 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Gene silencing using small interfering RNA (siRNA) is a promising new therapeutic approach for glioblastoma. The endocytic uptake and delivery of siRNA to intracellular compartments could be enhanced by complexation with polyamidoamine dendrimers. In the present work, the uptake mechanisms and intracellular traffic of siRNA/generation 7 dendrimer complexes (siRNA dendriplexes) were screened in T98G glioblastoma and J774 macrophages.
Methods
The effect of a set of chemical inhibitors of endocytosis on the uptake and silencing capacity of dendriplexes was determined by flow cytometry. Colocalization of fluorescent dendriplexes with endocytic markers and occurrence of intracellular dissociation were assessed by confocal laser scanning microscopy.
Results
Uptake of siRNA dendriplexes by T98G cells was reduced by methyl-β-cyclodextrin, and genistein, and cytochalasine D, silencing activity was reduced by genistein; dendriplexes colocalized with cholera toxin subunit B. Therefore, caveolin-dependent endocytosis was involved both in the uptake and silencing activity of siRNA dendriplexes. On the other hand, uptake of siRNA dendriplexes by J774 cells was reduced by methyl-β-cyclodextrin, genistein, chlorpromazine, chloroquine, cytochalasine D, and nocodazole, the silencing activity was not affected by chlorpromazine, genistein or chloroquine, and dendriplexes colocalized with transferrin and cholera toxin subunit B. Thus, both clathrin-dependent and caveolin-dependent endocytosis mediated the uptake and silencing activity of the siRNA dendriplexes. SiRNA dendriplexes were internalized at higher rates by T98G but induced lower silencing than in J774 cells. SiRNA dendriplexes showed relatively slow dissociation kinetics, and their escape towards the cytosol was not mediated by acidification independently of the uptake pathway.
Conclusion
The extent of cellular uptake of siRNA dendriplexes was inversely related to their silencing activity. The higher silencing activity of siRNA dendriplexes in J774 cells could be ascribed to the contribution of clathrin-dependent and caveolin-dependent endocytosis vs only caveolin-dependent endocytosis in T98G cells.
Keywords:
Introduction
Glioblastoma, the most common and aggressive malignant primary brain tumor in humans, is frequently accompanied by tumor-associated macrophages. These macrophages promote tumor growth and metastasis, in part by secreting a wide range of proangiogenic factors, growth factors, and metalloproteinases, and by suppressing the activity of CD8+ T cells. A high degree of macrophage infiltration is associated with a poor prognosis.Citation1 The current standard treatment for glioblastoma consists of maximal surgical resection and a combination of radiotherapy and chemotherapyCitation2 that at best leads to a median survival of approximately 1 year from the time of diagnosis.Citation3,Citation4 Recently, gene silencing by small interfering RNA (siRNA) of different therapeutic targets has emerged as a promising new approach that is able to inhibit glioblastoma growth in vitro and ex vivo.Citation5–Citation8 Because decreasing the number of tumor-associated macrophages leads to reduced tumor growth in vivo,Citation9 eliminating both glioblastoma cells and macrophages could result in a better therapeutic outcome.
SiRNA are hydrophilic polyanions with 21 base pairs that do not cross the cell membrane by diffusion and are not internalized upon docking to membrane receptors. By preparing nanoparticulate electrostatic complexes between siRNA and polycations, the uptake and subsequent delivery of siRNA to intracellular compartments is ensured.Citation10
Mammalian cells take up nanoparticles by endocytosis, that can be classified as phagocytosis (restricted to specialized cells) or pinocytosis (all cell types).Citation11 Pinocytosis is subdivided into macropinocytosis, and clathrin-dependent and clathrin-independent endocytosis. Furthermore, clathrin-independent endocytosis can be subdivided into dynamin-dependent caveolin-mediated and Rho-mediated endocytosis, and the less explored dynamin-independent flotillin, GEEC/CLIC (GPI-Enriched Endosomal Compartment/CLathrin Independent Carriers) and ARF6-mediated endocytosis.Citation12–Citation14 Each internalization mechanism leads to different intracellular trafficking and can yield different transfection efficiencies. For instance, nanoparticles taken up by clathrin-dependent endocytosis undergo endosomal acidification (pH 5–6) and end up in lysosomes (pH about 4.5) where acidic enzymatic degradation occurs.Citation15 Successful transfection via clathrin-dependent endocytosis relies on escape of nucleic acids from acidic compartments toward the cytoplasm. Because of this, titratable alkalinizing polycations, such as polyethylenimine,Citation16 chitosan,Citation17 and polyamidoamine dendrimers,Citation18 are used to induce endosomal/lysosomal disruption by the proton sponge mechanism.Citation16,Citation19–Citation21 In this sense, some authors have found the uptake of nanoparticles by clathrin-dependent endocytosis to be better than by clathrin-independent endocytosis for effective transfection.Citation22 Other authors have found caveolin-mediated endocytosis better suited than clathrin-dependent endocytosis for successful transfection because this pathway is thought to evade acid degradation.Citation23–Citation25 Caveolin-mediated endocytosis has been accepted to occur at neutral pH, and a number of pH-independent escape mechanisms by which neutral caveosomes can reach the cytoplasm have been proposed.Citation26 However, while caveosomes have been shown to fuse with early endosomes,Citation27 it was suggested recently that neutral caveosomes do not exist.Citation28 Polyethylenimine polyplexes, for instance, regardless of whether they are taken up by clathrin-dependent endocytosis or caveolin-mediated endocytosis, are exposed to the acidic environment of early endosomes during the first 4 hours following transfectionCitation23 and show efficient transfection uptake by both clathrin-independent endocytosis and clathrin-dependent endocytosis.Citation29 Caveolin-mediated endocytosis actually involves trafficking to early or late endosomes (pH about 6 vs 5, respectively) depending on the metabolic state of the cell, which underscores the complexity of this pathway.Citation28 Overall, nucleic acid/polycation complexes are taken up by different pathways, depending not only on their chemical and structural nature (size, charge, and stability) but also on cell type and cell metabolism. Thus far, an association between successful transfection and a determined pathway has not been demonstrated.Citation22
There has been growing interest in the use of complexes between nucleic acids and dendrimers (dendriplexes) as a new type of nanoparticle for gene therapy, because dendrimers differ from traditional cationic polymers vectors in their monodispersion and nanoscale architecture.Citation30 The uptake mechanism for dendriplexes of high molecular weight pDNA (1–1000 kbp, 650–650,000 kDa, and about 50 nm in size) is very much dependent on cell type and particle size.Citation31–Citation33 However, until now, the uptake mechanism and intracellular trafficking of dendriplexes comprised of small siRNA molecules (about 13 kDa and approximately 7 nm in size) has not been reported.
Previously we found that siRNA/polyamidoamine generation 7 (G7) dendrimer complexes (siRNA dendriplexes) prepared in low ionic strength medium produced the greatest inhibition of enhanced green fluorescent protein (EGFP) expression in both T98G human glioblastoma cells (35%) and in J774 macrophages (45%), as compared with dendriplexes prepared with lower dendrimer generations (G4, G5, and G6).Citation34 Similar results were recently reported by Waite and Roth, who found 40% silencing of the EGFP gene in U87 glioma cells using siRNA/G5 dendriplexes.Citation35
The delivery of siRNA to the cytoplasm of the cell is a critical step for the RNA-induced silencing assembly complex. Depending on the uptake mechanism, this step could be impaired if escape from endosomes, lysosomes, caveosomes, or less well described pinocytic vesicles is difficult. In the present work, the uptake mechanism of siRNA/G7 dendriplexes and fate in T98G cells and J774 cells (as models of glioblastoma cells and tumor-associated macrophages, respectively) were screened for the first time. The effect of a set of chemical inhibitors of endocytosis on the uptake of fluorescent dendriplexes and their silencing capacity was determined. In addition, colocalization of fluorescent dendriplexes with transferrin, cholera toxin, and lysotracker (used as markers of clathrin-dependent endocytosis, caveolin-mediated endocytosis, and endosomes/lysosomes, respectively) and intracellular dissociation of the dendriplexes were studied using confocal laser scanning microscopy. Gaining insights in this way could contribute to the rational design of cationic vectors to maximize gene silencing activity.
Materials and methods
Materials
Polyamidoamine G7 dendrimers (molecular weight 116,488.71 g/mol, 512 amine end groups), chloroquine diphosphate salt, cytochalasin D from Zygosporium mansonii, genistein, trypan blue, fluorescein isothiocyanate isomer I (FITC), and 3-(4,5-dimethythiazole-2-yl)-2,5- diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (Buenos Aires, Argentina). Nocodazole and methyl-β-cyclodextrin were acquired from Fluka (Buenos Aires, Argentina) and chlorpromazine was a gift from Laboratorios Ceballos, Argentina. Cy3-labeled siRNA was from Genbiotech (Buenos Aires, Argentina). Modified Eagle’s medium, G418, and Lipofectamine™ 2000 were purchased from Gibco (Buenos Aires, Argentina). Fetal bovine serum, antibiotic/antimycotic solution, glutamine, and trypsin/ethylenediamine tetra-acetic acid were from PAA Laboratories GmbH (Pasching, Austria). LysoTracker Red DND-99, Alexa Fluor 555 conjugate recombinant cholera toxin subunit B (CtxB-Alexa-555), and Alexa Fluor 555 conjugated human serum transferrin (Tf-Alexa-555) were purchased from Molecular Probes (Eugene, OR). T98G cells and T98G cells stably expressing EGFP (T98G-EGFP) were kindly provided by Dr David Silvestre (Universidad Nacional de Córdoba, Córdoba, Argentina), and J774-EGFP were obtained in our laboratory. Silencer® EGFP siRNA was from Ambion (Buenos Aires, Argentina). All solutions were treated with DEPC 0.1%. All other chemicals and reagents were of analytical grade and from Anedra (Buenos Aires, Argentina).
Preparation of siRNA/G7 dendriplexes
An aliquot of G7 from methanol stock was diluted in 10 mM Tris-HCl pH 7.4 buffer (Tris-HCl buffer) and siRNA was added to give a 10:1 N/P ratio (ratio of available amino groups in dendrimer to the RNA phosphate groups). The mixture was incubated for 20 minutes at room temperature prior to use. Size and zeta potential were determined by dynamic light scattering and phase analysis light scattering, respectively, using a nanosizer (ZEN 3600, Malvern, Worcestershire, UK).
Cell culture
T98G and J774 cells were cultured in modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum, 1% (v/v) antibiotic/antimycotic solution, and 2 mM glutamine, at 37°C in 5% CO2 and 95% humidity.
Kinetics of cellular uptake
T98G and J774 cells were seeded at a density of 3.5 × 105 and 5 × 105 cells/well, respectively, in six-well plates and allowed to attach overnight. Subsequently, the medium was replaced with fresh modified Eagle’s medium without fetal bovine serum containing siRNA-Cy3/G7 (50 nM siRNA, 38.4 nM G7, final concentrations used in all experiments) and the cells were incubated for 0.5, 1, 2, and 5 hours at 37°C. After each incubation period, the medium was removed, the cells were washed with phosphate-buffered saline (pH 7.4), and harvested by trypsinization. The fluorescence of the non-internalized dendriplexes attached to the plasma membrane was quenched by a 10-minute incubation with 0.2% trypan blue in phosphate-buffered saline solution. After washing with phosphate-buffered saline, a total of 10,000 cells were analyzed by flow cytometry (Becton Dickinson FACS-Calibur, San Jose, CA). Data were analyzed using WinMDI 2.9 software.
Meanwhile, T98G and J774 cells grown on six-well plates as previously stated were preincubated for 1 hour at 4°C prior to addition of the siRNA-Cy3/G7 dendriplexes. The cells were then incubated for 2 hours at 4°C and processed for flow cytometry. Additionally, cells incubated at 37°C with siRNA-Cy3/G7 dendriplexes for 2 hours were processed without trypan blue incubation.
Cytotoxicity
Cell viability upon treatment with inhibitors of endocytosis and siRNA/G7 dendriplexes was measured by MTT assay and lactate dehydrogenase leakage in culture supernatants. Cells were seeded at a density of 5 × 104 cells/well in 96-well plates and allowed to attach overnight. The medium was then replaced with fresh modified Eagle’s medium without fetal bovine serum containing the following inhibitors: chlorpromazine 5 μg/mL and 10 μg/mL, methyl-β-cyclodextrin 14 μM, 28 μM, and 10 mM for T98G cells; 0.1 mM and 10 mM for J774 cells; genistein 150, 200, and 300 μM for T98G cells and 200 and 300 μM for J774 cells; chloroquine 250, 500, and 600 μM; cytochalasin D 10, 30, and 50 mM; and nocodazole 30 and 50 μM. After 1 hour of incubation at 37°C, 10 μL of siRNA/G7 was added and the cells were incubated for another 2 hours. The supernatants were then transferred to fresh tubes, centrifuged at 250 × g for 4 minutes, and the lactate dehydrogenase content was measured using a CytoTox lactate dehydrogenase kit (Promega, Madison, WI). Percent lactate dehydrogenase leakage was expressed relative to treatment with Triton X-100. Cells attached to plates were processed for MTT assay. A 0.45 mg/mL MTT solution was added and incubated for 3 hours. The solution was then removed, the insoluble formazan crystals were dissolved with dimethylsulfoxide, and absorbance was measured at 570 nm using a microplate reader. Cell viability was expressed as a percentage of the viability of cells grown in medium.
Uptake in presence of inhibitors of endocytosis
T98G and J774 cells were seeded in six-well plates as stated earlier. The medium was then replaced with fresh modified Eagle’s medium without fetal bovine serum containing endocytic inhibitors at a nontoxic concentration. The cells were incubated for 1 hour at 37°C, then 10 μL of siRNA/G7 was added and the cells were incubated for another 2 hours. The medium was then removed, the cells were washed twice with phosphate-buffered saline and harvested by trypsinization, then processed for flow cytometry as stated before. The percentage of cellular internalization was calculated on the basis of the geometric mean (Gm), as follows:
where Gmobs is the Gm of cells incubated with inhibitors and siRNA-Cy3/G7 dendriplexes and Gmcontrol is the Gm of cells incubated with siRNA-Cy3/G7 dendriplexes.
If uptake of dendriplexes was not reduced, the effect of the same inhibitor concentration on uptake of Tf-Alexa-555 as a marker of clathrin-dependent endocytosisCitation36 or CtxB-Alexa- 555 as a marker of caveolin-mediated endocytosisCitation37,Citation38 was assayed (although a significant fraction is also taken up by clathrin-dependent endocytosisCitation39). Cells treated with inhibitors were incubated with 15 μg/mL of Tf-Alexa-555 or 1 μg/mL of CtxB-Alexa-555 for 15 minutes at 37°C, and the cells were then processed for flow cytometry as described earlier.
EGFP silencing activity in presence of inhibitors of endocytosis
T98G-EGFP and J774-EGFP cells seeded in six-well plates (1.2 × 105 cells per well) were allowed to attach overnight in modified Eagle’s medium containing 500 μg/mL of G418 and 10% of fetal bovine serum. The medium was then replaced by fetal bovine serum-free modified Eagle’s medium containing chlorpromazine 10 μg/mL, genistein 150 mM, or chloroquine 125 μM. The cells were incubated for 1 hour at 37°C, and then siRNA/G7 was added. After 5 hours of incubation at 37°C, the culture medium was replaced with fresh modified Eagle’s medium containing 5% fetal bovine serum. After 72 hours, the cells were prepared for flow cytometry as described earlier, and data for at least 50,000 cells were acquired. The level of EGFP inhibition in the control cells was set at 100%.
Colocalization of siRNA/G7 dendriplexes with transferrin and cholera toxin
Colocalization of siRNA/G7 with Tf-Alexa-555 and CtxB-Alexa- 555 was studied by confocal laser scanning microscopy. Due to emission superposition of the wavelength of Alexa fluor-555-labeled markers (λ em 565) and Cy3-labeled siRNA (λ em 570), G7 was labeled with FITC.Citation40 Briefly, G7 in Tris-HCl buffer was incubated overnight with FITC in acetone solution at a 1:20 molar ratio in the dark at 25°C. The G7-FITC was then separated from free FITC by size exclusion chromatography on a Sephadex-G25 column. The presence of G7 in the void volume was confirmed by 20% polyacrylamide gel electrophoresis stained with Coomassie Blue. The absence of free FITC in the conjugates was verified by thin layer chromatography, using chloroform and methanol (1:1) as solvents.Citation41
T98G and J774 cells were seeded at a density of 5 × 104 cells/well in 24-well plates with rounded cover slips on the bottom and allowed to attach overnight. The medium was then replaced with fresh modified Eagle’s medium without fetal bovine serum containing Tf-Alexa-555, CtxB-Alexa-555, siRNA/G7-FITC and Tf-Alexa-555 15 μg/mL, and siRNA/G7-FITC and CtxB-Alexa-555 1 μg/mL. After 2 hours of incubation at 37°C (Tf-Alexa-555 and CtxB-Alexa-555 were also incubated at 4°C), the medium was removed, and the cells were washed twice with phosphate-buffered saline and fixed with 4% formaldehyde in phosphate-buffered saline for 25 minutes at 4°C. The cells were then washed, the cover slips were mounted on slides, and samples were observed under a Nikon confocal laser microscope.
Pearson’s correlation coefficient, which describes the correlation of intensity distribution between channels, and is independent of the intensity of signals, was calculated using Mac Biophotonics MBF-Image J software (http://www.macbiophotonics.ca/imagej/). Pearson’s correlation coefficient values between 0.5 and 1 indicate colocalization, while values between -1 and 0.5 indicate no colocalization.Citation42
Intracellular association of siRNA and G7 and colocalization with LysoTracker Red
T98G and J774 cells were seeded on four 24-well plates with rounded cover slips on the bottom as stated before. Subsequently, the medium was replaced with fresh modified Eagle’s medium without fetal bovine serum containing siRNA-Cy3/G7-FITC. After 5 hours of incubation at 37°C, the medium was removed from all plates. Cells from two plates were immediately processed, one plate for confocal laser scanning microscopy as stated before, and the other plate for endosomal-lysosomal staining by 30 minutes of incubation with LysoTracker Red 3.8 μM. Cells from the remaining two plates were incubated for another 5 hours in fresh modified Eagle’s medium with 10% fetal bovine serum at 37°C, and the cells were then processed for confocal laser scanning microscopy and endosomal-lysosomal staining as stated before.
Alkalinizing capacity of siRNA/G7 dendriplexes
siRNA/G7-FITC (containing 10 μM G7) and siRNA/Lipofectamine 2000 complexes (1 μg siRNA:3.6 μg Lipofectamine 2000, 55 μg/mL Lipofectamine 2000) were incubated in the following buffers: 10 mM acetic acid-sodium acetate, pH 4; 10 mM acetic acid-sodium acetate, pH 5; 10 mM carbonic acid-sodium bicarbonate, pH 6; and 10 mM Tris-HCl pH 7.4. After 20 minutes of incubation at 25°C, the resulting pH values were measured, the fluorescence emission spectra were recorded using a LS 55 PerkinElmer luminescence spectrometer, and the release of siRNA was determined by 20% polyacrylamide gel electrophoresis in Tris/borate/ethylenediamine tetra-acetic acid buffer (pH 8.3). The siRNA bands were stained with 1 μg/mL of ethidium bromide for 5 minutes and detected using an ultraviolet transilluminator.
Statistical analysis
Statistical analyses were performed by one-way analysis of variance, followed by Dunnett’s test using Prisma software (v 4.00; Graphpad Software Corporation, San Diego, CA). Significance levels are shown in the figure legends.
Results
Kinetics, mechanisms of cellular uptake and gene silencing activity, effect of endocytosis inhibitors
First, the cellular uptake of 150 nm dendriplexes of 25 mV zeta potential was followed over 5 hours using trypan blue as a fluorescence quencher for the extracellular dendriplexes adsorbed to the plasma membrane. While internalization by T98G cells obeyed a linear fit, uptake by J774 cells fitted better to a nonlinear equation (). Uptake into T98G cells was higher than that into J774 cells after 5 hours. Curvilinear cellular uptake kinetics, due to saturation of membrane binding sites, is characteristic of adsorptive endocytosis.Citation43 On the other hand, Perumal et al found that uptake of cationic G4-NH3 dendrimers by A549 cells plateaus off after 1 hour, while uptake of anionic G3.5-COOH dendrimers and neutral G4-OH dendrimers increased more or less linearly over the duration of treatment.Citation44
Figure 1 Uptake of siRNA-Cy3/G7 dendriplexes by (A) J774 cells and (B) T98G cells at 37°C as a function of time.
Abbreviation: siRNA, small interfering RNA.
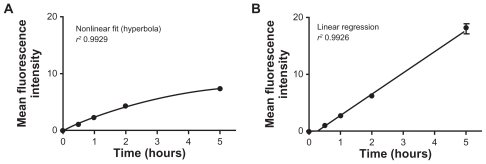
On the basis of the kinetic data, we defined 2 hours as the incubation time, ensuring uptake sufficient to allow further quantification. Additionally, upon 2 hours of incubation, we observed that cellular fluorescence was increased by 70% in the absence of trypan blue quenching, indicating surface adsorption of the dendriplexes. Cellular fluorescence was reduced by 50%–70% on incubation at 4°C, showing the energy dependence of dendriplex uptake.
Six endocytosis inhibitors were used to study the mechanism of dendriplex uptake: methyl-β-cyclodextrin (MβCD), a cholesterol-depleting cyclic oligomer of glycopiranoside used as a general endocytosis inhibitor;Citation45,Citation46 chlorpromazine, a cationic amphipathic molecule that inhibits formation of clathrin-coated pits;Citation47 genistein, a specific tyrosine kinase inhibitor that causes local disruption of the actin cytoskeleton and avoids recruiting of dynamin II, indispensable for both caveolin- mediated endocytosis and Rho-mediated endocytosis;Citation48,Citation49 chloroquine, a weak base that increases endosomal and lysosomal pH, causing lysosomal disruptionCitation50 and reduction of clathrin-dependent endocytosis;Citation51 cytochalasine D, which disrupts actin filaments and inhibits actin polymerization and membrane ruffling, affecting both phagocytosis and pinocytosis;Citation52,Citation53 and nocodazole, which disrupts microtubules and inhibits translocation of endosomes and lysosomes,Citation54 blocking clathrin-dependent endocytosis but not affecting SV40 uptake to caveosomes, and preventing its sorting from caveosomes to endoplasmic reticulum.Citation55
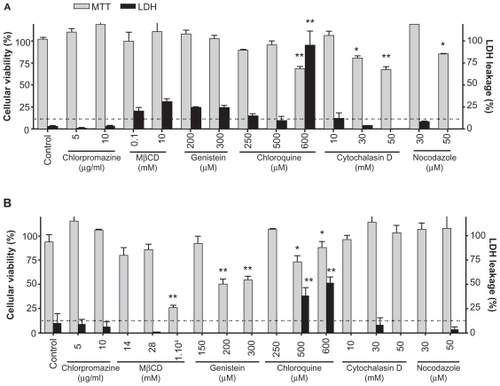
Cell viability in the presence of the above inhibitors was determined first in order to establish optimal concentrations for their use. Viability of J774 cells was reduced after incubation with 600 μM of chloroquine (30%), 30 and 50 μM of cytochalasine D (20% and 40%, respectively) and 50 μM of nocodazole (15%). Only treatment with 600 μM chloroquine produced 75% lactate dehydrogenase leakage (). In contrast, T98G cell viability was reduced upon incubation with 10 mM of MβCD (75%), 200 and 300 μM of genistein (50%), and 500 and 600 μM of chloroquine (10%–20%). Only treatments with 500 and 600 μM of chloroquine produced 40% and 50% lactate dehydrogenase leakage, respectively (). Based on these results, the following concentrations were selected for further experiments: chlorpromazine 10 μg/mL, MβCD 2 mM for J774 cells and 28 μM for T98G cells, genistein 150 μM, chloroquine 125 μM, cytochalasine D 10 μM, and nocodazole 10 μM.
Figure 2 Cytotoxicity of inhibitors of endocytosis and siRNA-Cy3/G7 dendriplexes on (A) J774 cells and (B) T98G cells. Viability of treated cells was determined by MTT assay and lactate dehydrogenase leakage.
Notes: Bars represent means ± standard deviations (n = 6). *P < 0.05, **P < 0.01.
Abbreviation: MβCD, methyl-β-cyclodextrin.
In the J774 cells, we found that MβCD (75%), chlorpromazine (35%), chloroquine (85%), cytochalasine D (70%), and nocodazole (75%) significantly reduced uptake of the siRNA dendriplexes. As seen in , uptake of siRNA dendriplexes was slightly reduced by genistein (25%) and the uptake of CtxB-Alexa-555, used as a control for genistein inhibition, was also slightly reduced (25%). In contrast, MβCD (75%), genistein (45%), and cytochalasine D (50%) reduced the uptake of siRNA dendriplexes in T98G cells. Chlorpromazine increased the uptake of siRNA dendriplexes. These results are consistent with those reported for HeLa, HepG2, and EA.hy 926 cells, where chlorpromazine was observed to increase the uptake of pDNA dendriplexes.Citation32 Chloroquine and nocodazole did not modify the uptake of siRNA dendriplexes. Uptake of Tf-Alexa-555, used as a control for chlorpromazine and chloroquine inhibition, was reduced by 50%.
Figure 3 Effect of inhibitors of endocytosis on (A) uptake and (B) silencing activity of siRNA-Cy3/G7 dendriplexes by J774 and T98G cells.
Notes: Bars represent means ± standard deviations (n = 6). *P < 0.05, **P < 0.01.
Abbreviations: EGFP, enhanced green fluorescent protein; siRNA, small interfering RNA; MβCD, methyl-β-cyclodextrin.
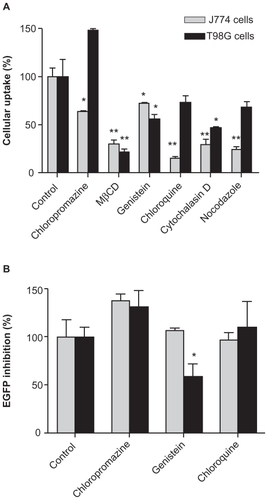
After the effects of six specific and nonspecific endocytosis inhibitors on dendriplex uptake were established, we examined the effect of specific inhibitors, ie, chlorpromazine (clathrin-dependent endocytosis), genistein (caveolin-mediated endocytosis), and chloroquine (endosome-lysosome acidification), on the silencing activity of the dendriplexes. No toxicity to either cell type was found after 5 hours of incubation of the inhibitors with siRNA/G7. In J774-GFP cells, genistein and chloroquine did not modify the silencing activity of dendriplexes but chlorpromazine slightly enhanced it (). On the other hand, in T98G-GFP cells, only genistein reduced the silencing activity and chlorpromazine slightly enhanced it. Other researchers have reported that chlorpromazine increases the gene delivery efficiency of pDNA/polyethylenimine polyplexes, because inhibiting clathrin-dependent endocytosis could increase uptake by caveolin-mediated endocytosis or other pathways.Citation23
Intracellular distribution
In addition to determining the effect of inhibitors of endocytosis on the uptake and silencing activity of siRNA/G7 dendriplexes, quantitative analysis of the colocalization of siRNA/G7-FITC with Tf-Alexa-555 and CtxB-Alexa-555 was also determined.
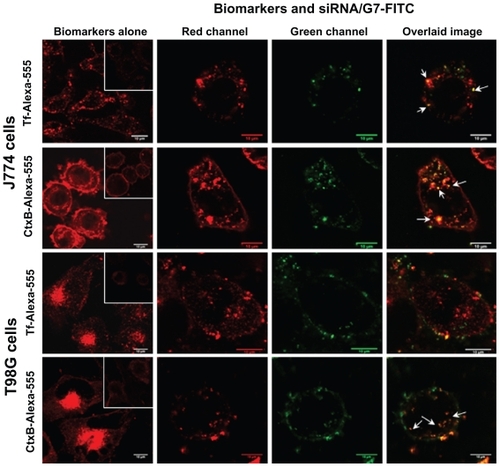
In J774 cells, Tf-Alexa-555 () was seen as homogeneously distributed cytoplasmic dots, while CtxB-Alexa- 555 ( inset) was found within the plasma membrane and heterogeneously distributed as cytoplasmic dots; these dots diminished in number and intensity at 4°C (). The distribution pattern of the markers did not change after coincubation with siRNA/G7-FITC dendriplexes, and high colocalization of both biomarkers was observed with G7-FITC (Pearson’s correlation coefficient >0.5). In T98G cells, both Tf-Alexa-555 and CtxB-Alexa- 555 were seen as perinuclear dots that faded at 4°C. After coincubation with siRNA/G7-FITC dendriplexes, the fluorescence of G7-FITC did not colocalize with Tf- Alexa-555 (Pearson’s correlation coefficient < 0.5), but did so with CtxB-Alexa-555 (Pearson’s correlation coefficient > 0.5).
Figure 4 Confocal laser scanning microscopic images of cells incubated with the Tf-Alexa-555 and CtxB-Alexa-555 biomarkers alone at 37°C and 4°C (insets) and coincubated with biomarkers and siRNA/G7-FITC.
Note: Arrows indicate colocalization dots (yellow-orange).
Abbreviations: siRNA, small interfering RNA; FITC, fluorescein isothiocyanate.
On the other hand, a key step in the siRNA interference mechanism is the association between siRNA and the RNA-induced silencing complex in the cytoplasm. The association between siRNA and G7 within the dendriplex and dendriplex entrapping within endosomes/lysosomes can impair release and activity in the cytoplasm. To estimate the extent of dendriplex dissociation and lysosomal escape, colocalization of siRNA-Cy3 with G7-FITC and G7-FITC with siRNA dendriplexes using LysoTracker Red as a lysosomal marker was determined by confocal laser scanning microscopy.
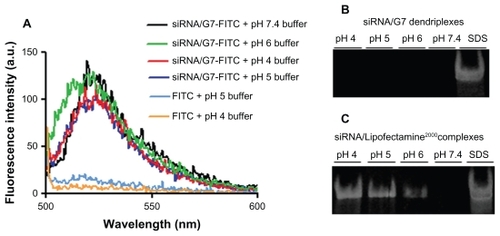
Our results showed that, in J774 cells, siRNA-Cy3/G7-FITC remained associated (Pearson’s correlation coefficient > 0.5) within the lysosomes, as indicated by colocalization of G7-FITC and the lysotracker (Pearson’s correlation coefficient > 0.5) at 5 and 10 hours of incubation (). However, in T98G cells, siRNA-Cy3/G7-FITC also remained associated (Pearson’s correlation coefficient > 0.5) within the lysosomes, as indicated by colocalization of G7-FITC with the lysotracker (Pearson’s correlation coefficient > 0.5) after 5 hours. After 10 hours, siRNA-Cy3/G7-FITC was still associated, but G7-FITC was no longer colocalized with the lysotracker (Pearson’s correlation coefficient < 0.5). FITC reversibly loses fluorescence below pH 5.5, so its fluorescence within lysosomes was expected to be quenched. However, we observed that the fluorescence of the G7-FITC-labeled dendriplexes was not diminished after internalization and persisted for 10 hours in both cell types. We also observed that, after incubation with siRNA/G7 dendriplexes, the pH of the set of buffers was shifted from 4, 5, and 6 to 5.6, 7.3, and 8.6, respectively, and furthermore, the fluorescence of the G7-FITC-labeled dendriplexes was not diminished (). Polyacrylamide gel electrophoresis showed that siRNA/G7 dendriplexes remained associated after incubation (), meaning that the dendriplexes induced medium alkalinization that could impair their acid-induced dismantling. The siRNA band was only observed upon addition of sodium dodecyl sulfate. In contrast, siRNA/Lipofectamine 2000 complexes had no alkalinizing effect and were dissociated when incubated with acidic buffers, releasing 38%, 68%, and 79% of siRNA at pH 6, 5, and 4, respectively ().
Figure 5 Confocal laser scanning microscopic images of cells incubated with siRNA-Cy3/G7-FITC and cells incubated with siRNA/G7-FITC where acidic endolysosomal compartments were stained with LysoTracker Red DND-99.
Note: Arrows indicate colocalization dots (yellow-orange).
Abbreviations: siRNA, small interfering RNA; FITC, fluorescein isothiocyanate.
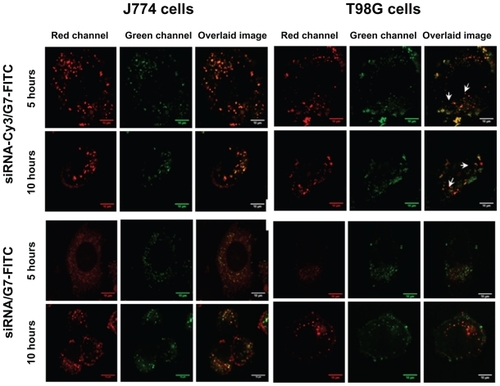
Figure 6 Emission spectra of siRNA/G7-FITC and free FITC incubated with a set of buffers (A), corresponding polyacrylamide gel electrophoresis of siRNA/G7 (B), and siRNA/Lipofectamine™ 2000 (C).
Abbreviations: siRNA, small interfering RNA; FITC, fluorescein isothiocyanate.
Overall, the results showed that caveolin, clathrin, vesicular acidification, cholesterol, actin filaments, and the microtubular cytoskeleton were involved in uptake of siRNA dendriplexes by J774 cells. Furthermore, while dendriplexes colocalized with Tf-Alexa-555 and CtxB-Alexa-555, their silencing activity was not decreased by chlorpromazine, genistein, or chloroquine. These results suggest that the dendriplexes were taken up by a combination of clathrin-dependent endocytosis and caveolin-mediated endocytosis, and that both these mechanisms were involved in the silencing activity. On the other hand, T98G cells, which are caveolin- 1-expressing nonphagocytic cells, internalized siRNA dendriplexes via pathways involving caveolin, cholesterol, and the actin cytoskeleton. Dendriplexes colocalized with CtxB-Alexa-555, and their silencing activity was decreased only by genistein, suggesting that the dendriplexes were taken up and expressed silencing activity only via caveolin-mediated endocytosis.
Discussion
The steps followed by a nanoparticle taken up via the main pinocytic pathways are now relatively well understood. However, our knowledge about the internalization pathway is insufficient to predict successful transfection by nucleic acids/polycationic complexes.
In the present work, we observed that T98G cells internalized siRNA dendriplexes at a higher rate than J774 cells. Previously, we have reported lower silencing activity of siRNA dendriplexes in T98G cells than in J774 cells (35% vs 45%).Citation34 This finding indicated that extent of cellular uptake of dendriplexes was not directly related to silencing. Similarly, O’Neill et al found that despite its higher uptake, cyclodextrin-mediated luciferase expression was significantly lower than for Lipofectamine 2000 and comparable with that of polyethylenimine in Caco-2 cells.Citation56
A potential explanation for this phenomenon is that dendriplexes enter the cells by a pinocytic pathway that can be more or less favorable for transfection. An example illustrating how different endocytic pathways can yield different transfection efficiencies was provided by Douglas et al, who reported significant uptake of alginate-chitosan nanoparticles by both 293T cells and Chinese hamster ovary cells.Citation22 However, while 293T cells were successfully transfected, no significant transgene expression was detected in Chinese hamster ovary cells. The authors concluded that, in 293T cells, clathrin-dependent endocytosis was responsible for uptake and expression of nanoparticles, while in Chinese hamster ovary cells, caveolin-mediated endocytosis was responsible for the uptake, but was not useful for gene expression. Our results indicate that in J774, a combination of clathrin-dependent endocytosis and caveolin-mediated endocytosis was involved in the uptake of dendriplexes, while in T98G, only caveolin-mediated endocytosis was involved. Silencing activity was mediated by both clathrin-dependent endocytosis and caveolin-mediated endocytosis in J774 cells and by caveolin-mediated endocytosis alone in T98G cells. The involvement of caveolin-mediated endocytosis in uptake and silencing activity by dendriplexes is consistent with a previous finding that caveolin-mediated endocytosis was involved in uptake and expression of large pDNA dendriplexes (800–900 nm) by EA.hy 926 endothelial cells and COS-7 fibroblasts, and that overexpression of caveolin-1 enhanced uptake and transfection efficiency.Citation32
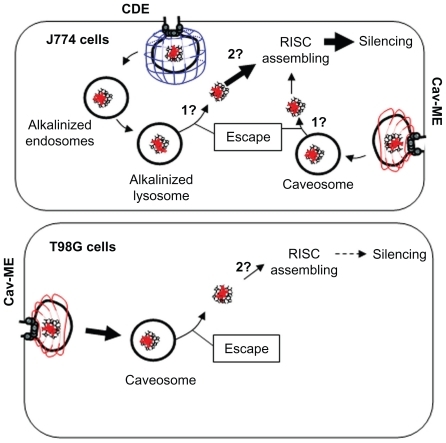
When the uptake mechanisms were determined, they were related to the amount of dendriplex uptake and subsequent silencing efficiency. T98G cells showed higher uptake but lower silencing activity (both involving caveolin-mediated endocytosis) than J774 cells, while in the latter cell type, clathrin-dependent endocytosis and caveolin-mediated endocytosis were responsible for higher silencing activity. These results show not only that the internalization rate was not directly related to the silencing activity, but also allow us to propose that clathrin-dependent endocytosis could favor the silencing activity of siRNA dendriplexes more than caveolin-mediated endocytosis. A schematic diagram of this model is shown in .
Figure 7 Internalization pathways for siRNA/G7 dendriplexes in J774 and T98G cells. (1) Unknown mechanism of cytoplasmic delivery. (2) Unknown mechanism of dendriplex dismantling.
Note: Thick arrows indicate favored uptake or silencing expression.
Abbreviations: cav-ME, caveolin-mediated endocytosis; CDE, clathrin-dependent endocytosis; RISC, RNA-induced silencing complex.
Whichever pathway is involved in uptake, endosomal/caveosomal escape is a significant barrier to polyplex-mediated transfection. Although the temporal scale of permeabilization and endosomal lysis caused by dendrimers is in the order of 30–75 minutes (induced by polyamidoamine G2 in Chinese hamster ovary cellsCitation19) we have found that siRNA/G7 dendriplexes remained within the lysosomes of J774 cells for up to 10 hours. Similar results were observed with a polyamidoamine G4 dendrimer that colocalized with lysosomal markers over a period of 3–12 hours, after which the signal decreased in the lysosomes and began to colocalize with the mitochondrial marker.Citation57 In contrast, the dendriplexes did not colocalize with the lysotracker after 10 hours in T98G cells. Even though the dendriplexes escaped into the cytosol of T98G cells more effectively than in J774 cells, the inhibition activity was lower. Therefore, association with the RNA-induced silencing complex and subsequent RNA degradation may affect inhibition activity.
We also observed that the two cell lines were expressing silencing activity 72 hours later, which leads us to conclude that siRNA was incorporated into the RNA-induced silencing complex, in spite of the dendriplexes still remaining associated at 10 hours. Relatively slow dissociation kinetics have also been shown for pDNA/polyethylenimine polyplexes.Citation58,Citation59 Although insufficient unpacking of polyplexes inside the cell is considered to be one of the major barriers in the transfection process (eg, pDNA has to be unwrapped in order to be expressed), it is uncertain if siRNA dendriplexes are affected by the same physical constraints. It is important to note that while the structure of pDNA/polycations corresponds to a model of pDNA wrapped around cationic cores, the siRNA/polycation structure is quite different,Citation60 because siRNA behaves as a rigid rod which, unlike pDNA, is not likely to condense further, resulting in weak complexes.Citation61,Citation62 Therefore, two scenarios could be hypothesized. Firstly, siRNA dendriplex dismantling is mediated by an as yet unknown mechanism which is pH-independent and slow, and no acidification-dependent process would be likely to be involved, given that the dendrimers are strongly alkalinizing molecules. Secondly, dendriplex dismantling may not be necessary for siRNA incorporation into the RNA-induced silencing complex.
Conclusion
Our results show that cellular uptake of siRNA dendriplexes was inversely related to the silencing activity of the siRNA dendriplex in J774 and T98G cells. Both caveolin-mediated endocytosis and clathrin-dependent endocytosis pathways contributed to the silencing activity in J774 cells, while only caveolin-mediated endocytosis was involved in T98G cells. siRNA dendriplexes showed relatively slow dissociation kinetics, and their escape towards the cytosol was probably not mediated by acidification, independent of the uptake pathway. Given that clathrin-dependent endocytosis was more effective than caveolin-mediated endocytosis, targeting of dendriplexes with clathrin-dependent endocytosis-specific ligands, such as transferrin, could increase the silencing activity in these two cell types.
Acknowledgments
This work was supported by the Secretaria de Investigaciones, Universidad Nacional de Quilmes. MJM and ELR are members of Carrera del Investigador Científico del Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina. APP has a fellowship from CONICET. MLC has a fellowship from the Comisión de Investigaciones Científicas de la Provincia de Buenos Aires.
Disclosure
The authors report no conflicts of interest in this work.
References
- HussainSFYangDSukiDAldapeKGrimmEHeimbergerABThe role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responsesNeuro Oncol20068326127916775224
- StuppRMasonWPBentMJRadiotherapy plus concomitant and adjuvant temozolomide for glioblastomaN Engl J Med200535298799615758009
- StylliSKayeAMacGregorLHowesMRajendraPPhotodynamic therapy of high grade glioma-long term survivalJ Clin Neurosci200512438939815925768
- DemuthTBerensMMolecular mechanisms of glioma cell migration and invasionJ Neurooncol200470221722815674479
- LingorPBährMTargeting neurological disease with RNAiMol BioSyst2007377378017940660
- ZhouZYuanXLiZRNA interference targeting EphA2 inhibits proliferation, induces apoptosis, and cooperates with cytotoxic drugs in human glioma cellsSurg Neurol200870656256918614215
- UlbrichtUEckerichCFillbrandtRWestphalMLamszusKRNA interference targeting protein tyrosine phosphatase zeta/receptor-type protein tyrosine phosphatase beta suppresses glioblastoma growth in vitro and in vivoJ Neurochem20069851497150616923162
- GrzelinskiMUrban-KleinBMartensTRNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenograftsHum Gene Ther200617775176616839274
- NagaiTTanakaMTsuneyoshiYTargeting tumor-associated macrophages in an experimental glioma model with a recombinant immunotoxin to folate receptor betaCancer Immunol Immunother200958105771586
- GaoYLiuXLLiXRResearch progress on siRNA delivery with nonviral carriersInt J Nanomedicine201161017102521720513
- SahayGAlakhovaDYKabanovAVEndocytosis of nanomedicinesJ Control Release2010145318219520226220
- GlebovOOBrightNANicholsBJFlotillin-1 defines a clathrin-independent endocytic pathway in mammalian cellsNat Cell Biol200681465416341206
- GongQHuntsmanCMaDClathrin-independent internalization and recyclingJ Cell Mol Med200812112614418039352
- KumariSSwethaMGMayorSEndocytosis unplugged: multiple ways to enter the cellCell Res201020325627520125123
- Dautry-VarsatACiechanoverALodishHFpH and the recycling of transferring during receptor-mediated endocytosisProc Natl Acad Sci U S A1983808225822626300903
- KichlerALeborgneCCoeytauxEDanosOPolyethylenimine-mediated gene delivery: a mechanistic studyJ Gene Med20013213514411318112
- BehrJPSynthetic gene-transfer vectorsAcc Chem Res199326274278
- HaenslerJSzokaFCPolyamidoamine cascade polymers mediate efficient transfection of cells in cultureBioconjugate Chem199345372379
- SonawaneNDSzokaFCJrVerkmanASChloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexesJ Biol Chem200327845448264483112944394
- ThomasTPMajorosIKotlyarAMullenDHollMMBakerJRCationic poly(amidoamine) dendrimer induces lysosomal apoptotic pathway at therapeutically relevant concentrationsBiomacromolecules200910123207321419924846
- AkincAThomasMKlibanovAMLangerRExploring polyethylenimine- mediated DNA transfection and the proton sponge hypothesisJ Gene Med20057565766315543529
- DouglasKLPiccirilloCATabrizianMCell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectorsEur J Pharm Biopharm200868367668717945472
- GabrielsonNPPackDWEfficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cellsJ Control Release20091361546119217921
- RejmanJBragonziAConeseMRole of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexesMolec Ther20051246847415963763
- van der AaMAHuthUSHäfeleSYCellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cellsPharm Research200724815901598
- ZuhornISEngbertsJBHoekstraDGene delivery by cationic lipid vectors: overcoming cellular barriersEur Biophys J2007364–534936217019592
- PartonRGCaveolae meet endosomes: a stable relationshipDev Cell200474445846015469832
- HayerAStoeberMRitzDEngelSMeyerHHHeleniusACaveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradationJ Cell Biol2010191361566221041450
- von GersdorffKSandersNNVandenbrouckeRDe SmedtSCWagnerEOgrisMThe internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex typeMolec Ther2006145574575316979385
- MenjogeARKannanRMTomaliaDADendrimer-based drug and imaging conjugates: design considerations for nanomedical applicationsDrug Discov Today2010155–617118520116448
- ManuntaMHongTPSagooPKasheKGeorgeAJGene delivery by dendrimers operates via a cholesterol dependent pathwayNucleic Acids Res20043292730273915148360
- ManuntaMNicholsBJHongTPSagooPHarperJGeorgeAJGene delivery by dendrimers operates via different pathways in different cells, but is enhanced by the presence of caveolinJ Immunol Methods20063141–213414616893551
- QiRMullenDGBakerJRBanaszak HollMMThe mechanism of polyplex internalization into cells: testing the GM1/Caveolin-1 lipid raft mediated endocytosis pathwayMol Pharm20097126727920025295
- PerezAPRomeroELMorillaMJEthylendiamine core PAMAM dendrimers/siRNA complexes as in vitro silencing agentsInt J Pharm20093801–218920019577619
- WaiteCLRothCMPAMAM-RGD Conjugates enhance siRNA delivery through a multicellular spheroid model of malignant gliomaBioconjug Chem200920101908191619775120
- WideraANorouziyanFShenWCMechanisms of TfR-mediated transcytosis and sorting in epithelial cells and applications toward drug deliveryAdv Drug Deliv Rev200355111439146614597140
- PartonRGSimonsKThe multiple faces of caveolaeNat Rev Mol Cell Biol20078318519417318224
- PelkmansLKartenbeckJHeleniusACaveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ERNat Cell Biol20013547348311331875
- TorgersenMLSkrettingGvan DeursBSandvigKInternalization of cholera toxin by different endocytic mechanismsJ Cell Sci2001114Pt 203737374711707525
- KolhePMisraEKannanRMKannanSLieh-LaiMDrug complexation, in vitro release and cellular entry of dendrimers and hyper-branched polymersInt J Pharm20032591–214316012787643
- JevprasesphantRPennyJAttwoodDMcKeownNBD’EmanueleAEngineering of dendrimer surfaces to enhance transepithelial transport and reduce cytotoxicityPharm Res200320101543155014620505
- ZinchukVZinchukOQuantitative colocalization analysis of confocal fluorescence microscopy imagesCurr Protocol Cell Biol2008Chapter 4Unit 419
- HuangMMaZKhorELimLYUptake of FITC–chitosan nanoparticles by A549 cellsPharm Res200219101488149412425466
- PerumalOPInapagollaRKannanSKannanRMThe effect of surface functionality on cellular trafficking of dendrimersBiomaterials20082924–253469347618501424
- SubtilAGaidarovIKobylarzKLampsonMAKeenJHMcGrawTEAcute cholesterol depletion inhibits clathrin coated pit buddingProc Natl Acad Sci U S A199996126775678010359788
- LakkarajuARahmanYEDubinskyJMLow-density lipoprotein receptor-related protein mediates the endocytosis of anionic liposomes in neuronsJ Biol Chem200227717150851509211830589
- WangLHRothbergKGAndersonRGMis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pits formationJ Cell Biol19931235110711178245121
- PartonRGJoggerstBSimonsKRegulated internalization of caveolaeJ Cell Biol19941275119912157962085
- NabiIRLePUCaveolae/raft-dependent endocytosisJ Cell Biol2003161467367712771123
- MichiharaATodaKKuboTFujiwaraYAkasakiKTsujiHDisruptive effect of chloroquine on lysosomes in cultured rat hepatocytesBiol Pharm Bull200528694795115930724
- PlessDDWellnerRBIn vitro fusion of endocytic vesicles: effects of reagents that alter endosomal pHJ Cell Biochem199662127398836873
- BoucrotESaffarianSMassolRKirchhausenTEhrlichMRole of lipids and actin in the formation of clathrin-coated pitsExp Cell Res200631204036404817097636
- QualmannBKesselsMMKellyRBMolecular links between endocytosis and the actin cytoskeletonJ Cell Biol20001504F11111610974009
- SubtilADautry-VarsatAMicrotubule depolymerization inhibits clathrin coated-pit internalization in non-adherent cell lines while interleukin 2 endocytosis is not affectedJ Cell Sci1997110Pt 19244124479410882
- PelkmansLKartenbeckJHeleniusACaveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ERNat Cell Biol20013547348311331875
- O’NeillMJGuoJByrneCDarcyRO’ DriscollCMMechanistic studies on the uptake and intracellular trafficking of novel cyclodextrins transfection complexes by intestinal epithelial cellsInt J Pharm20114131–217418321530624
- PaiCLShiehMJLouPJHuangFHWangTWLaiPSCharacterization of the uptake and intracellular trafficking of G4 polyamidoamine dendrimersAus J Chem201164302308
- OgrisMSteinleinPCarottaSBrunnerSWagnerEDNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expressionAAPS PharmSci200133E2111741272
- BieberTMeissnerWKostinSNiemannAElsasserHPIntracellular route and transcriptional competence of polyethylenimine–DNA complexesJ Control Release2002822–344145412175756
- VasumathiVMaitiPKComplexation of siRNA with dendrimer: a molecular modeling approachMacromolecules2010431082648274
- GaryDJPuriNWonYYPolymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA deliveryJ Control Release20071211–2647317588702
- SpagnouSMillerADKellerMLipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNABiochemistry20044342133481335615491141