlancoEShenHFerrariMPrinciples of nanoparticle design for overcoming biological barriers to drug deliveryNat Biotechnol201533994195126348965
- duXLiXXiongLZhangXKleitzFQiaoSZMesoporous silica nanoparticles with organo-bridged silsesquioxane framework as innovative platforms for bioimaging and therapeutic agent deliveryBiomaterials2016919012727017579
- TeimouriMKhosravi-NejadFAttarFGold nanoparticles fabrication by plant extracts: synthesis, characterization, degradation of 4-nitrophenol from industrial wastewater, and insecticidal activity – a reviewJ Clean Prod2018184740753
- TianJZhaoZKumarABoughtonRILiuHRecent progress in design, synthesis, and applications of one-dimensional TiO2 nanostructured surface heterostructures: a reviewChem Soc Rev201443206920693725014328
- LingDLeeNHyeonTChemical synthesis and assembly of uniformly sized iron oxide nanoparticles for medical applicationsAcc Chem Res20154851276128525922976
- ZhengHZhangYLiuLOne-pot synthesis of metal-organic frameworks with encapsulated target molecules and their applications for controlled drug deliveryJ Am Chem Soc2016138396296826710234
- PraveenPARamesh BabuRBalajiPMurugadasAAkbarshaMALaser assisted anticancer activity of benzimidazole based metal organic nanoparticlesJ Photochem Photobiol B201818021822429459313
- PatraNKarDPalABeheraAAntibacterial, anticancer, anti-diabetic and catalytic activity of bio-conjugated metal nanoparticlesAdv Nat Sci Nanosci Nanotechnol201893035001
- PérezNRuiz-RubioLVilasJLRodríguezMMartinez-MartinezVLeónLMSynthesis and characterization of near-infrared fluorescent and magnetic iron zero-valent nanoparticlesJ Photochem Photobiol A201631517
- LefevreEBossaNWiesnerMRGunschCKA review of the environmental implications of in situ remediation by nanoscale zero valent iron (nZVI): behavior, transport and impacts on microbial communitiesSci Total Environ201656588990126897610
- HuangK-JWuS-RShiehD-BZero-valent iron nanoparticles inhibited head and neck cancer cells growth: a pilot evaluation and mechanistic characterizationFree Radic Biol Med2017108S39
- ShiehD-BYangL-XLeeW-TZero-valent iron based nanoparticles selectively inhibit cancerous cells through mitochondria-mediated autophagyPaper presented at: IEEE 17th International Conference on Nanotechnology (IEEE-NANO)July 25–28, 2017Pittsburg, PA, USA
- NelAEMädlerLVelegolDUnderstanding biophysicochemical interactions at the nano-bio interfaceNat Mater20098754355719525947
- BaekMChungHEYuJPharmacokinetics, tissue distribution, and excretion of zinc oxide nanoparticlesInt J Nanomedicine20127308122811602
- ParkEJLeeGHYoonCTissue distribution following 28 day repeated oral administration of aluminum-based nanoparticles with different properties and the in vitro toxicityJ Appl Toxicol201737121408141928840595
- MajiABegMMandalAKSpectroscopic interaction study of human serum albumin and human hemoglobin with Marsilea quadrifolia leaves extract mediated silver nanoparticles having antibacterial and anticancer activityJ Mol Struct20171141584592
- KarthikaVArumugamAGopinathKGuazuma ulmifolia bark-synthesized Ag, Au and Ag/Au alloy nanoparticles: photocatalytic potential, DNA/protein interactions, anticancer activity and toxicity against 14 species of microbial pathogensJ Photochem Photobiol B201716718919928076823
- KhanZAl-ThabaitiSAGreen synthesis of zero-valent Fe-nanoparticles: catalytic degradation of rhodamine B, interactions with bovine serum albumin and their enhanced antimicrobial activitiesJ Photochem Photobiol B201818025926729477891
- Kragh-HansenUHuman serum albumin: a multifunctional proteinOtagiriMChuangVTGAlbumin in MedicineAmsterdam, NetherlandsSpringer2016124
- GaoXBiHJiaJTangLSpectroscopic and in silico study of binding mechanism of cynidine-3-O-glucoside with human serum albumin and glycated human serum albuminLuminescence201732464065127805306
- LauriniEMarsonDPosoccoPFermegliaMPriclSStructure and binding thermodynamics of viologen-phosphorous dendrimers to human serum albumin: a combined computational/experimental investigationFluid Phase Equilib20164221831
- LiuZChenXSimple bioconjugate chemistry serves great clinical advances: albumin as a versatile platform for diagnosis and precision therapyChem Soc Rev20164551432145626771036
- FallacaraALManciniAZamperiniCPyrazolo[3,4-d] pyrimidines-loaded human serum albumin (HSA) nanoparticles: preparation, characterization and cytotoxicity evaluation against neuroblastoma cell lineBioorg Med Chem Lett201727143196320028558969
- ValenteMJBastosMLFernandesECarvalhoFGuedes de PinhoPCarvalhoMNeurotoxicity of β-keto amphetamines: deathly mechanisms elicited by methylone and MDPV in human dopaminergic SH-SY5Y cellsACS Chem Neurosci20178485085928067045
- YuvakkumarRElangoVRajendranVKannanNPreparation and characterization of zero valent iron nanoparticlesDig J Nanomater Biostruct20116417711776
- AslBAMogharizadehLKhomjaniNProbing the interaction of zero valent iron nanoparticles with blood system by biophysical, docking, cellular, and molecular studiesInt J Biol Macromol201810963965029273525
- AbdlomajidEKharaziHChalakiMTitanium oxide nanoparticles fabrication, hemoglobin interaction, white blood cells cytotoxicity, and antibacterial studiesJ Biomol Struct Dyn Epub2018725
- AllabakshMBMandalBKKesarlaMKKumarKSReddyPSPreparation of stable zero valent iron nanoparticles using different chelating agentsJ Chem Pharm Res2010256774
- FooY-TChanJE-MNgohG-CAbdullahAZHorriBASalamatiniaBSynthesis and characterization of NiO and Ni nanoparticles using nanocrystalline cellulose (NCC) as a templateCeram Int201743181633116339
- PoureshghiFGhandforoushanPSafarnejadASoltaniSInteraction of an antiepileptic drug, lamotrigine with human serum albumin (HSA): application of spectroscopic techniques and molecular modeling methodsJ Photochem Photobiol B201716618719227951518
- DaneshNNavaee SedighiZBeigoliSSharifi-RadASaberiMRChamaniJDetermining the binding site and binding affinity of estradiol to human serum albumin and holo-transferrin: fluorescence spectroscopic, isothermal titration calorimetry and molecular modeling approachesJ Biomol Struct Dyn20183671747176328573922
- NaeeminejadSAssaran DarbanRBeigoliSSaberiMRChamaniJStudying the interaction between three synthesized heterocyclic sulfonamide compounds with hemoglobin by spectroscopy and molecular modeling techniquesJ Biomol Struct Dyn201735153250326727771986
- SinhaSTikarihaDLakraJInteraction of bovine serum albumin with cationic monomeric and dimeric surfactants: a comparative studyJ Mol Liq2016218421428
- ChenHRaoHYangJQiaoYWangFYaoJInteraction of diuron to human serum albumin: insights from spectroscopic and molecular docking studiesJ Environ Sci Health B201651315415926671830
- NunesNMPachecoAFCAgudeloÁJPInteraction of cinnamic acid and methyl cinnamate with bovine serum albumin: a thermodynamic approachFood Chem201723752553128764029
- ZeinabadHAKachooeiESabouryAAThermodynamic and conformational changes of protein toward interaction with nanoparticles: a spectroscopic overviewRSC Adv20166107105903105919
- ZeinabadHAZarrabianASabouryAAAlizadehAMFalahatiMInteraction of single and multi wall carbon nanotubes with the biological systems: tau protein and PC12 cells as targetsSci Rep201662650827216374
- AbdullahSMSFatmaSRabbaniGAshrafJMA spectroscopic and molecular docking approach on the binding of tinzaparin sodium with human serum albuminJ Mol Struct20171127283288
- PishkarLTaheriSMakaremSStudies on the interaction between nanodiamond and human hemoglobin by surface tension measurement and spectroscopy methodsJ Biomol Struct Dyn201735360361527151742
- EsfandfarPFalahatiMSabouryASpectroscopic studies of interaction between CuO nanoparticles and bovine serum albuminJ Biomol Struct Dyn20163491962196826555383
- HajsalimiGTaheriSShahiFAttarFAhmadiHFalahatiMInteraction of iron nanoparticles with nervous system: an in vitro studyJ Biomol Struct Dyn201836492893728271723
- SeeligJSchönfeldHJThermal protein unfolding by differential scanning calorimetry and circular dichroism spectroscopy two-state model versus sequential unfoldingQ Rev Biophys201649e927658613
- Abbasi-TajaragKDivsalarASabouryAAGhalandariBGhourchianHDestructive effect of anticancer oxali-palladium on heme degradation through the generation of endogenous hydrogen peroxideJ Biomol Struct Dyn201634112493250426651835
- SugioSKashimaAMochizukiSNodaMKobayashiKCrystal structure of human serum albumin at 2.5 Å resolutionProtein Eng199912643944610388840
- RafieiPHaddadiAPharmacokinetic consequences of PLGA nanoparticles in docetaxel drug deliveryPharm Nanotechnol20175132328948907
- LiuYLiJLiZTangXZhangZPharmacokinetics of a ternary conjugate based pH-responsive 10-HCPT prodrug nano-micelle delivery systemAsian J Pharm Sci2017126542549
- LacroixAEdwardsonTGWHancockMADoreMDSleimanHFDevelopment of DNA nanostructures for high-affinity binding to human serum albuminJ Am Chem Soc2017139217355736228475327
- NaveenrajSMangalarajaRVKrasulyaaOSyedAAmeenFAnandanSA general microwave synthesis of metal (Ni, Cu, Zn) selenide nanoparticles and their competitive interaction with human serum albuminNew J Chem201842857595766
- YamamotoSMaedaNNagashimaYA phase II, multicenter, single-arm study of tri-weekly low-dose nanoparticle albumin-bound paclitaxel chemotherapy for patients with metastatic or recurrent breast cancerBreast Cancer201724678378928439763
- MohanVSenguptaBAcharyyaAYadavRDasNSenPRegion-specific double denaturation of human serum albumin: combined effects of temperature and GnHCl on structural and dynamical responsesACS Omega2018381040610417
- NewberryRWRainesRTA prevalent intraresidue hydrogen bond stabilizes proteinsNat Chem Biol201612121084108827748749
- BardhanMMandalGGangulyTStateSSteady state, time resolved, and circular dichroism spectroscopic studies to reveal the nature of interactions of zinc oxide nanoparticles with transport protein bovine serum albumin and to monitor the possible protein conformational changesJ Appl Phys20091063034701
- WangooNSuriCRShekhawatGInteraction of gold nanoparticles with protein: a spectroscopic study to monitor protein conformational changesAppl Phys Lett20089213133104
- LiuSSuiYGuoKYinZGaoXSpectroscopic study on the interaction of pristine C60 and serum albumins in solutionNanoscale Res Lett20127143322856352
- YangQLiangJHanHProbing the interaction of magnetic iron oxide nanoparticles with bovine serum albumin by spectroscopic techniquesJ Phys Chem B200911330104541045819583232
- PremanathanMKarthikeyanKJeyasubramanianKManivannanGSelective toxicity of ZnO nanoparticles toward Gram-positive bacteria and cancer cells by apoptosis through lipid peroxidationNanomedicine20117218419221034861
- AhamedMAlhadlaqHAKhanMAMAkhtarMJSelective killing of cancer cells by iron oxide nanoparticles mediated through reactive oxygen species via p53 pathwayJ Nanopart Res20131511225
- ShafaghMRahmaniFDelirezhNCuO nanoparticles induce cytotoxicity and apoptosis in human K562 cancer cell line via mitochondrial pathway, through reactive oxygen species and P53Iran J Basic Med Sci2015181099326730334
- KeenanCRGoth-GoldsteinRLucasDSedlakDLOxidative stress induced by zero-valent iron nanoparticles and Fe (II) in human bronchial epithelial cellsEnviron Sci Technol200943124555456019603676
- LeeCKimJYLeeWINelsonKLYoonJSedlakDLBactericidal effect of zero-valent iron nanoparticles on Escherichia coliEnviron Sci Technol200842134927493318678028
- ZhangLJiangYDingYPoveyMYorkDInvestigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids)J Nanopart Res200793479489
- YamamotoOInfluence of particle size on the antibacterial activity of zinc oxideInt J Inorg Mater200137643646
- MakhlufSDrorRNitzanYAbramovichYJelinekRGedankenAMicrowave-assisted synthesis of nanocrystalline MgO and its use as a bacteriocideAdv Funct Mater2005151017081715
- KimJSKukEYuKNAntimicrobial effects of silver nanoparticlesNanomedicine2007319510117379174
- TranNMirAMallikDSinhaANayarSWebsterTJBactericidal effect of iron oxide nanoparticles on Staphylococcus aureusInt J Nanomedicine2010527720463943
- IsmailRASulaimanGMAbdulrahmanSAMarzoogTRAntibacterial activity of magnetic iron oxide nanoparticles synthesized by laser ablation in liquidMater Sci Eng C Mater Biol Appl20155328629726042717

![Figure 6 Far CD spectra of HSA (3 µM) in the presence of varying concentrations of ZVFe NP (0 [black], 3 [red], 10 [blue], and 30 [green] µM).Abbreviations: CD, circular dichroism; HSA, human serum albumin; ZVFe NPs, zero valent iron nanoparticles.](/cms/asset/6363c659-7635-4fc7-994c-193f3c508ab0/dijn_a_12190680_f0006_c.jpg)
![Figure 13 The antibacterial activity of ZVFe NPs aganist E.coli, P. aeruginosa and S. aureus bacterial strains tested by disc diffusion method. (Measuring inhibition zone diameter [mm]).](/cms/asset/23b1a144-20af-4775-a1e2-313eaf4f0c21/dijn_a_12190680_f0013_c.jpg)