Abstract
Background
Infectious diseases cause many molecular assemblies and pathways within cellular signaling networks to function aberrantly. The most effective way to treat complex, diseased cellular networks is to apply multiple drugs that attack the problem from many fronts. However, determining the optimal combination of several drugs at specific dosages to reach an endpoint objective is a daunting task.
Methods
In this study, we applied an experimental feedback system control (FSC) method and rapidly identified optimal drug combinations that inhibit herpes simplex virus-1 infection, by only testing less than 0.1% of the total possible drug combinations.
Results
Using antiviral efficacy as the criterion, FSC quickly identified a highly efficacious drug cocktail. This cocktail contained high dose ribavirin. Ribavirin, while being an effective antiviral drug, often induces toxic side effects that are not desirable in a therapeutic drug combination. To screen for less toxic drug combinations, we applied a second FSC search in cascade and used both high antiviral efficacy and low toxicity as criteria. Surprisingly, the new drug combination eliminated the need for ribavirin, but still blocked viral infection in nearly 100% of cases.
Conclusion
This cascade search provides a versatile platform for rapid discovery of new drug combinations that satisfy multiple criteria.
Introduction
Current drug discovery efforts have primarily focused on identifying agents that tackle specific preselected cellular targets.Citation1,Citation2 However, in most cases a single drug does not correct all of the aberrantly functioning pathways in a disease to produce an effective treatment. Drugs directed at an individual target often have limited efficacy and poor safety profiles due to various factors, including compensatory changes in cellular networks upon drug stimulation,Citation3 redundancy,Citation4 crosstalk,Citation5 and off-target activities.Citation6 The use of drug combinations which act on multiple targets has been shown to be a more effective treatment strategy and is being used more frequently.Citation7,Citation8 This approach has been supported by successful clinical applications to treat various diseases, such as AIDS, cancer, and atherosclerosis.Citation9–Citation11 Often, the studies used high dosages of individual drugs to ensure treatment efficacy.Citation12 Unfortunately, the high doses necessary to provide efficacy often come with toxic side effects. Therefore, treatments with a drug combination at the lowest optimal dosages will achieve the goal of high efficacy and low toxicity, resulting in the most desirable drug cocktail.Citation13,Citation14 However, identifying the combination of effective drug molecules, and determining the proper dosage of each constitute is a challenging task. For example, even a small number of different drugs (six drugs) each tested at a few concentrations (seven dosages) results in 7Citation6 = 117,649 combinations. Screening all 117,649 combinations for the most desirable combination is an enormous task in terms of labor and time. Furthermore, another problem with combination medicine is that the highly efficacious drug combination may include drug(s) that are toxic or have side effects.
Viral infections have stood out as an interesting candidate for combination drug therapy. HIV, hepatitis C virus, and influenza infections have been shown to be effectively treated by combinations of antiviral drugs. The pathogenesis of viral infections is caused by a coordinated reprogramming of cellular pathways and protein complexes by viral factors to favor the replication and spread of the virus. Within these pathways and protein complexes, single targets have been found that upon drug manipulation can disrupt viral replication. However, intervention against a single drug target usually results in the selection of escape mutants that are ineffectively suppressed by the single drug. The preferred method is to target multiple viral pathways simultaneously, so that the drugs target distinct steps of viral replication to more effectively block replication and limit the likelihood that a multiple drug-resistant mutant will arise.
Herpes simplex virus-1 (HSV-1) is one of the most pervasive infections worldwide, causing genital, skin, and eye infections in millions of people.Citation15,Citation16 Common treatments for HSV-1, including virus-specific drugs such as acyclovir, are effective but exhibit limited long-term efficacy due to the development of drug resistant strains. Thus, more effective therapeutic methods are needed to combat the increasing spread of drug-resistant HSV-1. Based on an intensive literature search, six drugs associated with antiviral gene regulation, viral proliferation, cell growth, and cell death were selected in our experiments as candidates for establishing a new combination drug therapy. First, the standard HSV-1 antiviral drug acyclovir,Citation17 which is effective for the treatment of most herpes virus infections, acts as a chain terminator of DNA polymerase in virus infected cells. Acyclovir is also an effective control to measure efficacy. The second drug we included was ribavirin,Citation18 which has well established antiviral activity against RNA virus infections such as poliovirus and hepatitis C virus but the mechanism for antiviral activity against DNA viruses, such as HSV-1, remains unknown. Next, we included three cellular produced interferons (IFNs), IFN-α,Citation19 IFN-β,Citation20 and IFN-γ,Citation21 that have potent antiviral effects through the induction of cellular innate immune pathways. Finally, we also included tumor necrosis factor (TNF)-α,Citation22 a cellular protein that induces activation of nuclear factor kappa B (NF-κB) and cellular death pathways. Each of these compounds can potentially block HSV-1 replication by modulating distinct viral or cellular protein complexes and pathways, and thus represent distinct potential therapies. Therefore, a combination of these drugs should be a highly efficacious drug therapy.
Instead of testing all possible combinations of these drugs at different dosages by a high-throughput screen, an experimental feedback system control (FSC) approach can identify optimal drug combinations by testing approximately 0.1% or less of all possible combinations.Citation23–Citation25 Previously, we focused only on rapid searching for highly efficacious drug combinations.Citation23–Citation25 Here, we have successfully applied the FSC approach in our experiments to search for drug combinations that have high antiviral efficacy and then applied FSC in cascade to lower the doses of a toxic drug (ribavirin) for the treatment of HSV-1 using an in vitro infection model.
Methods
Algorithm
DE algorithm was coded with MATLAB software (Mathworks Inc, Natick, MA). Each drug combination was represented as a vector in the software. Coded dosage was used rather than absolute concentration. The dosages of 16 combinations in the first iteration were chosen arbitrarily. The code computed the objective function value of each combination, and suggested the new group of drug combinations to test in the following iteration.
Reagents
IFN-α, IFN-β, and IFN-γ were purchased from PBL Interferon Source (Piscataway, NJ). Ribavirin and acyclovir were purchased from Calbiochem (San Diego, CA). TNF-α was purchased from R&D Systems (Minneapolis, MN). Dulbecco’s Modified Eagle’s Medium (DMEM) was purchased from CELLGRO (Manassas, VA) and Fetalplex from Gemini Bio-Products (Woodland, CA). Penicillin/streptomycin and Trypsin-ethylenediaminetetraacetic acid (EDTA) were obtained from GIBCO (Grand Island, NY). Paraformaldehyde (PFA) was purchased from Electron Microscopy Sciences (Hatfield, PA). Phosphate buffered saline (PBS) was from EMD (Rockland, MA). All other plates and tubes were from BD Falcon (San Jose, CA).
Cell culture
NIH 3T3 cells were grown on 15 cm plates in DMEM supplemented with 5% Fetalplex and 1% penicillin/streptomycin and kept in a 37°C incubator with 5% CO2. To propagate cells, we plated 107 on each 15 mm plate and split the cells every 24 hours. For each experimental iteration, we plated 2 × 105 cells/well in a 24-well plate. To minimize variance generated from different batches of cells, the trial group and crossover group were tested and compared using the same batch of cells for each iteration.
Viral infection
HSV-1 KOS strain expressing green fluorescent protein (GFP) in frame with the ICP0 protein between amino acids 104 and 105 were a gift from Dr William Halford (Southern Illinois University School of Medicine, Springfield, IL). The virus was prepared by propagation of virus on a confluent monolayer of Vero cells.Citation26 Supernatants from infected cells were collected and centrifuged to separate cell debris. The cell pellet in residual medium was frozen and thawed three times at −80°C and 37°C, respectively. The residual supernatant was then pooled together with the original supernatant and viral titers were determined by a standard plaque assay on Vero cell monolayers. Multiplicity of infection (MOI) of 0.1 was used throughout except as indicated. To control MOI, we added cells, virus, and drug combinations at the same time and incubated at 37°C. After 17 hours, culture medium was aspirated and cells were detached with PBS-EDTA treatment at 37°C for 5 minutes. Detached cells were transferred to flow cytometry tubes, pelleted, and resuspended in 1.6% PFA and kept at 4°C until analysis. A BD FACS Canto II was used for flow cytometry analysis.
Results
HSV-1 infectious disease model
HSV-1 infection on an NIH 3T3 fibroblast cell line was used as an in vitro model system to search for new therapeutic drug combinations. The antiviral drugs that we have used in our therapeutic model include three antiviral cytokines (IFN-α, IFN-β, and IFN-γ), ribavirin, acyclovir, and TNF-α. Virus-infected cells were treated with single drugs or drug combinations and cultured for 16 hours. The HSV-1 strain used to infect the NIH 3T3 cells encodes a GFP reporter in infected cells, enabling flow cytometric analysis of cells to measure the rate and extent of infection, because the fluorescence intensity of GFP correlated to the presence of virus.Citation27 We determined efficacy of drug treatments by comparing the number of GFP-negative non-infected cells in the absence or presence of drug treatment. This value was considered the antiviral readout of a drug treatment.
The success of antiviral drug combinations depends on two essential factors: the drug combination used and the dose of each drug used. In the present study, seven dose concentrations for each of the six drugs were evaluated. Consequently, the total possible combinations of drugs and dosages are 7Citation6 = 117,649. The dosage levels were coded with numbers from 0 to 6, where 0 stands for a dosage of zero, 6 is the highest dose used for that drug as reported in the literature, and 5 to 1 are four-fold dilutions from the highest dose. The absolute concentrations, as well as the antiviral readouts (percentage of infected cells following treatment), are shown in and . This pilot study shows that ribavirin is an effective drug, inhibiting HSV-1 infection by approximately 95% at very high doses. Treatment with any of the IFNs or acyclovir reduced the infection rate, though a large percentage of cells were infected despite drug treatment. In contrast, TNF-α treatment actually potentiated HSV-1 infection, resulting in more infected cells than the non-treated control. Despite the fact TNF-α enhanced the infection rate, it was kept in the combination drug test for two reasons. First, TNF-α could have an antiviral effect if used in combination with other drugs. Second, if TNF-α had no antiviral effect or enhanced HSV-1 infection, we sought to determine whether it would be screened out of the possible drug combinations by the FSC technique.
Table 1 Concentration of drugs (ng/mL)
Figure 1 Setup of the experiment. (A) Plot of single drug dosages against efficacy. (B) Plot of infection percentage against MOI. (C) Scheme of FSC: virus attempts to infect normal cells, while drug combinations try to inhibit virus infection. For a non-optimal drug combination, a majority of cells would become infected. More effective drug combinations, suggested in later iterations, lead to fewer infected cells. Iteratively, algorithm stops when optimal drug combination is reached.
Abbreviations: FSC, feedback system control; GFP, green fluorescent protein; HSV-1, herpes simplex virus-1; MOI, multiplicity of infection.
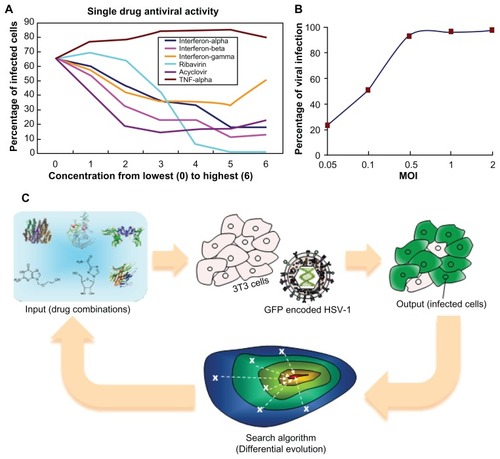
The infectious dose of HSV-1 used (MOI: number of infectious virions per cell) is a critical parameter when evaluating the outcome of potential therapies. Using a very high MOI resulted in rapid cell death, but a low MOI did not sufficiently reflect the antiviral effectiveness of different drug combinations for inhibiting HSV-1 infection. In pilot studies, we found that the viral infection level was a monotonic function of MOI and reached a plateau around MOI of 0.5 (). In general, HSV-1 infection with an MOI of 0.1 in the absence of any drug resulted in an infection rate of approximately 60% (GFP-positive cells) at 16 hours post-infection. An MOI of 0.1 was used throughout these studies.
The FSC platform technology
The FSC platform technology consisted of four modules. The first module was the input stimulations, namely, the drug combinations. The second module was the biocomplex system of interest, which in this case was the virus and host cell. The third module was the objective function readouts, which were the goals for optimization, such as efficacy, toxicity, drug resistance, etc. The fourth module was the search algorithm, which provided the next set of stimulant dosages for directing the biocomplex system toward the desired phenotype ().
For the FSC approach, we started with a set of drugs at arbitrarily chosen concentrations to stimulate the cells infected with HSV-1. The percentage of the host cells that become infected was used as the endpoint readout of the objective function in the third FSC module, and will most likely not be satisfactory in the first permutation. The fourth module of the FSC used a search algorithm to determine a selection of drug concentrations with potentially better performance, which was used in the next iteration of the experiment and fed back into the biocomplex system. Iterations of this feedback continued until the optimal drug combination was reached (ie, when the system objective function(s) became satisfactory).
The search algorithm was the module of FSC that directed the tested drug combinations towards an optimal treatment for the biocomplex system. In this study, a differential evolution (DE) algorithm was applied.Citation28 DE is a parallel search algorithm in which several drug combinations are tested in each iteration of the algorithm. A diagram of the process for implementing DE in the HSV-1 inhibition study is shown in Figure S1.
The search for high-efficacy drug combinations
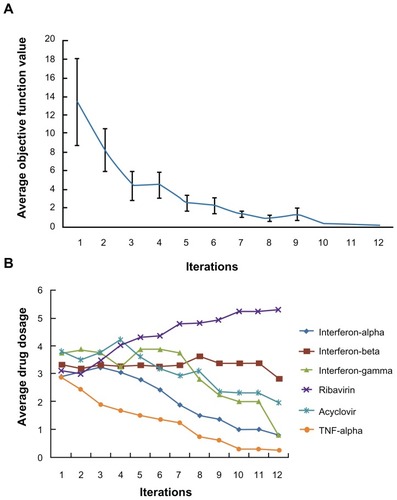
In the first part of the study, inhibition of viral infection was the sole objective function used in the FSC screening for drug combinations. To initiate the FSC process, 16 parallel drug combinations with arbitrarily chosen concentrations were generated using the numerical analysis software MATLAB. As FSC progressed, the 16 drug combinations were updated in such a way that the combination drug treatment reduced the percentage of HSV-1-infected cells. shows the average objective function value of the 16 combinations as the iterations progress. This value reached a plateau at the 8th iteration. As FSC continued, the average dosage levels for each of the six drugs in the 16 combinations were reduced, except for the dose of ribavirin (). At the 12th iteration, FSC suggested a drug combination consisting of 0.2 ng/mL IFN-β, 80 ng/mL acyclovir and 25 ng/mL ribavirin. Treatment of HSV-1-infected cells for 16 hours with this drug combination resulted in less than 0.1% GFP-positive cells, suggesting that it completely blocked HSV-1 infection. This drug combination is designated DE1. For comparison, treatment with the highest dose of ribavirin resulted in 5% of cells becoming HSV-1-infected.
Figure 2 Apply FSC to search for high efficacy drug combinations. (A) The average objective function value in 16 drug combinations reduced as iteration moves on. (B) After twelve iterations, the average dosage of ribavirin in 16 combinations increased, while the average dosages of the other antivirals reduced.
Abbreviations: FSC, feedback system control; TNF, tumor necrosis factor.
In order to verify the efficacy of DE1, we also tested DE1 on a more vicious viral strain, HSV-1 strain 17. The optimal drug combinations DE1 and a non-optimal drug combination of (0.78, 0.78, 0.2, 0, 0, 5) ng/mL (IFN-α, IFN-β, IFN-γ, ribavirin, acyclovir, TNF-α) developed in the present paper were tested. Cells were co-treated with drug combinations and HSV-1 strain 17 (MOI = 1) for 1 hour, followed by two times wash with regular cell culture medium (DMEM with 5% FBS and 1% Pen-Strep). The cells were then left in fresh culture medium for 24 hours. Supernatant of each sample was then subjected to the plaque assay in order to assess the viral titer in the supernatant. Our results indicated that the optimal drug combination DE1 (optimized for KOS strain) is still very effective, inhibiting about ten-fold of the strain 17 infection. Meanwhile, the non-optimal drug combination did not exhibit much inhibition of either KOS or strain 17. This positive result indicates the same trend in efficacy for two HSV-1 strains (data not published in this paper).
The search for high efficacy and low toxicity antiviral combinations by cascading FSC search
The drug combination DE1 includes a high dose of ribavirin. However, side effects for high doses of ribavirin are a drawback of this drug. Ribavirin has been reported to cause anemia, to be teratogenic in some animal tests, and to inhibit DNA synthesis in a dose dependent manner.Citation29,Citation30 Therefore, we attempted to determine whether FSC can search for a drug combination that simultaneously satisfies two requirements: (1) high antiviral efficacy and (2) low toxicity (ie, lower ribavirin dosage).
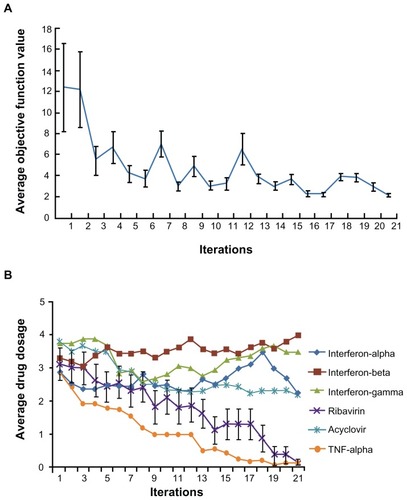
For this search we defined a new objective function, OF = αVi + βRc, where Vi stands for the percentage of infected cells after drug treatment, Rc stands for the coded dosage of ribavirin, from 0 to 5. α and β are called penalty factors. With the introduction of penalty factors, a hybrid objective function for the fourth module of the FSC was created with these multiple requirements applied to the FSC optimization process. The values of α and β reflect the relative importance of Vi and Rc. To ensure high efficacy, we set α = 0.9 and set β = 0.1 to screen out drug combinations with higher dosages of ribavirin. Thus, the new objective function is OF = 0.9Vi + 0.1Rc. To verify whether this addition to the cascade FSC drug screening technique could direct the biocomplex system to satisfy this objective function for low toxicity and high efficacy, the same 16 initial combinations were applied in a second search. As FSC proceeded through the iterations, the average objective function value approached a plateau after approximately 12 iterations (). Strikingly, at the 21st iteration, the average concentration of ribavirin in the 16 combinations was close to 0 (). The FSC suggested a ribavirin-free combination consisting of 3.12 ng/mL IFN-β, 3.12 ng/mL IFN-γ, and 80 ng/mL acyclovir. Surprisingly, this ribavirin-free drug cocktail inhibited 95% HSV-1 infection of the treated culture. This combination is designated DE2 in the rest of the study.
Figure 3 Cascade FSC based search for low ribavirin high efficacy combinations. (A) The average objective function value in 16 drug combinations reduced as iteration moves on. (B) After 21 iterations, the average dosage of ribavirin in 16 combinations reduced to close to 0.
Abbreviation: FSC, feedback system control.
Comparison between FSC identified combinations and single antiviral drugs
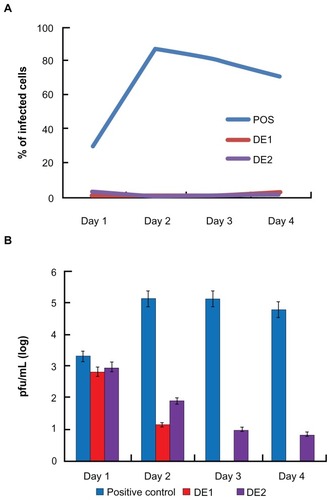
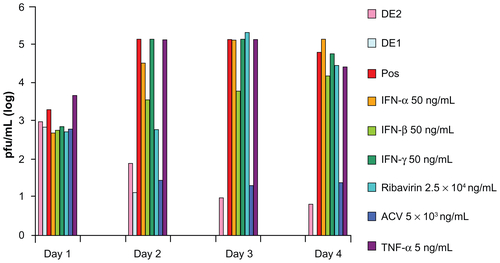
Both drug combinations DE1 and DE2 were able to inhibit viral infection by approximately 100%, which could not be achieved by using any of the single drugs alone. Compared to single drug treatment, both DE1 and DE2 offer lower doses of the drugs and greater antiviral efficacy (). Additionally, we cultured HSV-1-infected cells for longer time points, ranging from 1 day to 4 days, in the presence of DE1 or DE2. Both DE1- and DE2-treated samples sustained low levels of viral infection through day 4 (). We found that treatment with the different IFNs had decreased efficacy as time increased and ribavirin showed a similar decrease in efficacy over time (Figure S2). In contrast, antiviral activity of acyclovir remained constant as time increased. To independently confirm the flow cytometry results for the time course experiment, the HSV-1 virus yield in the culture supernatants was determined by plaque assay of the time course. The infectious titers of the supernatants were consistent with the flow cytometry results, confirming the antiviral effect of the DE1 and DE2 drug combinations ( and Figure S3).
Figure 4 Comparison between drug combinations from cascade FSC search and single antivirals.
Notes: After single drug treatment, only ribavirin could achieve near 100% viral inhibition at the highest concentration used. Acyclovir did not require high dosage when used as single drug, but had a plateau in efficacy, leaving approximately 15% of cells infected. IFNs could not achieve perfect antiviral effectiveness even when used in high dosage. DE1 and DE2 combinations represent the optimal drug combinations from two rounds of drug screening. Both combinations had better antiviral effects and lower individual drug concentration than the individual component drugs.
Abbreviations: FSC, feedback system control; IFNs, interferons; TNF, tumor necrosis factor.
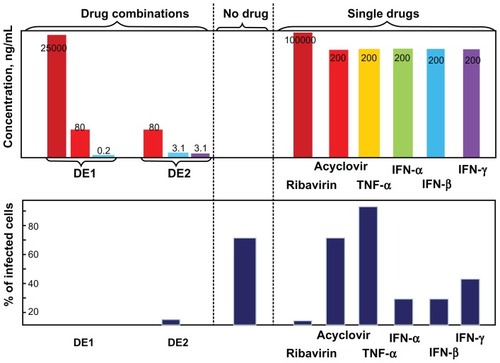
Figure 5 FSC identified drug combinations are more robust against changes in incubation time. (A) After HSV-1 infection, DE1 and DE2 were tested against incubation time ranging from 1 day to 4 days. Both combinations showed robustness to time change. (B) Plaque assay for extracellular supernatant showed that cells treated with both DE1 and DE2 release little virus through 4 days post infection.
Notes: Data for individual drugs are available in Figures S2 and S3. Error bars represent the standard error of two experiments.
Abbreviation: FSC, feedback system control; HSV-1, herpes simplex virus-1.
Comparison between FSC identified combinations and random combinations
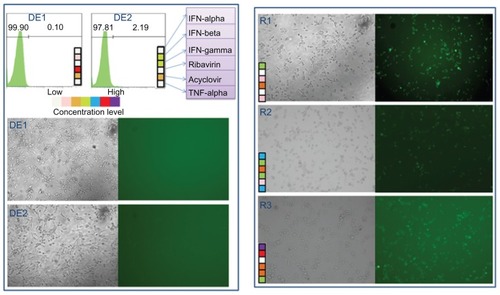
We next compared the drug efficacy of the FSC identified drug combinations with three random combinations of the six antiviral drugs. Both flow cytometry analysis and GFP fluorescent images are shown in . In , both DE1 and DE2 treatment almost completely blocked infection, resulting in <5% GFP-positive cells; however, there were 50% to 80% virus-infected cells when treated with the random combinations.
Figure 6 Comparison between cascade FSC identified drug combinations and random drug combinations DE1 and DE2 drug combinations from the two FSC drug screens.
Notes: Three randomly generated drug combinations, named R1, R2, and R3 are compared to DE1 and DE2. Both phase contrast pictures and fluorescent microscopy pictures are shown. The random drug combinations did not completely inhibit HSV-1 infection, while DE1 and DE2 nearly completely inhibited infection.
Abbreviations: FSC, feedback system control; HSV-1, herpes simplex virus-1.
Discussion
This study demonstrates that the cascade FSC scheme is a very versatile method in identifying optimal drug combinations to achieve multiple desired biological endpoint results.Citation23–Citation25 In previous studies, only one objective function was considered, but here we show that cascade FSC can be used successfully to rapidly search combinations of multiple drugs for optimal doses to satisfy both high efficacy and low toxicity. In this work, we first used drug efficacy as the only endpoint objective criterion. In the cascade screen, we introduced penalty factors against high doses of ribavirin, and found a distinct, effective, drug combination that did not include ribavirin. This is important because high doses of ribavirin could be toxic, including possible teratogenic effects. Further, this study shows the flexibility of the cascade FSC method in that it allows greater freedom to design screens for optimal drug combinations based on various criteria. In theory, even more parameters may be added to the FSC-based search for drug combinations, including degree of off-target effects or other important factors that determine the clinical significance of drug combinations.
For drugs that have no positive contribution to viral infection, such as TNF-α in the current study, each iteration of the FCS suggested a decreased TNF-α dose, with the dose eventually dropping to and remaining at zero. Together, these results show that the FSC method is an effective drug screening process.
DE1 and DE2 are both effective at blocking HSV-1 infection. An interesting but extremely challenging question is how these drug combinations work synergistically to affect a group of genes, which eventually leads towards the inhibition of infection. The first step is to identify the target genes influenced by a single drug. Assisted by high throughput screening technique, the interactions among the pathways and mechanisms under stimulations of combinatorial drug can then be studied step by step. This question is actively pursued by many research groups including the author’s laboratories.
DE1 and DE2 represent two distinct drug combinations that work much more efficiently at blocking HSV-1 infection/replication than the individual drugs alone. DE1 is a combination of acyclovir, ribavirin, and a low level of IFN-β, while DE2 is a combination of acyclovir and both IFN-α and IFN-β. However, acyclovir, a common HSV-1 antiviral, by itself does not block HSV-1 replication as effectively as DE1 or DE2 treatment. We attribute the high antiviral efficacy of DE1 and DE2 to the combinations acting on multiple cellular signaling networks simultaneously. In DE1, ribavirin is present at a concentration high enough to engage other unclear signaling pathways, working in concert with acyclovir and IFN-β to direct a global antiviral activity. In addition, these drug combinations could potentiate new pathways that disrupt HSV-1 replication that are not triggered by single drugs alone. For example, IFN-β and/or ribavirin could potentiate the effect of acyclovir to induce apoptosis in HSV-1-infected cells. Similarly, in the absence of ribavirin in DE2, we suspect that the combined effects of IFN-α and IFN-β synergize with acyclovir to block HSV-1 replication. Current research in our group is aimed at elucidating the mechanisms of how these antiviral drugs work in combination and this will likely lead to greater insight on HSV-1 inhibition strategies.
In conclusion, we have developed a platform for rapidly screening drug combinations to determine the optimal drug combinations and doses from a vast search space with multiple search parameters. The cascade FCS allowed us to screen for drug combinations that are highly effective against HSV-1 infection and potentially limit or eliminate the toxic effects of some drugs by lowering the dose. This will open new avenues into treatment of HSV-1 infection by providing drug combinations that are much more effective than standard acyclovir treatment. In the searches that resulted in combinations DE1 and DE2, the two searches started with the same initial 16 drug combinations, but the different objective functions operating in the cascade FSC resulted in the identification of two distinct, though equally effective, drug combinations. This is especially important as we only needed to test approximately 180 drug combinations, representing only approximately 0.1% of the 117,649 possible drug and dose combinations.
Acknowledgments
Presented in part: NIH Nanomedicine Development Centers Awardee Meeting, Washington, DC, March 2011. The work was supported by National Institutes of Health Nanomedicine Development Center, grant number PN2EY018228.
Disclosure
The authors have no conflict of interest to declare.
Supplementary figures
Figure S1 Illustration of differential evolution (DE) search algorithm. DE is divided into four main steps, which can me summarized as production of the original drug combinations, mutation stage, crossover stage, and production of the new drug combinations.
Abbreviation: TNF, tumor necrosis factor.
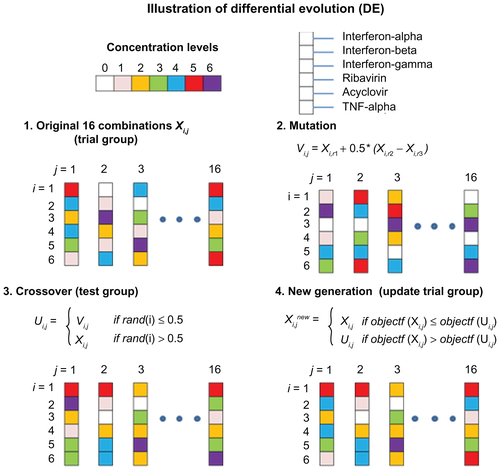
Figure S2 Long-term test between optimized drug combinations and individual drugs. Both optimal drug combinations DE1 and DE2 show low percentage of infection from day 1 to day 4, while individual drugs in general lost their antiviral efficacy after day 3.
Abbreviations: ACV, acyclovir; IFN, interferon; TNF, tumor necrosis factor.
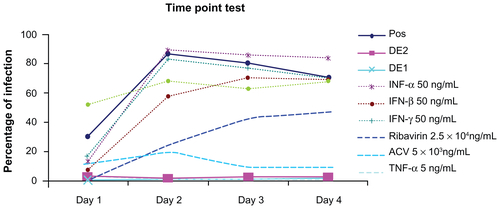
Figure S3 Plaque assay analysis of the viral titer in the supernatant. The supernatant of each sample from Figure S2 was tested for the absolute viral titer using plaque assay. Viral titer gradually clears up by optimized drug combination DE1 and DE2 after 2 days.
Abbreviations: ACV, acyclovir; IFN, interferon; TNF, tumor necrosis factor.
References
- ImmingPSinningCMeyerADrugs, their targets and the nature and number of drug targetsNat Rev Drug Discov200651082183417016423
- ZhengCJHanLYYapCWJiZLCaoZWChenYZTherapeutic targets: Progress of their exploration and investigation of their characteristicsPharmacol Rev200658325927916714488
- PappBPalCHurstLDMetabolic network analysis of the causes and evolution of enzyme dispensability in yeastNature2004429699266166415190353
- PilpelYSudarsanamPChurchGMIdentifying regulatory networks by combinatorial analysis of promoter elementsNat Genet200129215315911547334
- PengXHKarnaPCaoZHJiangBHZhouMXYangLCross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1 alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expressionJ Biol Chem200628136259032591416847054
- OverallCMKleifeldOTumour microenvironment – Opinion – Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapyNat Rev Cancer20066322723916498445
- KeithCTBorisyAAStockwellBRMulticomponent therapeutics for networked systemsNat Rev Drug Discov200541711015688074
- KitanoHInnovation – a robustness-based approach to systems-oriented drug designNat Rev Drug Discov20076320221017318209
- HuangSRational drug discovery: what can we learn from regulatory networks?Drug Discov Today2002720S16316912546900
- RyanQIbrahimACohenMHFDA drug approval summary: Lapatinib in combination with capecitabine for previously treated metastatic breast cancer that overexpresses HER-2Oncologist200813101114111918849320
- BorisyAAElliottPJHurstNWSystematic discovery of multicomponent therapeuticsP Natl Acad Sci U S A20031001379777982
- O’HaraMKieferDFarrellKKemperKA review of 12 commonly used medicinal herbsArch Fam Med1998765235369821826
- TugwellPPincusTYocumDCombination therapy with cyclosporine and methotrexate in severe rheumatoid arthritisNew Engl J Med199533331371417791814
- LawMRWaldNJMorrisJKJordanREValue of low dose combination treatment with blood pressure lowering drugs: analysis of 354 randomised trialsBrit Med J200332674041427143112829555
- GuptaRWarrenTWaldAGenital herpesLancet200737096052127213718156035
- KoelleDMCoreyLHerpes simplex: Insights on pathogenesis and possible vaccinesAnnu Rev Med20085938139518186706
- BurnsWHSantosGWSaralRIsolation and characterization of resistant herpes-simplex virus after acyclovir therapyLancet1982182694214236121091
- De ClercqEAntiviral drugs in current clinical useJ Clin Virol200430211513315125867
- RongQAlexanderTSKoskiGKRosenthalKSMultiple mechanisms for HSV-1 induction of interferon alpha production by peripheral blood mononuclear cellsArch Virol2003148232934412556996
- ChenSHOakesJELauschRNSynergistic anti-HSV effect of tumor-necrosis-factor-alpha and interferon-gamma in human corneal fibroblasts is associated with interferon-beta inductionAntivir Res199322115298250541
- FeduchiECarrascoLMechanism of inhibition of HSV-1 replication by tumor-necrosis-factor and interferon-gammaVirology199118028228251846503
- DempseyPWDoyleSEHeJQChengGHThe signaling adaptors and pathways activated by TNF superfamilyCytokine Growth F R2003143–4193209
- WongPKYuFQShahangianAChengGHSunRHoCMClosed-loop control of cellular functions using combinatory drugs guided by a stochastic search algorithmP Natl Acad Sci U S A20081051351055110
- TsutsuiHValamehrBHindoyanAAn optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cellsNat Commun2011216721266967
- SunCPUsuiTYuFIntegrative systems control approach for reactivating Kaposi’s sarcoma-associated herpesvirus (KSHV) with combinatory drugsIntegr Biol (Camb)20091112313019851479
- LiuMYRakowskiBGershburgEICP0 antagonizes ICP4-dependent silencing of the herpes simplex virus ICP0 genePlos One201051e883720098619
- ElliottGO’HarePLive-cell analysis of a green fluorescent protein-tagged herpes simplex virus infectionJ Virol19997354110411910196307
- StornRPriceKDifferential evolution – a simple and efficient heuristic for global optimization over continuous spacesJ Global Optim1997114341359
- KakudaTNBrinkmanKMitochondrial toxic effects and ribavirinLancet200135792701802180311407388
- De FranceschiLFattovichGTurriniFHemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damageHepatology2000314997100410733558