?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Purpose:
Clinical applications of curcumin (Cur) have been greatly restricted due to its low solubility and poor systemic bioavailability. Three-arm amphiphilic copolymer tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol (Tri-CL-mPEG) nanoparticles (NPs) were designed to improve the solubility and bioavailability of Cur. The present study adopted a microchannel system to precisely control the preparation of self-assembly polymeric NPs via liquid flow-focusing and gas displacing method.
Methods:
The amphiphilic three-arm copolymer Tri-CL-mPEG was synthesized and self-assembled into nearly spherical NPs, yielding Cur encapsulated into NP cores (Cur-NPs). The obtained NPs were evaluated for physicochemical properties, morphology, toxicity, cellular uptake by A549 cells, release in vitro, biodistribution, and pharmacokinetics in vivo.
Results:
Rapidly fabricated and isodispersed Cur-NPs prepared by this method had an average diameter of 116±3 nm and a polydispersity index of 0.197±0.008. The drug loading capacity and entrapment efficiency of Cur-NPs were 5.58±0.23% and 91.42±0.39%, respectively. In vitro release experiments showed sustained release of Cur, with cumulative release values of 40.1% and 66.1% at pH 7.4 and pH 5.0, respectively, after 10 days post-incubation. The results of cellular uptake, biodistribution, and in vivo pharmacokinetics experiments demonstrated that Cur-NPs exhibited better biocompatibility and bioavailability, while additionally enabling greater cellular uptake and prolonged circulation with possible spleen, lung, and kidney targeting effects when compared to the properties of free Cur.
Conclusion:
These results indicate that Tri-CL-mPEG NPs are promising in clinical applications as a controllable delivery system for hydrophobic drugs.
Introduction
Curcumin (Cur), a natural phenolic compound from the rhizome of turmeric, has been attracting increasing attention due to its ability to regulate multiple cell signaling pathways,Citation1 as well as for its anti-inflammatory,Citation2 anti-microbial,Citation3 anti-oxidant,Citation4 wound-healing,Citation5,Citation6 hypoglycemic,Citation7 bone regenerative,Citation8 and anti-cancer properties.Citation9,Citation10 It is generally recognized as safe by the US Food and Drug Administration (FDA).Citation11 Moreover, several clinical trials have shown that the maximum tolerable dose of Cur could be as high as 8–12 g/day.Citation12 However, poor aqueous solubility, low absorption, rapid clearance, as well as poor bioavailability greatly hinder its clinical applications.Citation13
To overcome the above limitations, various nanocarriers have been exploited for the delivery of Cur, including liposomes,Citation14 nanocrystals,Citation15 nanotubes,Citation16 micelles,Citation17 polymer–drug conjugates,Citation18 lipid-based nanoparticles (NPs),Citation19 magnetic NPs,Citation20,Citation21 and synthetic polymer NPs.Citation22 Among these vectors, synthetic polymer NPs are widely investigated, owing to their small size, excellent hydrotropy, controlled-release ability, outstanding chemical modifying ability, and remarkable biocompatibility. Synthetic polymers with branched architectures, such as dendrimers, cyclic brush polymers, and star-shaped polymers, have been demonstrated to distinctly promote NPs stability.Citation23 Amphiphilic star-shaped copolymers show lower critical micelle concentrations than linear copolymers with the same molecular weight and composition. This circumstance makes corresponding NPs much more stable, which prevents premature drug release.Citation24 Poly ε-caprolactone (PCL), a semi-crystalline and biodegradable polymer, is widely used for drug delivery purposes due to its biocompatibility and capability to enhance hydrophobic drug water-solubility and control drug release.Citation25,Citation26 Combination of PCL with other polymers allows the user to change physicochemical properties and degradation kinetics to suit the needs of a specific application. Polyethylene glycol (PEG) is a hydrophilic, non-ionic polyether with excellent biocompatibility, which is approved by the FDA in food, cosmetics, and drug formulations.Citation26,Citation27 PEG could be added to the drug delivery system via covalent bonding, blending during preparation, or surface adsorption, in order to reduce immunogenicity, stabilize the drug delivery system, and prolong its systemic circulation time.Citation28,Citation29
Synthetic polymer NPs could be fabricated by many approaches, such as nano-deposition,Citation28 salting-out,Citation26 dialysis,Citation30 high-pressure homogenization,Citation31 supercritical fluid technology,Citation32 and emulsion-solvent evaporation.Citation33 However, the salting out agent may increase NP size, emulsion-solvent evaporation is time consuming and possible coalescence of the NPs may affect particle size and morphology, whereas dialysis is costly and not suitable for upscaling.Citation26 High-pressure homogenization and supercritical fluid technology require complicated operating conditions and cost-added apparatus, making their application more difficult. In addition, those methods fail to regulate particle nucleation and aggregation processes,Citation34 leading to uncontrolled and non-uniform NPs with widely varying polydispersity index (PDI) values. A new and more controllable fabrication method is necessary to prepare synthetic polymer NPs with a uniform particle size and distribution. An earlier attempt has been reported,Citation34 in which synthetic polymer NPs were fabricated in a microchannel system via liquid flow-focusing and gas displacing method (LFGDM), successfully achieving narrow particle size distribution.
Therefore, in the present study we aimed to 1) synthesize an amphiphilic three-arm copolymer tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol (Tri-CL-mPEG), with tricarballylic acid-poly (ε-caprolactone) (Tri-CL) as the hydrophobic branch and methoxypolyethylene glycol (mPEG) as the hydrophilic branch, 2) fabricate Cur-loaded polymeric NPs (Cur-NPs) via LFGDM in a microchannel system with a cross-junction, encapsulating Cur into the core of the self-assembly NPs, and 3) characterize the obtained Cur-NPs in vitro and in vivo.
Materials and methods
Materials
Cur was donated by Hangzhou Guang Lin Biological Pharmaceutical Co. (Hangzhou, China). mPEG (MW=1,900), ε-caprolactone, tin (II) 2-ethylhexanoate (Sn(OCt)2), N,N′-dicyclohexylcarbodiimide (DCC), 4-dimethylaminopyridine (DMAP), Poloxamer 188, and acetonitrile were purchased from Aladdin Industrial Corporation (Shanghai, China). Tricarballylic acid was bought from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). Kunming mice and Sprague-Dawley (SD) rats were supplied by Zhejiang Academy of Medical Sciences (Hangzhou, China). Mouse embryonic fibroblast L929 cells and human lung adenocarcinoma A549 cells were provided by the Cell Bank of the Chinese Academy of Sciences (Beijing, China). Dichloromethane was obtained from Shanghai Lingfeng Chemical Reagent Co., Ltd (Shanghai, China). Acetone was purchased from Hangzhou Shuanglin Chemical Reagent Co., Ltd (Hangzhou, China). Hydrochloric acid was obtained from Xilong Scientific Co., Ltd (Shantou, China). Anhydrous Mg2SO4 was bought from Shanghai Hongguang Chemical Factory Co., Ltd (Shanghai, China). Dulbecco minimum essential medium (DMEM) and Roswell Park Memorial Institute 1640 medium (RMPI 1640) were purchased from Thermo Fisher Scientific Suzhou Co., Ltd (Suzhou, China).
Synthesis and characterization of Tri-CL-mPEG
Amphiphilic three-arm Tri-CL-mPEG copolymer was synthesized from tricarballylic acid, ε-caprolactone, and mPEG ().
Figure 1 Synthesis scheme of Tri-CL-mPEG.
Abbreviations: DCC, N,N′-dicyclohexylcarbodiimide; DMAP, 4-dimethylaminopyridine; RT, room temperature; Sn(Oct)2, tin (II) 2-ethylhexanoate; Tri-CL, tricarballylic acid-poly (ε-caprolactone); Tri-CL-mPEG, tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol.
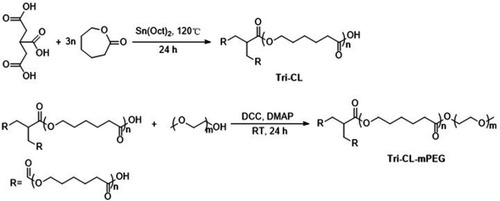
First, tricarballylic acid (0.7043 g, 4 mmol) and ε-caprolactone (13.697 g, 120 mmol) were heated at 120°C in a three-neck round-bottom flask until complete fusion. Sn(Oct)2 (0.2917 g, 0.72 mmol) was then added, and the mixture was maintained with slow stirring under nitrogen atmosphere for 24 hours at 120°C. After cooling to room temperature, the crude product was dissolved in dichloromethane and precipitated in cold diethyl ether (1:10, v/v). Purified Tri-CL was obtained by filtration and dried under vacuum at 25°C for 48 hours.
Secondly, mPEG (0.1994 g, 0.105 mmol), Tri-CL (1.0830 g, 0.1 mmol), DCC (0.1032 g, 0.5 mmol), and DMAP (0.0122 g, 0.1 mmol) were dissolved in 10 mL dichloromethane in a three-neck round-bottom flask.Citation35 The mixture was stirred under nitrogen atmosphere at room temperature for 24 hours. The crude product was precipitated in acetone to remove the precipitated dicyclohexylurea and DCC. After evaporating to dryness, the solid was dissolved in dichloromethane, and the obtained solution was washed with diluted hydrochloric acid solution (pH=5), followed by water, and then dried over anhydrous Mg2SO4. Purified Tri-CL-mPEG was obtained through evaporation.
The structure of Tri-CL-mPEG was confirmed by Fourier transform infrared spectroscopy (FT-IR, Nicolet 6,700, Thermo Fisher Scientific, Waltham, MA, USA), 1H-nuclear magnetic resonance spectroscopy (1H NMR, ANANCE-III, 500 MHz, Bruker Daltonics, Bremen, Germany), and 13C-nuclear magnetic resonance spectroscopy (13C NMR, AscendTM, 600 MHz, Bruke Daltonics, Bremen, Germany). The weight-average molecular weight (Mw) and PDI values for Tri-CL-mPEG were determined by gel permeation chromatography (GPC, LC-20AT, Shimadzu, Tokyo, Japan) using tetrahydrofuran as elution solvent (flow rate: 1 mL/min, at 40°C) and polystyrene as standard.
Preparation of blank NPs and Cur-NPs
Blank NPs and Cur-NPs were prepared as described in a previous report.Citation34 The microchannel device used in this study provides a cross-junction for the focus of organic solution and aqueous solution and a T-junction for the injection of gas bubbles (). NPs are formed due to local supersaturation of drug and copolymers induced by the diffusion of the solvent from the water-miscible organic solution stream into the aqueous phase. The Taylor bubble created by the continuous injection of nitrogen prevented the deposition of and blockage by NPs, ensuring the continuous process.Citation36
Figure 2 The size parameters of the microchannel device and the schematic illustration of NPs formation in the microchannel system.
Abbreviation: NPs, nanoparticles.
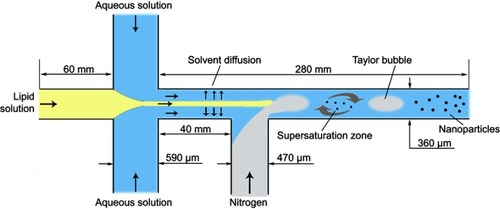
Briefly, Tri-CL-mPEG copolymer (16 mg/mL) and Cur (0.8 mg/mL) were dissolved in acetone as the organic phase, whereas Poloxamer 188 was dissolved simultaneously in distilled water as the aqueous phase (0.1 mg/mL). Then, 2 mL of the organic phase and 10 mL of the aqueous phase was pumped into the main channel and the two branch channels of the cross-junction by precision syringe pumps, respectively. The velocities of organic and aqueous phase delivery were 0.5 and 1.25 mL/min, respectively. Nitrogen was injected into the branch of the T junction (). To ensure that gas bubbles formed steadily, the gas valve was adjusted to 0.01 mPa for each batch. The operation was conducted at ambient temperature. The effluents were collected by stirring with a magnetic stirrer and then centrifuged at 6,000 rpm for 20 minutes to obtain homogeneous NPs suspension. Thereafter, the suspension of NPs was centrifuged at 15,000 rpm for 60 minutes. The precipitate was lyophilized to obtain NPs.
Entrapment efficiency (EE) and drug loading capacity (DL)
Cur concentration was determined by ultraviolet-visible spectroscopy (UV-vis) with a TU-1900 spectrophotometer (Beijing Purkinje General Instrument Co., Ltd, Beijing, China) at 420 nm. EE and DL of Cur-NPs were calculated according to the following equations:
Physicochemical characterization of NPs
Morphological analysis
A drop of Cur-NPs solution was placed on a copper grid (50 mesh) and negatively stained with 2 wt% sodium phosphotungstate. Excess solution was absorbed with filter paper, and the samples were dried at room temperature before morphological analysis was carried out by transmission electron microscopy (TEM, JEM-1010, Jeol Ltd, Tokyo, Japan) working at 80 kV.
Determination of particle size and zeta potential
The particle size and zeta potential of blank NPs and Cur-NPs were investigated using a dynamic light scattering (DLS) laser particle size analyzer (Malvern Zetasizer Nano-ZS 90, Malvern Instruments Ltd, UK) equipped with a He-Ne laser (633 nm) at 25°C. Data were evaluated based on intensity distribution. Each sample was measured in deionized water, the test was repeated three times, and all data were expressed as the mean±standard deviation.
X-ray powder diffraction (XRD)
Samples were characterized by XRD (X’ Pert PRO, PNAlytical, Holland) with the following parameters: output voltage=40 kV, output current=40 mA, and wavelength=0.1546 nm.
In vitro stability of NPs
Cur-NPs solution (2 mL) was diluted 4-fold with 0.1 M phosphate buffered saline (PBS, pH 7.4), DMEM, and fetal bovine serum, respectively, and incubated at 37°C. Then, 1 mL of the sample solution was collected at 1, 3, 8, and 24 hours, and the particle diameter, PDI, and zeta potential of Cur-NPs were measured. Besides, Cur-NPs solution was kept at 4°C for 60 days; 1 mL of the sample solution was collected at 1, 8, 15, 22, 29, and 60 days, and the particle diameter, PDI, and zeta potential of Cur-NPs were measured. The test was repeated three times, and all data were expressed as the mean±standard deviation.
In vitro drug release
In vitro release of Cur from Cur-NPs was investigated by the dialysis method. Briefly, 5 mL of free Cur (Cur-DMSO) or Cur-NPs solutions was placed into dialysis bags (MW cut-off=14 kDa). The dialysis bags were incubated in 100 mL of pre-warmed PBS (pH=5.0 or 7.4) solution with gentle shaking (100 rpm) at 37°C. At specific times, 5 mL of the released medium was collected and replaced with the same volume of fresh medium. Cur concentration in the 5 mL sample was determined by UV-vis at 420 nm. The test was repeated three times, and all data were expressed as the mean±standard deviation.
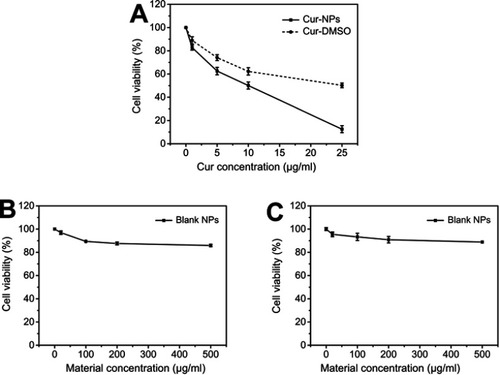
Anti-proliferative effect of Cur-NPs
The anti-proliferative effects of Cur-NPs, Cur-DMSO, and blank NPs were assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method. A549 cells were seeded in DMEM supplemented with 10% (v/v) fetal bovine serum in 96-well plates at a density of 1×104 cells/well, and incubated for 24 hours in a CO2 incubator at 37°C. A549 cells were treated with Cur-NPs, Cur-DMSO, and blank NPs at different concentrations (Cur concentrations of Cur-NPs and Cur-DMSO: 1, 5, 10, and 25 μg/mL, corresponding material concentrations of blank NPs: 20, 100, 200, 500 μg/mL) for 48 hours at 37°C. After incubating for 48 hours, a 50-μL volume of MTT solution (5 mg/mL) was added, and cells were co-cultured for 4 hours at 37°C. Then, MTT solutions were discarded and 100 µL DMSO was added to each well to dissolve MTT formazan crystals. The absorbance was immediately measured at 570 nm in a Microplate Absorbance Reader (Multiskan FC, Thermo Fisher Scientific, Waltham, MA, USA). The anti-proliferative effect of blank NPs against L929 was assessed in the same way. L929 cells were cultured in RMPI 1640 medium. Each sample was measured in triplicate. Cell viability (%) was calculated as the following formula:
where ODx is the absorption of test sample; ODb is the absorption of blank control (medium without cells only); and ODc is the absorption of negative control (medium with cells only).
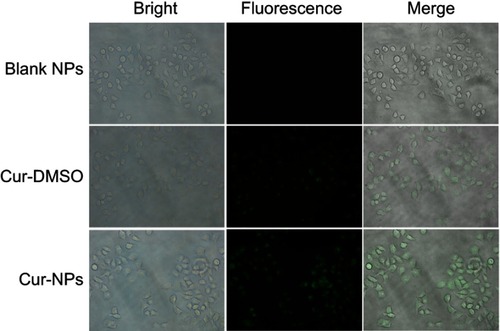
Evaluation of cellular uptake by fluorescence microscopy
Analysis of Cur-NPs cellular uptake was carried out in A549 cells. Cells were seeded into a 6-well plate at a density of 2×105 cells/well and incubated in a CO2 incubator at 37°C for 24 hours. Subsequently, Cur-DMSO, Cur-NPs, and blank NPs were incubated with cells for 4 hours at 37°C in the CO2 incubator. The concentration of Cur was 25 μg/mL. Then the cells were washed with PBS and observed with fluorescence microscopy at 20× magnification (Eclipse Ti-S, Nikon, Tokyo, Japan).
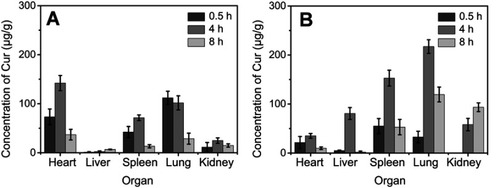
Biodistribution experiments
Kunming mice (18–22 g) were fasting (free access to water only) for 12 hours before the experiments. They were randomly divided into two groups (n=36 each). One group of mice received Cur-NPs at a dose equivalent to 2 mg/kg Cur by tail vein injection. The same dose of Cur-DMSO was injected into another group of mice for comparison. Mice from each group were sacrificed by cervical dislocation at 0.5, 4, or 8 hours after intravenous administration (n=6 for each time point). Tissue samples (ie, the heart, liver, spleen, lung, and kidney) were collected, washed with PBS, blotted with filter paper, weighed, and homogenized in 200 µL of PBS. Then 1 mL acetonitrile was added to each sample, followed by vortexing for 30 seconds and sonication for 5 minutes at 37°C. The above mixtures were centrifuged for 10 minutes at 8,000 rpm and 4°C. The supernatant was dried under a stream of nitrogen gas at 40°C. The obtained solid samples were re-dissolved with 50 µL acetonitrile, vortexed for 1 minute, and centrifuged at 10,000 rpm for 5 minutes. A 20 µL aliquot of the supernatant was collected and analyzed by HPLC (Ultimate 3,000, Thermo Scientific, Germering, Germany) at 420 nm. The mobile phase consisted of 0.6% (v/v) acetic acid solution and acetonitrile (40:60, v/v) at a flow-rate of 1.0 mL/min.
In vivo pharmacokinetics
SD rats (n=6; weighing 200±20 g) were randomly divided into two groups and fasted (free access to water only) for 12 hours before experiment. Cur-NPs and Cur-DMSO solution were injected intravenously via the tail vein of the rats at a dose of 2 mg/kg, respectively. Serial blood samples (0.5 mL) were collected via eye sockets at 0.0167, 0.0833, 0.25, 0.5, 1, 2, 4, 6, 8, 12, and 24 hours after intravenous (IV) administration. The blood sample was centrifuged for 15 minutes at 6,000 rpm and 4°C. A 200 µL aliquot of the supernatant was treated with 1 mL of acetonitrile, vortexed for 30 seconds, sonicated for 5 minutes at 37°C, and then centrifuged for 10 minutes at 8,000 rpm and 4°C. The supernatant was dried under a stream of nitrogen gas at 40°C. The obtained solid samples were re-dissolved with 50 µL acetonitrile, vortexed for 1 minute, and centrifuged at 10,000 rpm for 5 minutes. A 20 µL aliquot of the supernatant was collected and analyzed by HPLC, as described in the section “Biodistribution experiments”. All animal experiments were approved by the local ethics committee of the Zhejiang University of Technology (Hangzhou, China). All animal experiments were conducted in conformity with institutional guideline for the care and use of laboratory animals in Zhejiang University of Technology and conformed to the National Institutes of Health Guide for Care and Use of Laboratory Animals (Publication No. 85–23, revised 1996).
Results
Characterization of Tri-CL-mPEG structure
As observed in the FT-IR spectra of Tri-CL-mPEG (), the copolymer exhibited characteristic peaks of both Tri-CL and mPEG segments. The signals at 1,721.6 and 1,185.8 cm−1 were assigned to the characteristic C=O stretching vibration and characteristic O-C-O asymmetrical stretching vibration of ɛ-CL used in ring-opening polymerization. The peak at 1,105.1 cm−1 corresponded to the characteristic absorption of C-O-C stretching bonds of repeated -OCH2CH2- units in mPEG. Absorption bands at 2,926.2, 2,864.4, 1,468.1, and 730.6 cm−1 were attributed to the characteristic C-H asymmetrical stretching vibration, symmetrical stretching vibration, bending vibration, and rocking vibration of repeated -CH2- units of Tri-CL and mPEG,Citation37 respectively. The signals at 3,434.0 cm−1, 2,887.4 cm−1, and 1,114.5 cm−1 were assigned to the characteristic absorption of -OH, -CH2-, and C-O-C stretching bonds of mPEG, respectively. The absence of 3,434.0 cm−1 and 2,887.4 cm−1 in the FT-IR spectrum of Tri-CL-mPEG suggested the successful formation of Tri-CL-mPEG ().
Figure 3 Tri-CL-mPEG characterization: (A) FT-IR spectra of (a) Tri-CL-mPEG, (b) Tri-CL, and (c) mPEG; (B) 1H NMR spectrum; (C) GPC curve; (D) 13C NMR spectrum.
Abbreviations: mPEG, methoxypolyethylene glycol; Tri-CL, tricarballylic acid-poly (ε-caprolactone); Tri-CL-mPEG, tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol.
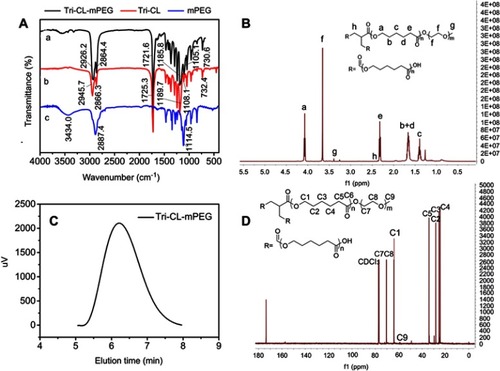
As illustrated in the 1H NMR spectra of Tri-CL-mPEG (), the peaks at 1.40 (c), 1.66 (b+d), 2.32 (e) and 4.07 (a) ppm were assigned to the methylene protons of -O-CH2-CH2-CH2-CH2-CH2-CO-, -O-CH2-CH2-CH2-CH2-CH2-CO-, -CH2-CO-, and -O-CH2- in Tri-CL repeating units, respectively, which was concordant with previous evidence.Citation38 The single peaks at 3.66 (f) and 3.39 (g) ppm were attributed to the methylene protons of -CH2CH2O- and protons of –OCH3 in mPEG units,Citation39 respectively. The structure of Tri-CL-mPEG was also fully identified by 13C NMR in (C1: 64.12 ppm; C2: 28.32 ppm; C3: 25.50 ppm; C4: 24.55 ppm; C5: 34.09 ppm; C6: 173.52 ppm; C7, C8: 70.40 ppm; C9: 58.89 ppm). As listed in , the molecular weights of Tri-CL-mPEG characterized by 1H-NMR and GPC were similar to the theoretical molecular weight and under a narrow distribution (PDI=1.21). The GPC curve of Tri-CL-mPEG had a symmetrical single peak, as illustrated in . All the FT-IR, 1H NMR, and GPC results indicated that Tri-CL-mPEG copolymer was prepared successfully.
Table 1 Molecular weights of Tri-CL and Tri-CL-mPEG
Characterization of NPs
As indicated in , the hydrodynamic diameters of blank NPs and Cur-NPs were 109±2 nm and 116±3 nm, respectively, with a narrow size distribution. Zeta potentials of blank NPs and Cur-NPs were −18.6±0.666 mV and −12.2±0.404 mV, respectively. TEM imaging of Cur-NPs () revealed that NPs were nearly spherical in shape with a diameter around 110 nm, which was consistent with the DLS result (116 nm, ). The EE% and DL% of Cur-NPs were 91.48% and 5.59%, respectively.
Table 2 Characterization of blank NPs and Cur-NPs
Figure 4 Characterization of NPs: (A) TEM photograph of Cur-NPs, (B) histogram of size distribution of DLS data, (C) XRD curves of curcumin, blank NPs, and Cur-NPs, (D) in vitro stabilities of Cur-NPs in PBS, (E) in vitro stabilities of Cur-NPs in DMEM, (F) in vitro stabilities of Cur-NPs in FBS, (G) in vitro stabilities of Cur-NPs in purified water in 4°C, (H) the release curves of Cur-DMSO and Cur-NPs in PBS (pH 7.4/5.0) at 37°C.
Abbreviations: blank NPs, blank Tri-CL-mPEG nanoparticles, Cur, curcumin; Cur-DMSO, free curcumin; Cur-NPs, Cur-loaded Tri-CL-mPEG nanoparticles; PDI, polydispersity index.
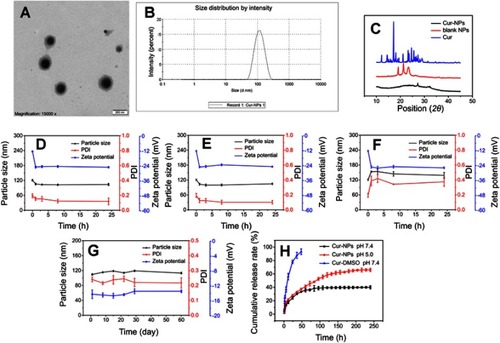
The XRD patterns of Cur, blank NPs, and Cur-NPs are shown in . The peaks at 2θ values of 19.40º and 23.71º could be ascribed to mPEG. The intense peak 2θ=21.57º in blank NPs was contributed by the crystallinity of Tri-CL, which was similar to the previously reported results.Citation40 In comparison with the XRD curves of Cur, blank NPs, and Cur-NPs, specific diffraction peaks of Cur in the Cur-NPs preparation could not be observed, indicating successful encapsulation of Cur into Cur-NPs.
In vitro stability of NPs
indicated that Cur-NPs displayed prominent stability in 0.01 M PBS (pH=7.4) and DMEM for 24 hours with particle size ranging between 104−105 nm. However, the protein adsorption effect was observed in fetal bovine serum (). The size of Cur-NPs increased in the first 3 hours. Thereafter, a particle size of 141 nm was maintained. In particular, 3 hours after incubation, the size of Cur-NPs was constant, suggesting that Cur-NPs could be generally stable in the body. Besides, the absolute values of zeta potential of Cur-NPs increased, accompanied by the dilution with PBS, DMEM, and fetal bovine serum, and stayed stable afterwards. As shown in , after storage at 4°C for 60 days, the particle size, PDI, and zeta potential did not change significantly, suggesting Cur-NPs could remain physicochemically stable at 4°C for 60 days.
In vitro release of Cur-NPs
shows kinetics data on Cur release from Cur-DMSO and Cur-NP preparations. A lower release rate of Cur (40.1%) was observed from Cur-NPs at 240 hours than from Cur-DMSO (93.24%) at 48 hours. Therefore, Cur release from Cur-NPs was sustained. To imitate the conditions of blood circulation and intracellular release (eg, from lysosomal environment), in vitro release of Cur-NPs was carried out at 37°C in PBS solutions with pH 7.4 and 5.0, respectively.Citation41 As illustrated in , both Cur release profiles exhibited an initial fast release in the first 12 hours, followed by a sustained release that achieved a plateau. Overall, cumulative release of Cur at pH 7.4 was slower (40.1%) than at the lower pH condition (66.1%) after incubation for 240 hours.
Kinetic models, including zero and first order, Higuchi, Ritger-Peppas, and Weibull models, were applied to simulate the release mechanism. The equations and obtained regression coefficient R2 values are shown in . According to the largest R2 value (ie, 0.921 and 0.992 in pH 7.4 and pH 5.0 PBS solutions, respectively), the Weibull model (b=0.332 in pH 7.4 PBS solution and b=0.535 in pH 5.0 PBS solution, b<0.7) showed the best correlation with drug release from the sustained release matrix, implying the release was aptly modeled by Fickian diffusion.Citation42
Table 3 Mathematical models of Cur release from Cur-NPs
Interestingly, according to the Ritger-Peppas model, the release indices were 0.273 and 0.387 in PBS at pH 7.4 and pH 5.0 (R2 value=0.899 and 0.983), respectively, indicating that the release of Cur abided by the “Fickian diffusion”,Citation43 which is consistent with the result deduced by the Weibull model.
Anti-proliferative effect of Cur-NPs
The anti-proliferative effect of Cur-NPs, Cur-DMSO, and blank NPs against A549 cells is summarized in . Cell viabilities comprised 12.51% and 50.25% in the presence of, respectively, Cur-NPs and Cur-DMSO, both at a concentration of 25 μg/mL. IC50 values of Cur-NPs and Cur-DMSO against A549 cells were 11.39 μg/mL and 21.94 μg/mL, respectively. Interestingly, blank NPs had almost no negative effect on A549 cells. The viability of L929 cells was concentration-dependently modulated by blank NPs, but it did not decrease significantly, even at a dose as high as 25 μg/mL (). These results confirmed satisfactory biocompatibility of the copolymers.
Cellular uptake of Cur-NPs
Cellular uptake of Cur-NPs by A549 cells was examined by fluorescence microscopy. As shown in , stronger fluorescence intensity was observed in the case of Cur-NPs, whereas there was less fluorescence in the Cur-DMSO group, indicating that Cur-NPs were easier internalized by A549 cells.
Biodistribution of Cur-NPs
Tissue distributions of Cur-NPs and Cur-DMSO after IV administration are illustrated in . As shown in , in experiments with Cur-NPs, Cur concentrations in the heart, liver, spleen, and lung increased from 0.5 hours to 4 hours and decreased from 4 hours to 8 hours after IV administration. Maximal Cur concentrations following application of Cur-NPs in the heart, liver, spleen, and lung (35.07, 80.67, 152.81, and 217.22 μg/g, respectively) were at 4 hours, whereas the maximal Cur concentration in the kidneys (93.38 μg/g) was at 8 hours, which indicated that most of Cur-NPs accumulated in organs other than the heart. In experiments with Cur-DMSO (), the highest Cur concentrations following application of Cur-DMSO in the heart, spleen, lung, and kidney (142.14, 71.40, 101.75, and 24.81 μg/g, respectively) were at 4 hours, whereas the maximal Cur concentration in the liver (2.56 μg/g) was at 8 hours. In contrast, samples from Cur-NPs group had lower levels of Cur distribution in the heart, but higher levels in the liver, spleen, lung, and kidney. Cur concentration of Cur-NPs in the heart at 4 hours was 4.05-times lower than that in the Cur-DMSO group, suggesting that uptake of Cur-NPs into the heart was lower than in the Cur-DMSO group. In addition, Cur concentrations of Cur-NPs in the liver, spleen, lung, and kidney at 4 hours were 31.15-, 2.14-, 2.13-, and 2.34-fold higher than those in the Cur-DMSO group, respectively.
In vivo pharmacokinetics of Cur-NPs
Cur-NPs were formulated to improve Cur bioavailability. The profiles of Cur concentration in plasma as functions of time are illustrated in . As can be seen, Cur plasma concentrations in both Cur-NPs and Cur-DMSO groups rapidly decreased within the first 30 minutes, followed by a slow decrease afterwards. Plasma concentration of Cur-NPs at 1 minute (1447.02 ng/mL) was 12.60-fold of that in the Cur-DMSO group (114.84 ng/mL).
Figure 8 Plasma concentration-time profiles of Cur in SD rats after IV administration of Cur-DMSO and Cur-NPs.
Abbreviations: Cur, curcumin; Cur-DMSO, free curcumin; Cur-NPs, Cur-loaded Tri-CL-mPEG nanoparticles; IV, intravenous.
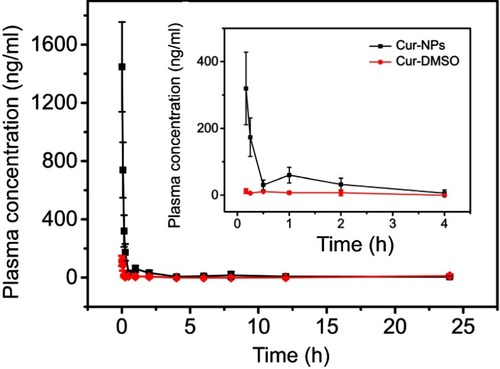
Pharmacokinetic parameters of Cur-NPs and Cur-DMSO in rats after IV administration were calculated by DAS 2.0 Practical Pharmacokinetics Program (). The elimination half-life value of Cur-NPs (t1/2, 4.99±2.45 h) was approximately 2.07-fold higher than that of the Cur-DMSO group (2.41±1.11 h). The area under curve (AUC) value in the Cur-NPs group was 4.14-times greater than that in the Cur-DMSO group (AUC0–24h, 456.42±26.39 h×ng/mL vs 110.27±11.05 h×ng/mL). The maximal plasma concentration (Cmax) of Cur-NPs (1447.02±71.62 ng/mL) was 12.40-fold higher than in the Cur-DMSO group (116.74±14.70 ng/mL). The total body clearance (Cl) value of Cur-NPs (4054.74±302.59 mL/h/kg) was much lower than in the Cur-DMSO group (28480.93±703.89 mL/h/kg), which is consistent with plasma concentration change tendency. Pharmacokinetic analysis showed that the relative bioavailability (F%)Citation44 of Cur-NPs was 413.91%.
Table 4 The pharmacokinetic parameters of Cur-NPs and Cur-DMSO after IV administration in rats
Discussion
Cur has a remarkable antitumor activity against various cancers. However, its clinical applications are greatly restricted by its low aqueous-solubility, poor absorption, and low bioavailability. Thus, new strategies are needed to overcome these limitations and to avail the entire benefits of Cur. In this study, we presented a nanoformulation to improve the solubility and bioavailability of Cur.
The amphiphilic three-arm copolymer Tri-CL-mPEG was successfully synthetized and characterized. Tri-CL-mPEG with a narrow PDI value was beneficial to the stability of the nanoparticles preparation. Cur-loaded Tri-CL-mPEG NPs were fabricated by LFGDM in a crossed-shaped microchannel system. Tri-CL-mPEG polymer self-assembled to form biodegradable NPs (Cur-NPs and blank NPs) with a hydrophilic shell (mPEG segment) and a hydrophobic core (Tri-CL segment). Besides, XRD analysis revealed that Cur specific diffraction peaks disappeared in Cur-NPs, indicating that Cur molecules were successfully entrapped into the core of the NPs rather than just adsorbed onto their surfaces. Moreover, TEM imaging and DLS results demonstrated that Cur-NPs had an average size around 110 nm, which could be advantageous for passive accumulation within tumor cells by the enhanced permeability and retention effect.Citation45 The evaluation of stability of Cur-NPs in the presence of PBS, DMEM, and FBS suggested that Cur-NPs are stable inside the body.
Characteristics of Cur-NP release in vitro were evaluated at neutral and slightly acidic PBS solutions to imitate the conditions of blood and intracellular (eg, lysosomal) environments, respectively. Whereas the release of Cur from Cur-DMSO formulation was rapid, Cur release from NPs was more sustained. Curves of Cur release from Cur-NPs at pH 7.4 and 5.0 demonstrated a fast release phase in the first 12 hours followed by a sustained release phase (). Compared to the observations at pH 7.4, enhanced Cur release at pH 5.0 was likely due to the degradation of the Tri-CL core at lower pH.Citation17 Differences in release properties at distinct pH values provide a very interesting possibility for minimizing the exposure to Cur of healthy tissues, where pH is ~7.4, while increasing the accumulation of Cur in intracellular lysosomes (pH 5.0). This circumstance may lead to enhanced bioavailability and lower systemic side-effects.
A dose-dependent antiproliferative effect of Cur-NPs against A549 cells was observed, which was greater than that of Cur-DMSO at the same concentration (). However, no obvious antiproliferative effect was observed at a low concentration of 1 μg/mL. Thus, for efficient treatment, Cur concentration should be 5 μg/mL or higher. Cur-NPs cellular uptake was assessed by fluorescence microscopy, and because stronger fluorescence intensity was observed for Cur-NPs than for Cur-DMSO, it can be concluded that Cur-NPs were easier internalized by A549 cells ().
Examination of biodistribution parameters of Cur-NPs and Cur-DMSO (control group) () showed that Cur accumulation in the heart was lower in the case of Cur-NPs, whereas Cur concentration was higher in the lung, spleen, and kidney for this formulation. This suggests that Cur-NPs may be applied in treating lesions in the latter organs, whereas the harm to the heart would be minimized.Citation46 In the case of the Cur-DMSO formulation, Cur concentrations in the liver, spleen, and kidney decreased from 1 hour to 8 hours. Simple passive diffusion and rapid clearance of Cur are likely explained by the poor bioavailability of Cur in Cur-DMSO formulation. In the case of Cur-NPs, which provided sustained release of Cur in vivo, Cur concentrations in the liver, spleen, and lung increased from 1 hour to 4 hours after IV administration and then decreased up to 8 hours.
In vivo pharmacokinetics results indicated that plasma concentration and bioavailability of Cur from Cur-NPs were significantly better than those of Cur from Cur-DMSO solution ( and ). The likely reason for this difference was that Cur in Cur-DMSO underwent rapid clearance once it entered the blood due to its poor bioavailability and poor targeting. In contrast, Cur-NPs formulation was stabilized by hydrophilic PEG shell, which attenuated clearance of Cur-NPs from blood and prolonged systemic circulation time of Cur-NPs.Citation27 Thus, our experimental data indicated that Cur-NPs could act as a drug reservoir from which the drug could be released in sustained fashion, leading to markedly increased bioavailability.
Conclusion
In conclusion, we synthesized amphiphilic three-arm copolymer Tri-CL-mPEG, and Cur-NPs were successfully fabricated in a microchannel system with a uniform particle size and distribution. Cur-NPs had improved biocompatibility and enhanced cellular uptake, suggesting that Cur-NPs could be safe and effective in vivo. Furthermore, Cur-NPs tended to accumulate at higher levels in the spleen, lung, and kidney compared to Cur-DMSO. The improved pharmacokinetic parameters of Cur-NPs in rats suggested that Cur bioavailability was significantly enhanced. Therefore, microchannel-fabricated Tri-CL-mPEG NPs could be a promising candidate as a delivery system for curcumin.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 21376223) and Zhejiang Provincial Natural Science Foundation of China (LY19B060012). We are also thankful to the Research Center of Analysis & Measurement of Zhejiang University of Technology for providing the NMR, FT-IR, and XRD analytical services used in this work. We would like to thank Zachary Gouveia for helping with manuscript editing.
References
- Li M, Gao M, Fu Y, et al. Acetal-linked polymeric prodrug micelles for enhanced curcumin delivery. Colloids Surf B Biointerfaces. 2016;140:11–18. doi:10.1016/j.colsurfb.2015.12.02526731193
- Kant V, Gopal A, Pathak NN, Kumar P, Tandan SK, Kumar D. Antioxidant and anti-inflammatory potential of curcumin accelerated the cutaneous wound healing in streptozotocin-induced diabetic rats. Int Immunopharmacol. 2014;20(2):322–330. doi:10.1016/j.intimp.2014.03.00924675438
- Krausz AE, Adler BL, Cabral V, et al. Curcumin-encapsulated nanoparticles as innovative antimicrobial and wound healing agent. Nanomedicine. 2015;11(1):195–206. doi:10.1016/j.nano.2014.09.00425240595
- Hussain Z, Thu HE, Amjad MW, Hussain F, Ahmed TA, Khan S. Exploring recent developments to improve antioxidant, anti-inflammatory and antimicrobial efficacy of curcumin: a review of new trends and future perspectives. Mat Sci Eng C-Mater. 2017;77:1316–1326. doi:10.1016/j.msec.2017.03.226
- Chereddy KK, Coco R, Memvanga PB, et al. Combined effect of PLGA and curcumin on wound healing activity. J Control Release. 2013;171(2):208–215. doi:10.1016/j.jconrel.2013.07.01523891622
- Hussain Z, Thu HE, Ng SF, Khan S, Katas H. Nanoencapsulation, an efficient and promising approach to maximize wound healing efficacy of curcumin: A review of new trends and state-of-the-art. Colloids Surf B Biointerfaces. 2017;150:223–241. doi:10.1016/j.colsurfb.2016.11.03627918967
- Das KK, Razzaghi-Asl N, Tikare SN, et al. Hypoglycemic activity of curcumin synthetic analogues in alloxan-induced diabetic rats. J Enzyme Inhib Med Chem. 2016;31(1):99–105. doi:10.3109/14756366.2015.100406125683079
- Dong JL, Tao L, Abourehab MAS, Hussain Z. Design and development of novel hyaluronate-modified nanoparticles for combo-delivery of curcumin and alendronate: fabrication, characterization, and cellular and molecular evidences of enhanced bone regeneration. Int J Biol Macromol. 2018;116:1268–1281. doi:10.1016/j.ijbiomac.2018.05.11629782984
- Gaikwad D, Shewale R, Patil V, Mali D, Gaikwad U, Jadhav N. Enhancement in in vitro anti-angiogenesis activity and cytotoxicity in lung cancer cell by pectin-PVP based curcumin particulates. Int J Biol Macromol. 2017;104(Pt A):656–664. doi:10.1016/j.ijbiomac.2017.05.17028602990
- Khan S, Imran M, Butt TT, et al. Curcumin based nanomedicines as efficient nanoplatform for treatment of cancer: new developments in reversing cancer drug resistance, rapid internalization, and improved anticancer efficacy. Trends Food Sci Technol. 2018;80:8–22. doi:10.1016/j.tifs.2018.07.026
- Ravichandiran V, Masilamani K, Senthilnathan B, Maheshwaran A, Wong TW, Roy P. Quercetin-decorated curcumin liposome design for cancer therapy: in-vitro and in-vivo studies. Curr Drug Deliv. 2017;14(8):1053–1059. doi:10.2174/156720181366616082910045327572089
- Gupta SC, Patchva S, Aggarwal BB. Therapeutic roles of curcumin: lessons learned from clinical trials. Aaps J. 2013;15(1):195–218. doi:10.1208/s12248-012-9432-823143785
- Heger M, van Golen RF, Broekgaarden M, Michel MC. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol Rev. 2014;66(1):222–307. doi:10.1124/pr.111.00511624368738
- Jiang K, Shen M, Xu W. Arginine, glycine, aspartic acid peptide-modified paclitaxel and curcumin co-loaded liposome for the treatment of lung cancer: in vitro/vivo evaluation. Int J Nanomedicine. 2018;13:2561–2569. doi:10.2147/IJN.S17762729731631
- Guo R, Lan Y, Xue W, et al. Collagen-cellulose nanocrystal scaffolds containing curcumin-loaded microspheres on infected full-thickness burns repair. J Tissue Eng Regen Med. 2017;11(12):3544–3555. doi:10.1002/term.227228326684
- Rao KM, Kumar A, Suneetha M, Han SS. pH and near-infrared active; chitosan-coated halloysite nanotubes loaded with curcumin-Au hybrid nanoparticles for cancer drug delivery. Int J Biol Macromol. 2018;112:119–125. doi:10.1016/j.ijbiomac.2018.01.16329378273
- Cai M, Cao J, Wu Z, Cheng F, Chen Y, Luo X. In vitro and in vivo anti-tumor efficiency comparison of phosphorylcholine micelles with PEG micelles. Colloids Surf B Biointerfaces. 2017;157:268–279. doi:10.1016/j.colsurfb.2017.05.05328601755
- Fang XB, Zhang JM, Xie X, et al. pH-sensitive micelles based on acid-labile pluronic F68-curcumin conjugates for improved tumor intracellular drug delivery. Int J Pharm. 2016;502(1–2):28–37. doi:10.1016/j.ijpharm.2016.02.03726784981
- Duan R, Li C, Wang F, Yangi JC. Polymer-lipid hybrid nanoparticles-based paclitaxel and etoposide combinations for the synergistic anticancer efficacy in osteosarcoma. Colloids Surf B Biointerfaces. 2017;159:880–887. doi:10.1016/j.colsurfb.2017.08.01928892872
- Nosrati H, Salehiabar M, Attari E, Davaran S, Danafar H, Manjili HK. Green and one-pot surface coating of iron oxide magnetic nanoparticles with natural amino acids and biocompatibility investigation. Appl Organomet Chem. 2018;32(2). doi:10.1002/aoc.4069
- Nosrati H, Rashidi N, Danafar H, Manjili HK. Anticancer activity of tamoxifen loaded tyrosine decorated biocompatible Fe3O4 magnetic nanoparticles against breast cancer cell lines. J Inorg Organomet P. 2018;28(3):1178–1186. doi:10.1007/s10904-017-0758-7
- Guo F, Wu J, Wu W, et al. PEGylated self-assembled enzyme-responsive nanoparticles for effective targeted therapy against lung tumors. J Nanobiotechno. 2018;16(1):57. doi:10.1186/s12951-018-0384-8
- Zuo C, Peng J, Cong Y, et al. Fabrication of supramolecular star-shaped amphiphilic copolymers for ROS-triggered drug release. J Colloid Interface Sci. 2018;514:122–131. doi:10.1016/j.jcis.2017.12.02229248814
- Shi C, Guo X, Qu Q, Tang Z, Wang Y, Zhou S. Actively targeted delivery of anticancer drug to tumor cells by redox-responsive star-shaped micelles. Biomaterials. 2014;35(30):8711–8722. doi:10.1016/j.biomaterials.2014.01.02625002267
- Feng R, Deng P, Song Z, et al. Glycyrrhetinic acid-modified PEG-PCL copolymeric micelles for the delivery of curcumin. React Funct Polym. 2017;111:30–37. doi:10.1016/j.reactfunctpolym.2016.12.011
- Danafar H. Applications of copolymeric nanoparticles in drug delivery systems. Drug Res (Stuttg). 2016;66(10):506–519. doi:10.1055/s-0042-10986527403578
- Grossen P, Witzigmann D, Sieber S, Huwyler J. PEG-PCL-based nanomedicines: A biodegradable drug delivery system and its application. J Control Release. 2017;260:46–60. doi:10.1016/j.jconrel.2017.05.02828536049
- Gharebaghi F, Dalali N, Ahmadi E, Danafar H. Preparation of wormlike polymeric nanoparticles coated with silica for delivery of methotrexate and evaluation of anticancer activity against MCF7 cells. J Biomater Appl. 2017;31(9):1305–1316. doi:10.1177/088532821769806328447548
- Suk JS, Xu Q, Kim N, Hanes J, Ensign LM. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv Drug Deliv Rev. 2016;99(Pt A):28–51. doi:10.1016/j.addr.2015.09.01226456916
- Kostag M, Kohler S, Liebert T, Heinze T. Pure cellulose nanoparticles from trimethylsilyl cellulose. Macromol Sy. 2010;294-Ii:96–106. doi:10.1002/masy.200900095
- Deshmukh R, Mujumdar A, Naik J. Production of aceclofenac-loaded sustained release micro/nanoparticles using pressure homogenization and spray drying. Dry Technol. 2018;36(4):459–467. doi:10.1080/07373937.2017.1341418
- Nuchuchua O, Nejadnik MR, Goulooze SC, Ljeskovic NJ, Every HA, Jiskoot W. Characterization of drug delivery particles, produced by supercritical carbon dioxide technologies. J Supercrit Fluid. 2017;128:244–262. doi:10.1016/j.supflu.2017.06.002
- Deng R, Yang L, Bain CD. Combining inkjet printing with emulsion solvent evaporation to pattern polymeric particles. ACS Appl Mater Interfaces. 2018;10(15):12317–12322. doi:10.1021/acsami.8b0201729595241
- Guo F, Guo D, Zhang W, et al. Preparation of curcumin-loaded PCL-PEG-PCL triblock copolymeric nanoparticles by a microchannel technology. Eur J Pharm Sci. 2017;99:328–336. doi:10.1016/j.ejps.2017.01.00128062259
- Maglio G, Nese G, Nuzzo M, Palumbo R. Synthesis and characterization of star-shaped diblock poly(epsilon-caprolactone)/poly(ethylene oxide) copolymers. Macromol Rapid Comm. 2004;25(12):1139–1144. doi:10.1002/marc.200400113
- Yun JX, Lei QA, Zhang SH, Shen SC, Yao KJ. Slug flow characteristics of gas-miscible liquids in a rectangular microchannel with cross and T-shaped junctions. Chem Eng Sci. 2010;65(18):5256–5263. doi:10.1016/j.ces.2010.06.031
- Eatemadi A, Daraee H, Aiyelabegan HT, Negahdari B, Rajeian B, Zarghami N. Synthesis and characterization of Chrysin-loaded PCL-PEG-PCL nanoparticle and its effect on breast cancer cell line. Biomed Pharmacother. 2016;84:1915–1922.27847208
- Kumar A, Lale SV, Naz F, Choudhary V, Koul V. Synthesis and biological evaluation of dual functionalized glutathione sensitive poly(ester-urethane) multiblock polymeric nanoparticles for cancer targeted drug delivery. Polym Chem-Uk. 2015;6(43):7603–7617. doi:10.1039/C5PY00898K
- Gong CY, Wei XW, Wang XH, et al. Biodegradable self-assembled PEG-PCL-PEG micelles for hydrophobic honokiol delivery: I. Preparation and characterization. Nanotechnology. 2010;21(21). doi:10.1088/0957-4484/21/21/215103.
- Song L, Shen YY, Hou JW, Lei L, Guo SR, Qian CY. Polymeric micelles for parenteral delivery of curcumin: preparation, characterization and in vitro evaluation. Colloid Surf A. 2011;390(1–3):25–32. doi:10.1016/j.colsurfa.2011.08.031
- Palange AL, Di Mascolo D, Carallo C, Gnasso A, Decuzzi P. Lipid-polymer nanoparticles encapsulating curcumin for modulating the vascular deposition of breast cancer cells. Nanomedicine. 2014;10(5):991–1002. doi:10.1016/j.nano.2013.12.00124566270
- Papadopoulou V, Kosmidis K, Vlachou M, Macheras P. On the use of the Weibull function for the discernment of drug release mechanisms. Int J Pharm. 2006;309(1–2):44–50. doi:10.1016/j.ijpharm.2005.11.03716376033
- Khoee S, Kavand A. Preparation, co-assembling and interfacial crosslinking of photocurable and folate-conjugated amphiphilic block copolymers for controlled and targeted drug delivery: smart armored nanocarriers. Eur J Med Chem. 2014;73:18–29. doi:10.1016/j.ejmech.2013.11.03324374349
- Wang F, Chen J, Dai W, He Z, Zhai D, Chen W. Pharmacokinetic studies and anticancer activity of curcumin-loaded nanostructured lipid carriers. Acta Pharm. 2017;67(3):357–371. doi:10.1515/acph-2017-002128858837
- Cabral H, Matsumoto Y, Mizuno K, et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat Nanotechnol. 2011;6(12):815–823. doi:10.1038/nnano.2011.16622020122
- Tsai YM, Chien CF, Lin LC, Tsai TH. Curcumin and its nano-formulation: the kinetics of tissue distribution and blood-brain barrier penetration. Int J Pharm. 2011;416(1):331–338. doi:10.1016/j.ijpharm.2011.06.03021729743