?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
To solve the efficiency versus cytotoxicity and tumor-targeting problems of polyethylenimine (PEI) used as a nonviral gene delivery vector, a degradable PEI derivate coupled to a bifunctional peptide R13 was developed.
Methods
First, we synthesized a degradable PEI derivate by crosslinking low-molecular-weight PEI with pluronic P123, then used tumor-targeting peptide arginine-glycine-aspartate-cysteine (RGDC), in conjunction with the cell-penetrating peptide Tat (49–57), to yield a bifunctional peptide RGDC-Tat (49–57) named R13, which can improve cell selection and increase cellular uptake, and, lastly, adopted R13 to modify the PEI derivates so as to prepare a new polymeric gene vector (P123-PEI-R13). The new gene vector was characterized in terms of its chemical structure and biophysical parameters. We also investigated the specificity, cytotoxicity, and gene transfection efficiency of this vector in αvβ3-positive human cervical carcinoma Hela cells and murine melanoma B16 cells in vitro.
Results
The vector showed controlled degradation, strong targeting specificity to αvβ3 receptor, and noncytotoxicity in Hela cells and B16 cells at higher doses, in contrast to PEI 25 KDa. The particle size of P123-PEI-R13/DNA complexes was around 100–250 nm, with proper zeta potential. The nanoparticles can protect plasmid DNA from being digested by DNase I at a concentration of 6 U DNase I/μg DNA. The nanoparticles were resistant to dissociation induced by 50% fetal bovine serum and 600 μg/mL sodium heparin. P123-PEI-R13 also revealed higher transfection efficiency in two cell lines as compared with PEI 25 KDa.
Conclusion
P123-PEI-R13 is a potential candidate as a safe and efficient gene-delivery carrier for gene therapy.
Introduction
Cancer is a scourge of the human race. Its classical treatments such as surgery, chemotherapy, and radiotherapy are still insufficiently effective in many cases; thus, new anticancer strategies are urgently required.Citation1 Gene therapy provides a promising tool to eradicate this disease by treating it at its source. The key to gene therapy is finding an ideal gene-delivery vector. Delivery of nucleic acids into cells using cationic polymers has recently attracted remarkable interest in the field of nonviral gene therapy, due to their structural diversity, easy production, nonimmunogenicity, and safety.Citation2 One of the most effective and widely studied synthetic nonviral gene-delivery vectors is the polycation polyethylenimine (PEI), which has recently been employed for the design of DNA-delivery vehicles. By adding this cationic polymer to DNA, the condensed complexes can be spontaneously formed through electrostatic interactions, resulting in efficient transport of intact DNA into the nucleus.Citation3–Citation5 Gene delivery using PEI involves condensation of DNA into compact particles, uptake into the cells, release from the endosomal compartment into the cytoplasm, and uptake of the DNA into the nucleus. The high transfection efficiency of PEI/DNA complexes should be ascribed to the “proton sponge” effect of PEI.Citation6,Citation7 However, polyplexes of PEI/DNA have shown two outstanding problems. First of all, long PEI chains are highly effective in gene transfection but are cytotoxic. Conversely, short PEI chains display lower cytotoxicity but lower efficiency.Citation8 The major drawback with gene delivery using PEI is the lack of satisfactory specificity toward tumor cells, because there is no binding selectivity between the positively charged polycation and the negatively charged body cells.Citation9
Pluronic block copolymers, which consist of hydrophilic ethylene oxide (EO) and hydrophobic propylene oxide (PO) blocks arranged in a basic A-B-A structure (EO x -PO y -EO x), are amphiphilic molecules that can be used as structural elements of the polycation-based gene-delivery systems.Citation10 Kabanov et alCitation10–Citation13 previously reported that pluronic can enhance cell interactions, DNA transport, and transgene expression, and possesses the unique ability to be incorporated into cell membranes as a result of the presence of the hydrophobic poly-PO chain. In contrast, hydrophilic EO chains do not interact with lipid membranes but can be used to prevent binding of other polymers with the membranes.Citation10 The systems are stabilized in dispersion by the EO corona in a manner similar to regular pluronic micelles. The micelles of these copolymers are sufficiently potent in the ability to bind cellular membrane, and a relatively high concentration of micelles can be reached in solution.Citation14 Among these copolymers, pluronic notation P123 with formula EO20-PO70-EO20 exhibits higher stability in terms of critical micelle concentration.Citation15 In this paper we link low-molecular-weight PEI (LMW PEI) with P123 by chemical crosslinking to obtain cladodromous or cancellated PEI derivates with high molecular weight, which can guarantee higher transfection efficiency and degrade into lower-cytotoxic LMW PEI because of crosslinked chemical bond cleavage by hydrolysis after uptake into the cells. Hence, this makes it possible to greatly reduce the cytotoxicity of PEI on the premise of ensuring higher transfection efficiency and thereby solves the “catch 22” problem of efficiency versus cytotoxicity.
Receptor-mediated tumor targeting is another increasingly important research field of nonviral gene-delivery systems. Integrins are a family of transmembrane glycoproteins with relatively small cytoplasmic domains. They consist of two subunits, α and β. Generally, as receptors, an individual integrin may recognize several distinct proteins. Among the integrins, the αvβ3 receptor is involved in tumor cell migration and angiogenesis. The αvβ3 receptors are highly expressed on tumor cells and tumor angiogenic blood vessels, whereas they are not detectable on quiescent blood vessels. Thus, the αvβ3 integrin participates in tumor metastasis and tumor-related angiogenesis. Citation16 The highly selective expression of αvβ3 integrin in the neovascular tissue and various tumors allows its use as a suitable target. Arginine-glycine-aspartate (RGD) peptide is a key binding moiety that has been shown to bind specifically to αvβ3 integrin receptors.Citation17 Cell-penetrating peptides (CPPs) are 7–30 amino acid long peptides capable of translocating various macromolecules across the plasma membrane and targeting to the cell nucleus. CPPs have been applied to gene delivery. Kilk et alCitation18 demonstrated that CPP transportan 10 (TP10) could enhance PEI-mediated transfection at relatively low concentrations and help to develop future gene-delivery systems with reduced toxicity. The HIV Tat-derived peptide is a small basic peptide for successfully delivering a large variety of cargoes into cells such as nanoparticles, proteins, and nucleic acids. The transduction domain or region conveying CPPs appears to be confined to a small stretch of basic amino acids with the sequence RKKRRQRRR and is known as Tat (49–57).Citation19,Citation20 Therefore, we have used a linear RGD peptide, RGDC, in conjunction with Tat (49–57) to yield a new chimeric peptide, RGDC-Tat (49–57), named R13. The bifunctional R13 can target to αvβ3 receptors expressed abundantly on tumor cells and carry cargo molecules inside the cells. Adopting R13 to modify PEI derivates can improve cell selection, promote cargo transport, and enhance transfection efficiency.
Above all, in this work, we first linked LMW PEI with P123 to obtain degradable PEI derivates with high molecular weight, then used tumor-targeting peptide RGD, in conjunction with the CPP Tat (49–57) to yield a bifunctional peptide R13, and, lastly, adopted R13 to modify the PEI derivates so as to prepare a new nonviral gene delivery vector (P123-PEI-R13). The new gene vector was characterized in terms of its chemical structure and biophysical parameters. We also investigated the specificity, cytotoxicity, and gene transfection efficiency of this vector in αvβ3-positive human cervical carcinoma Hela cells and murine melanoma B16 cells in vitro. The purpose of the present study was to reduce cytotoxicity of PEI, improve its tumor targeting, and increase cellular uptake of genes, enhancing the therapeutic effect of gene therapy on cancer.
Materials and methods
Materials
Branched PEI (MW 2000), fluorescein isothiocyanate (FITC), N-succinimidyl-4-(N-maleimido-methyl) cyclo-hexane- 1-carboxylate (SMCC), horseradish peroxidase (HRP), heparin, and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) were obtained from Sigma-Aldrich (St Louis, MO). P123 was kindly provided by BASF Corporation (Mount Olive, NJ). DNase I was purchased from the Worthington Company (Lakewood, NJ). RPMI 1640 culture medium and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA). Luciferase assay system for in vitro transfection assay and pGL-3 control vector with SC-40 promoter and enhancer encoding firefly (Photinus pyralis) luciferase were obtained by Promega (Madison, WI). The plasmid encoding the enhanced green fluorescent protein (pEGFP-N2) was kindly provided by the Institute of Life Science and Technology of Tongji University (Shanghai, People’s Republic of China). The plasmids were amplified using Escherichia coli DH5α and prepared using the Qiagen End-free Plasmid Mega Kit (Qiagen GmbH, Hilden, Germany). The purity of the purified and concentrated DNA was determined by measuring its ultraviolet absorbance at 260 nm and 280 nm respectively. The oligopeptide Arg-Gly-Asp-Cys-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg (R13, MW 1771.10 Da) was synthesized by GL Biochem (Shanghai, China), and the amino acid sequence was confirmed by mass spectroscopy.
Binding of R13 to integrin receptor
Integrin αvβ3 receptors are highly expressed on Hela cells and B16 cells and almost not expressed on MCF-7 cells. Three kinds of cells were cultivated under standard conditions at 37°C in a humidified 95% air/5% CO2 atmosphere in RPMI 1640 in the presence of 10% FBS. Before the experiment, the cells were trypsinized and centrifuged to remove culture medium, and then 1% bovine serum albumin solution was added to block the protein-binding site. After incubation for 20 minutes, the cells were centrifuged to remove the bovine serum albumin solution, resuspended, and cultivated with FITC-conjugated R13 (FITC-R13) for 60 minutes at 37°C (approximately 1 × 106 cells and 0.5 nmol FITC-R13 in 1 mL phosphate-buffered saline [PBS]). The cells were washed with 3 mL of 0.01M PBS four times and resuspended in PBS to determine the binding affinity of R13 to integrin receptor using flow cytometry (BD FaCSAria; BD, NJ).
Synthesis of P123-PEI-R13
Synthesis of P123-PEI
Activated P123 should be obtained first in order to prepare P123-PEI. P123 (0.6 mmol) was dried by coevaporation with anhydrous toluene in a vacuum at 50°C, then dissolved in toluene/dichloromethane (3:1, 40 mL) and treated with bis-(trichloromethyl)-carbonate (0.356 g, 1.2 mmol) overnight. After the solution was evaporated to dryness under a vacuum, the residue was redissolved in toluene/dechloromethane (2:1, 30 mL) and treated with solid N-hydroxysuccinimide (0.240 g, 2.0 mmol), followed by anhydrous triethylamine (0.28 mL, 2.0 mmol). After 4 hours of stirring, the solution was filtered and evaporated to dryness. The product was purified on a silica gel column using stepwise elution with trichloromethane methanol (10%–15%, v/v) mixtures.
LMW PEI (0.20 g, 0.10 mmol) was dissolved in 10 mL anhydrous dichloromethane as Solution A, and the predetermined amount of activated P123 (0.01 mmol) was dissolved in 10 mL anhydrous ethanol as Solution B. Then, Solution A and Solution B were slowly added to a base solution of 10 mL anhydrous dichloromethane with constant stirring. The reaction mixture was left overnight at room temperature. After completion of reaction, the conjugate was dialyzed using Spectra/Por 1 dialysis membrane (Spectrum, CA) against distilled water at 4°C for 2 days and then lyophilized.
Conjugation of P123-PEI with R13
P123-PEI was conjugated with R13 using SMCC as cross-linker. After the appropriate amount of SMCC solution (3.33 mg/mL in dimethyl sulfoxide) was added dropwise to P123-PEI solution (9 mg/mL in 0.1M PBS) at a molar ratio of 10:1, 5:1, and 2:1, the reaction mixture was incubated for 30 minutes at room temperature with gentle shaking. The excess nonreacted crosslinker was removed by gel chromatography (Sephadex G-25; Pharmacia, Milton Keynes, UK). Thus, N-hydroxysuccinimide (NHS) eaters of SMCC have reacted with primary amines of P123-PEI to form amide bonds and produce the maleimide-activated P123-PEI. Then, R13 (10 mg/mL in 0.1M PBS) was reacted with the maleimide-activated P123-PEI at a molar ratio of 10:1, 5:1, and 2:1. The reaction mixture was stirred in the dark overnight at 4°C and then lyophilized after the nonreacted R13 was removed by ultrafiltration with Amicon Ultra-4 centrifugal filter devices (Millipore, Billerica, MA). Then, three types of cationic polymer were obtained and named P123-PEI-R13-h, P123-PEI-R13-m, and P123-PEI-R13-l, respectively.
Characterizations of P123-PEI-R13 (NMR, GPC)
A total of 10 mg of P123-PEI and P123-PEI-R13 was dissolved in 0.6 mL of deuterium oxide (D2O) in a nuclear magnetic resonance (NMR) tube, and the 1H NMR spectra were recorded using a Varian 300 MHz spectrometer at room temperature.
The molecular weight and its distribution of the polymers were measured by gel permeation chromatography with multiangle laser light scattering (GPC-MALLS) (LC-20 AD; Shimadzu, Kyoto, Japan) (690 nm laser wavelength) using TSK-GEL G5000PWXL column (temperature 40°C) operated at a flow rate of 0.4 mL/minute. Ammonium acetate (0.2M) was used as a mobile phase.
Degradation of P123-PEI-R13
Degradation of P123-PEI-R13 was estimated by the measurement of molecular weight. Of the polymers, 0.5 g was dissolved in 10 mL of PBS (0.1M, pH = 7.4) and incubated at 37°C with shaking at 100 rpm. After incubation for different lengths of time, solutions of the polymers were lyophilized and the molecular weights of lyophilized samples were measured by GPC-MALLS with 690 nm laser wavelength.
Preparation of P123-PEI-R13/DNA complexes
Charge ratio (w/w) of P123-PEI-R13/DNA complex was expressed as the ratio of the weight of P123-PEI-R13 and DNA. Complexes were induced to self-assemble by mixing DNA solution with polymer solution (0.1M PBS, pH = 7.4) at a desired charge ratio. The complexes were allowed to stand at room temperature for 30 minutes.
Measurement of particle size and zeta potential
The sizes and zeta potential of polymer/DNA complexes in PBS buffer at room temperature were measured using an electrophoretic light-scattering spectrophotometer (Zetasizer Nano ZS90, MAN0317 Issue 5.0; Malvern Instruments, Malvern, UK), with 90° scattering angles. The complexes were prepared at desired w/w ratios and incubated at room temperature for 30 minutes before the sizes and zeta potential measurements were carried out. All the experiments were performed in triplicate.
Agarose gel retardation assay
After preparation of polymer/DNA complexes, 10 μL of the complex solution (250 ng of plasmid DNA) was added to 1 μL of a 10 × loading buffer. The sample was electrophoresed on 1% (w/v) agarose gel for 40 minutes at 120 V. The gel was stained with ethidium bromide (0.5 μg/mL) for about 20 minutes at room temperature and illuminated on an ultraviolet illuminator to show the location of the DNA.
Resistance to DNase I digestion
Different amounts of DNase I were added to 10 μL of the complex solution (250 ng of plasmid DNA) in 0.5 mL Eppendorf tubes and incubated at 37°C for 15 minutes. Then, 2 μL of 250 mM EDTA solution was added to each tube and incubated at room temperature for 10 minutes to inactivate DNase I. Next, 10 μL of 2 mg/mL sodium heparin was added to each tube and incubated at room temperature for 2 hours to dissociate the complex completely. After that, the electrophoresis was performed to evaluate the resistance of P123-PEI-R13 to DNase I digestion.
Resistance to serum and heparin
A total of 10 μL per tube of the complex solution (250 ng of plasmid DNA) was added to 0.5 mL Eppendorf tubes. Different volumes of FBS were added and incubated at 37°C for 60 minutes. Similarly, different concentrations of 2 μL sodium heparin solution were added to 0.5 mL tubes containing 10 μL of the complex solution and incubated at 37°C for 30 minutes. Then, the electrophoresis was performed to determine the sensitivity of the polymer/DNA complex to serum and heparin.
Cytotoxicity assay
The cytotoxicity of the polymers on the B16 and Hela cells was measured using the MTT assay. B16 and Hela cells were seeded at a density of 5000 cells per well in 200 μL of growth medium in 96-well plates and incubated for 24 hours to reach 80% confluence. Then, the culture medium was replaced with fresh serum-free mediums containing serial dilutions of P123-PEI-R13, PEI 2 kDa, and PEI 25 kDa at various concentrations (4 μg/mL, 8 μg/mL, 16 μg/mL, 24 μg/mL, 32 μg/mL, and 48 μg/mL). The cells were incubated for another 24 hours before the medium was replaced with 180 μL of fresh growth medium and 20 μL MTT (5 mg/mL) was added. After 4 hours of incubation, unreacted dye was removed by aspiration and 150 μL of dimethyl sulfoxide was added as a solvent for formazan crystals. The absorbance value at 570 nm was determined using an ELISA plate reader (Model 680; Bio- Rad, Hercules, CA). Cell viability was calculated from these data. The cell viability (%) was calculated according to the following equation:
where Atest is the PEI-P123-R13 or PEI-treated cells and Acontrol is the absorbance of the untreated cells.
In vitro gene transfection
We examined the ability of P123-PEI-R13 to transfect Hela cells and B16 cells using the plasmid pEGFP-N2 and pGL3-control. The cells were seeded in 24-well plates at a density of 1 × 105 cells per well in RPMI 1640 medium containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, and incubated for 18–24 hours to reach 80% confluence. Before transfection, the medium was replaced with 450 μL fresh RPMI 1640 supplemented with 10% FBS. Then, 50 μL of the polymer/DNA complex solution containing 0.8 μg of pDNA at various weight ratios ranging from 2 to 30 were added to 24-well plates and incubated for 4 hours at 37°C in a 5% CO2 atmosphere. Then, the polymer concentration was calculated according to the w/w ratio, and the final volume of culture medium per well ranged from 3.2 μg/mL to 48 μg/mL. After exchanging with a fresh medium with 10% FBS, cells were further incubated for 48 hours. After transfection for 48 hours, inverted fluorescent microscope (AE-31, Motic Corporation, Wetzlar, Germany) was used to observe the EGFP expression of the polyplexes in Hela cells and B16 cells. Then, the cells were trypsinized, centrifuged to remove culture medium, and resuspended in PBS to determine transfection efficiency using flow cytometry (BD FaCSAria). The data analysis was performed using the CellQuest software (Becton Dickinson, Franklin Lakes, NJ).
The luciferase assay was carried out according to the manufacturer’s instructions (Promega, Madison, WI). The growth medium was removed, and the cells were shaken for 30 minutes at room temperature in 200 μL of cell culture lysis reagent (CCLR). Luciferase activity was measured with a luminometer (Turner Designs Luminometer Model TD-20/20; Promega). The relative light units were normalized against protein concentration in the cell extracts, which was measured using a BCA protein assay kit (Pierce, Rockford, IL).
Results and discussion
Binding of FITC-R13 to αvβ3-positive cells
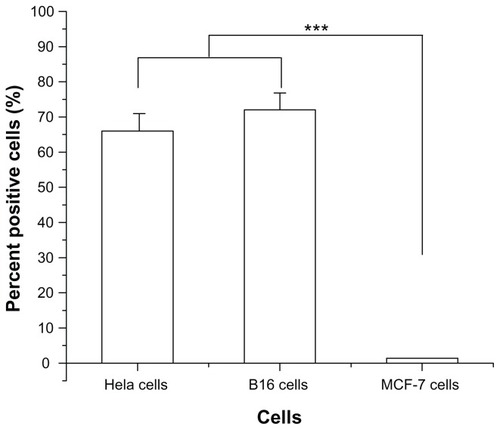
In order to evaluate the specificity of R13 for αvβ3 receptor, αvβ3-positive cells (Hela cells and B16 cells) and αvβ3-negative cells (MCF-7 cells) were chosen to perform comparative experiments. Hela cells, B16 cells, and MCF-7 cells were incubated with FITC-R13 and then analyzed using flow cytometry. As is shown in , the percentage of positive Hela cells and B16 cells is much higher than that of MCF-7 cells. The results indicate that R13 showed specificity for αvβ3-positive cells in vitro and in Hela cells and B16 cells.
Synthesis and characterization of P123-PEI-R13
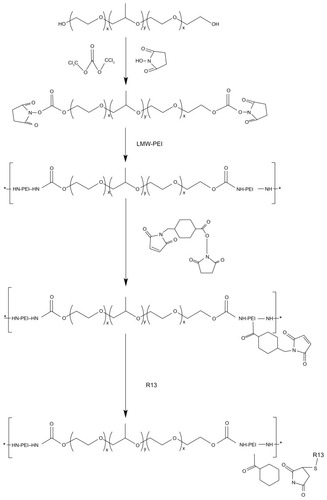
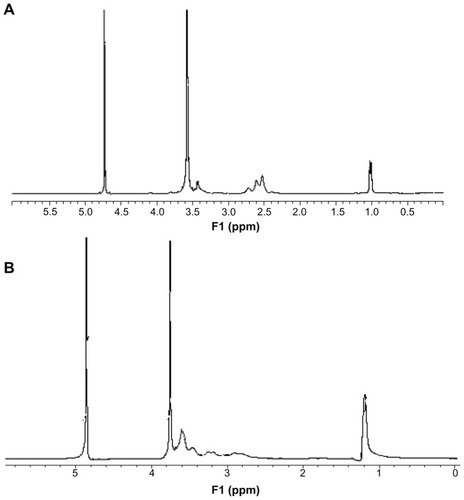
The degradable PEI derivates of P123-PEI were synthesized by linking PEI 2 kDa with P123 and then conjugated with a bifunctional peptide R13 to prepare a new nonviral gene delivery vector P123-PEI-R13 (). As for P123-PEI, the free hydroxyl groups of P123 were activated by succinimidyl carbonate in advance and then linked to the amino groups of PEI.Citation21 SMCC was used as crosslinker to conjugate P123-PEI with R13. The structures of P123-PEI and P123-PEI-R13 were confirmed by 1H NMR spectroscopy (). shows the 1H-NMR spectra of P123-PEI in D2O, where -CH2CH2O-proton peaks appear at δ3.55 ppm and -CH2CH2 NH-proton peaks appear at δ2.51–2.7 ppm. shows the 1H-NMR spectra of P123-PEI-R13 in D2O, where the proton peaks move toward the lower magnet field as compared with those in , due to the production of groups with electronic screening effect from R13, such as the amide group and carbonyl group. At the same time, the proton peaks at δ2.51–2.7 ppm applanatus, and the peak value at δ1.00 ppm increases, indicating that R13 has been coupled to P123-PEI successfully.
Degradation studies
After a nondegradable carrier (eg, PEI 25 kDa) transfers pDNA into target cells, it cannot be degraded or eliminated from the cells. The intracellular accumulation leads to potential cytotoxicity. So degradation of gene-delivery polymers in vivo is very important for safe gene transfection.Citation22,Citation23 The ester bonds of the synthetic P123-PEI-R13 are susceptible to hydrolysis at physiological conditions so that the polymer can degrade into poloxamer oligomers and LMW PEI, which can be secreted to the extracellular environment by exocytosis, and shows little toxic effect on cells. The average results of the test for in vitro degradation are shown in with standard deviation bars. P123-PEI-R13 was degraded slowly and the degradation was nearly completed after about 60 hours.
Figure 4 Degradation of P123-polyethylenimine-R13. The polymers were dissolved in 0.1 M phosphate-buffered saline (pH = 7.4) and incubated at 37°C and 100 rpm. Determination of molecular weight (MW) was measured by gel permeation chromatography with multiangle laser light scattering (n = 3).
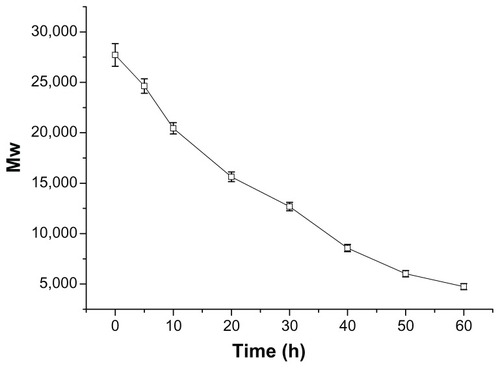
In order to obtain meaningful information for the degradation model, the degradation profile was fitted to three different kinetic models, and the goodness of fit of the degradation data was assessed. The degradation profile of P123-PEI-R13 could be well described by a zero-order model. The half-life of P123-PEI-R13 was found to be about 30 hours, according to the model.
Particle size and zeta potential measurements
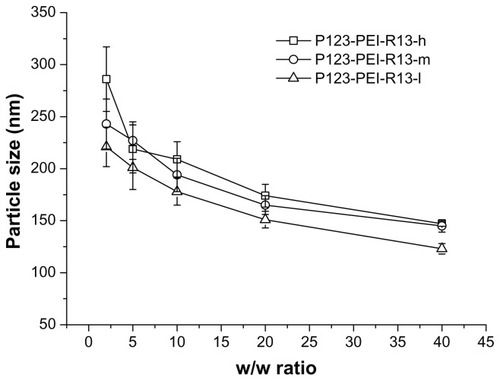
The particle size and zeta potential of polymer/DNA complexes were examined at different w/w ratios. As is shown in , with an increase of polymer/DNA weight ratios the particle size decreased. This showed that the polymers with more charge could condense DNA more effectively. All polymers can concentrate pDNA into nanoparticles, and their average particle sizes vary from 100 nm to 250 nm, depending on the composition of the complexes, which are suitable for efficient gene delivery in vivo.Citation24 In the range of concentration studied, no precipitation was observed at any w/w ratios.
Figure 5 Particle sizes (nm) of P123-polyethylenimine (PEI)-R13/DNA complexes at various w/w ratios.
Note: The data were expressed as mean values (±standard deviations, n = 3).
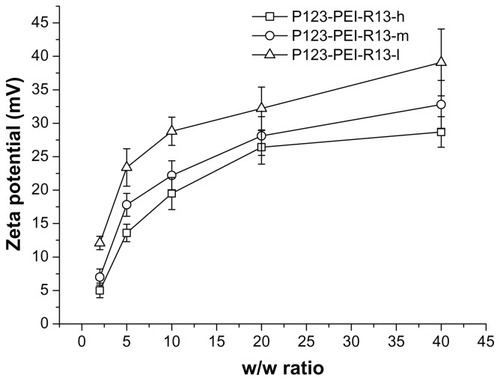
Positively charged surfaces of the complexes are thought to be important for binding to negatively charged cellular membranes and cellular uptake via endocytosis.Citation25 However, strong cationic charges of the complexes also lead to cytotoxicity. Citation21 shows zeta potentials of the complexes according to w/w ratio. The zeta potentials of all complexes were gradually increased in accordance with the increase of charge ratios. At low w/w ratios, the zeta potential of the complexes was a slightly positive value. The zeta potentials rapidly increased up to w/w ratio 10, whereas they slowly increased from w/w ratio 10.
Condensation status of plasmid DNA by P123-PEI-R13
The DNA condensation capacity of P123-PEI-R13 was determined by gel retardation experiments at various w/w ratios. As is shown in , the movement of the plasmid DNA in the gel is retarded as the amount of the polymers increases, demonstrating that the polymers bind to the DNA and neutralize its charge. When the w/w ratios of the polymer and DNA exceed the neutralization composition, the complexes will have a positive charge and stop migrating toward the anode. In this experiment, all the polymers of P123-PEIR13 were able to condense DNA effectively and neutralized its charge at a w/w ratio of 2. Moreover, for unconjugated P123-PEI, DNA was retarded at a w/w ratio of 0.4 (data not shown). Correspondingly, more P123-PEI-R13 was needed to condense pDNA completely than P123-PEI, possibly because more primary amines of P123-PEI were substituted by R13 so that less positively charged primary amines interacted with negatively charged pDNA phosphates.Citation24
Protection of P123-PEI-R13 on plasmid DNA
After the complex solution was incubated with DNase I, partial DNA was still protected from digestion by DNase I, even at a concentration of 6 U DNase I/μg DNA, and was released from the complex (). In fact, under the same experimental conditions, DNA could be completely digested by DNase I at a concentration of 0.08 U DNase I/μg DNA.Citation26
Figure 8 Protection of P123-polyethylenimine (PEI)-R13 on plasmid DNA. (A) Protection of plasmid DNA from degradation by DNase I at varying concentrations of 0 DNase I/μg DNA, 0.15 DNase I/μg DNA, 0.375 DNase I/μg DNA, 0.75 DNase I/μg DNA, 1.5 DNase I/μg DNA, 2.25 DNase I/μg DNA, 3 DNase I/μg DNA, 3.75 DNase I/μg DNA, 4.5 DNase I/μg DNA, 5.25 DNase I/μg DNA, 6 DNase I/μg DNA, 6.75 DNase I/μg DNA, and 7.5 U DNase I/μg DNA. (B) Protection of plasmid DNA from dissociation by serum at varying concentrations of 10%, 25%, and 50%. The lanes 10%, 25%, and 50% without “+” refer to the presence of only 10%, 25%, and 50% serum. The lanes 10%, 25%, and 50% with “+” refer to the presence of P123-PEI-R13/DNA complexes at w/w ratio 20 with different concentration of serum. (C) Protection of plasmid DNA from dissociation by sodium heparin at varying concentrations of 0 μg/mL, 120 μg/mL, 140 μg/mL, 160 μg/mL, 180 μg/mL, 200 μg/mL, 220 μg/mL, 240 μg/mL, 260 μg/mL, 280 μg/mL, 300 μg/mL, 400 μg/mL, 500 μg/mL, and 600 μg/mL.
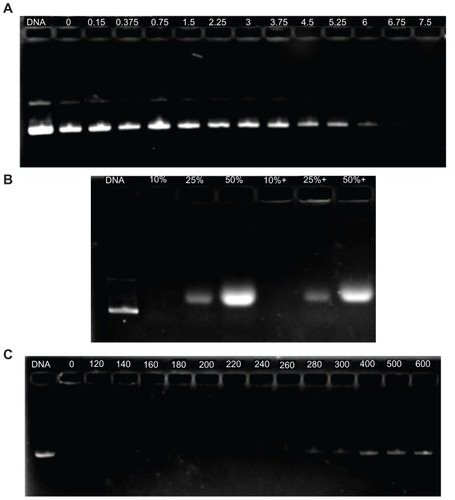
The stability evaluation of the P123-PEI-R13/DNA complex in vivo was simulated by treating the complex with serum and sodium heparin in vitro. As is shown in , serum cannot dissociate the complex even at a concentration of 50%. To infer, the P123-PEI-R13/DNA complex may remain stable in blood circulation.
Sodium heparin with a potent negative charge can be used to simulate macromolecules carrying negative charges in the blood and evaluate the antidissociation ability of a vector system in vitro. Sodium heparin can dissociate the complexes in a concentration-dependent manner.Citation26 Sodium heparin could dissociate the P123-PEI-R13/DNA complexes when its concentration was 260 μg/mL at a w/w ratio of 20:1. The concentration of sodium heparin was more than 600 μg/mL before complete dissociation of the complexes ().
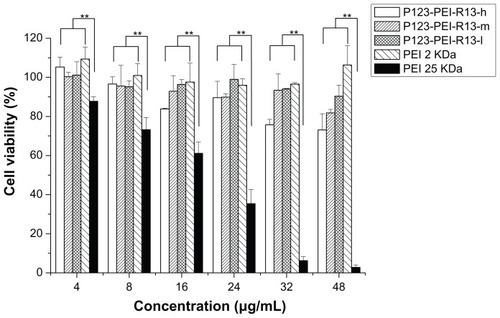
Cytotoxicity assay
There was a correlation between cytotoxicity and molecular weight. The high-molecular-weight PEI was limited as a transfection reagent for its relatively high cytotoxicity. Many attempts were made to reduce the cytotoxicity of the gene carrier, of which the most favorable one is an introduction of degradable polymer through some linkers. Citation23,Citation27–Citation29 Here, the cytotoxicity of the degradable P123-PEI-R13 was evaluated in Hela cells and B16 cells using the MTT assay in comparison with PEI 25 kDa. As is shown in and , P123-PEI-R13 and PEI 2 KDa showed significantly higher cell viability as compared with PEI 25 kDa at any concentration (P < 0.01). It is worth-while noting that P123-PEI-R13 showed low cytotoxicity at various concentrations, even at a high concentration, indicating that the polymer is very suitable for gene delivery because we can administrate more P123-PEI-R13 to improve transfection efficiency if necessary.
Figure 9 Cytotoxicity of P123-polyethylenimine (PEI)-R13 at various concentrations in Hela cell lines using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide assay.
Notes: Each data point represents the mean ± standard deviation. n = 6, **P < 0.01.
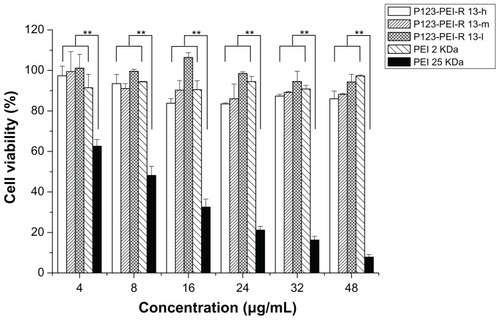
Figure 10 Cytotoxicity of P123-polyethylenimine (PEI)-R13 at various concentrations in B16 cell lines using the 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide assay.
Notes: Each data point represents the mean ± standard deviation; n = 6, **P < 0.01.
The lower cytotoxicity may be due to the lower amino group density and low toxic building blocks.Citation9,Citation22 As far as P123-PEI-R13 is concerned, the ester bonds of the polymer can degrade into poloxamer oligomers and LMW PEI at physiological conditions, which can be rapidly excluded from the cell, then show little cytotoxicity. Meanwhile, the amino group density compared with high-molecular-weight PEI may also be reduced, resulting in lower cytotoxicity to the cells.
In vitro transfection efficiency
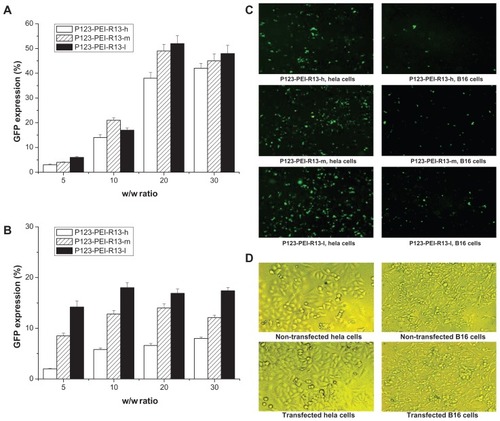
As for the pEGFP-N2 reporter gene, transfection efficiency of P123-PEI-R13 polymers in Hela and B16 cells is shown in . In , transfection efficiency of P123-PEI-R13/DNA complexes increased in correlation with the w/w ratio in Hela cells and B16 cells. On the whole, the EGFP reporter gene expression in Hela cells exceeded that in B16 cells, showing that the gene transfection was selective to cells. Representative fluorescence images for the transfection of Hela cells and B16 cells using P123-PEI-R13 at the optimal conditions are shown in . The microscopic images in a bright field of nontransfected cells and transfected cells are included in . The transfection of P123-PEI-R13/DNA complexes did not alter the morphology of the cells, indicating that P123-PEI-R13 was suitable for gene delivery.
Figure 11 Green fluorescent protein (GFP) reporter gene transfection in Hela cells and B16 cells by P123-polyethylenimine (PEI)-R13. (A) Percentage of GFP transfection in Hela cells by flow cytometry analysis (the mean ± standard deviation, n = 3). (B) Percentage of GFP transfection in B16 cells by flow cytometry analysis (the mean ± standard deviation, n = 6). (C) Representative fluorescence images for the transfection of Hela cells and B16 cells using P123-PEI-R13 at optimal conditions. (D) Microscopic images in bright field of nontransfected cells and transfected cells.
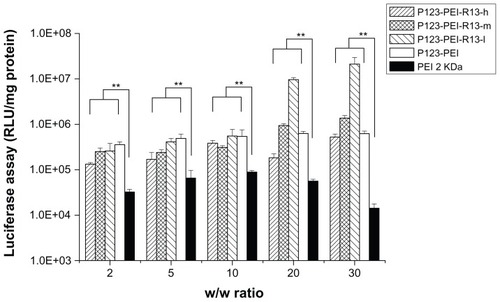
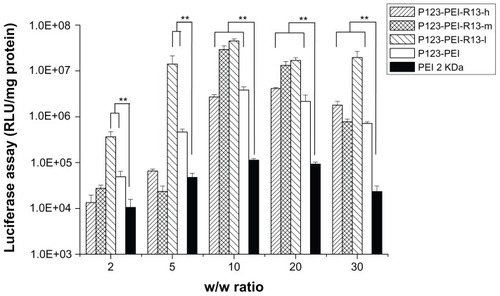
shows the results of the gene transfection efficiency of P123-PEI-R13/pGL3-control complexes in Hela cells in comparison with P123-PEI and PEI 2 KDa polyplexes. The P123-PEI-R13-h, P123-PEI-R13-m, and P123-PEI-R13-l polyplexes exhibited the maximal transfection efficiency at the weight ratio of 30. Based on cytotoxicity assay, the cell viability was over 85% at the w/w (polymer/ DNA) ratio of 30. All synthesized P123-PEI-R13 showed much higher gene transfer ability compared with PEI 2 KDa, and the highest luciferase expression level obtained at P123-PEI-R13-l was more than 230 times higher compared with that of PEI 2 KDa, and even about two times higher than PEI 25 KDa at the optimal conditions (w/w = 0.8, data not shown). It should be noted that the bifunctional R13 peptide not only could make the P123-PEI-R13/DNA complexes target the cells but also could promote cargo transport. Therefore, the P123-PEI-R13/DNA complexes showed about 34 times higher gene transfection than P123-PEI/DNA complexes at optimal conditions. In addition, transfection efficiency of P123-PEI-R13 decreased with an increase of the R13 rate, which can be explained by the high modification degree of R13 leading to a less positive surface. shows the results of the gene transfection efficiency of P123-PEI-R13/ pGL3-control complexes in B16 cells in comparison with P123-PEI and PEI 2 KDa polyplexes. The P123-PEI-R13-h, P123-PEI-R13-m, and P123-PEI-R13-l polyplexes exhibited the maximal transfection efficiency at the weight ratio of 20, 10, and 10, respectively. Accordingly, the cell viabilities were 75.73% ± 2.81%, 92.84% ± 8.06%, and 96.40% ± 2.41%, based on cytotoxicity assay. Similar to the transfection of P123-PEI-R13 polyplexes in Hela cells, they also showed much higher gene transfection than P123-PEI, PEI 2 KDa, and PEI 25 KDa polyplexes at optimal conditions.
Conclusion
We developed a new nonviral gene vector, PEI-P123-R13, by crosslinking LMW PEI with P123 and further coupled a bifunctional peptide R13 to the polymer for targeting tumor and increasing cellular uptake. Through various physico-chemical methods, we confirmed that PEI-P123-R13 had the ability to form complexes with DNA and suitable physico-chemical properties for gene delivery. Furthermore, the new gene vector showed much lower cytotoxicity and higher gene transfection efficiency compared with PEI 25 kDa in two different cell lines (Hela and B16). In conclusion, this new polymer might be a potential candidate in gene delivery with low cytotoxicity and high transfection efficiency. However, it is worth investigating whether PEI-P123-R13 can exhibit the same excellent properties such as cyclical stability, tumor targeting, noncytotoxicity, and effective transfection in vivo. Also, the delivery of therapeutic genes mediated by this gene vector and the therapeutic efficiency need to be studied in animal models. We are currently conducting more comprehensive studies with the aim to answer these questions. We will present our findings in another manuscript.
Acknowledgment
This study was supported by the National Natural Science Foundation, People’s Republic of China (81001024), and the Leading Academic Discipline Project of Shanghai Municipal Education Commission (J50704).
Disclosure
The authors report no conflicts of interest in this work.
References
- MorilleMPassiraniCVonarbourgAClavreulABenoitJPProgress in developing cationic vectors for non-viral systemic gene therapy against cancerBiomaterials2008293477349618499247
- WuXDingBGaoJSecond-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapyInt J Nanomedicine201161747175621980237
- ChoKCChoiSHParkTGLow molecular weight PEI conjugated pluronic copolymer: useful additive for enhancing gene transfection efficiencyMacromolecular Research2006143348353
- LeeKBaeKHLeeYLeeSHAhnCHParkTGPluronic/polyethylenimine shell crosslinked nanocapsules with embedded magnetite nanocrystals for magnetically triggered delivery of siRNAMacromol Biosci201010323924519924685
- LungwitzUBreunigMBlunkTGöpferichAPolyethylenimine-based non-viral gene delivery systemsEur J Pharm Biopharm20056024726615939236
- LiuTYuXKanBGuoQWangXShiSEnhanced gene delivery using biodegradable poly(ester amine)s (PEAs) based on low-molecular-weight polyethylenimine and poly(epsilon-caprolactone)-pluronic-poly(epsilon-caprolactone)J Biomed Nanotechnol20106435135921323108
- NeuMFischerDKisselTRecent advances in rational gene transfer vector design based on poly (ethylene imine) and its derivativesJ Gene Med20057992100915920783
- DengRYueYJiFChenYKungH-FLinMCMRevisit the complexation of PEI and DNA-how to make low cytotoxic and highly efficient PEI gene transfection non-viral vectors with a controllable chain length and structureJ Control Release2009140404619625007
- HuangHYuHTangGWangQLiJLow molecular weight polyethylenimine cross-linked by 2-hydroxypropyl-gamma-cyclodextrin coupled to peptide targeting HER2 as a gene delivery vectorBiomaterials2010311830183819942284
- KabanovAVLemieuxPVinogradovSAlakhovVPluronic block copolymers: novel functional molecules for gene therapyAdv Drug Deliv Rev20025422323311897147
- GuoQShiSWangXKanBGuYShiXSynthesis of a novel biodegradable poly(ester amine) (PEAs) copolymer based on low-molecular-weight polyethyleneimine for gene deliveryInt J Pharm20093791828919539737
- KoYTKaleAHartnerWCPapahadjopoulos-SternbergBTorchilinVPSelf-assembling micelle-like nanoparticles based on phospholipid-polyethyleneimine conjugates for systemic gene deliveryJ Control Release2009133213213818929605
- KabanovAVBatrakovaEVSriadibhatlaSYangZKellyDLAlakovVYPolymer genomics: shifting the gene and drug delivery paradigmsJ Control Release200510125927115588910
- SahayGBatrakovaEVKabanovAVDifferent internalization pathways of polymeric micelles and unimers and their effects on vesicular transportBioconjug Chem2008192023202918729494
- ChooESGYuBXueJMSynthesis of poly(acrylic acid) (PAA) modified pluronic P123 copolymers for pH-stimulated release of doxorubicinJ Colloid Interf Sci2011358462470
- OgawaMHatanoKOishiSDirect electrophilic radiofluorination of a cyclic RGD peptide for in vivo αvβ3 integrin related tumor imagingNucl Med Biol2003301912493537
- LeeDEHongYDChoiKHLeeS-YParkPHChoiSJPreparation and evaluation of 99mTc-labeled cyclic arginine-glycine-aspartate (RGD) peptide for integrin targetingAppl Radiat Isotopes20106818961902
- KilkKEl-AndaloussiSJärverPEvaluation of transportan 10 in PEI mediated plasmid delivery assayJ Control Release200510351152315763630
- Santos-CuevasCLFerro-FloresGMurphyCADesign, preparation, in vitro and in vivo evaluation of 99mTc-N2S2-Tat49–57-bombesin: a target-specific hybrid radiopharmaceuticalInt J Pharm2009375758319393305
- BrooksHLebleuBVivésETat peptide-mediated cellular delivery: back to basicsAdv Drug Deliv Rev20055755957715722164
- YinDFDingXYGaoJChuCZouHGaoSNonionic amphiphilic surfactant conjuncted polyethyleneimine as a new and highly efficient non-viral gene carrierMacromol Res2009171925
- KimTHCookSEAroteRBChoMHNahJWChoiYJChoCSA degradable hyperbranched poly(ester amine) based on poloxamer diacrylate and polyethylenimine as a gene carrierMacromol Biosci2007761161917457939
- AroteRKimTHKimYKA biodegradable poly(ester amine) based on polycaprolactone and polyethylenimine as a gene carrierBiomaterials20072873574417034844
- MaKHuMXQiYPAMAM-triamcinolone acetonide conjugate as a nucleus-targeting gene carrier for enhanced transfer activityBiomaterials2009306109611819656564
- KimT-IBaekJ-UYoonJKChoiJSKimKParkJSSynthesis and characterization of a novel arginine-grafted dendritic block copolymer for gene delivery and study of its cellular uptake pathway leading to transfectionBioconjug Chem20071830931717315976
- HaoJGShaXYTangYJEnhanced transfection of polyplexes based on pluronic-polypropylenimine dendrimer for gene transferArch Pharm Res2009321045105419641886
- AndersonDGAkincAHossainNLangerRStructure/property studies of polymeric gene delivery using a library of poly(b-amino esters)Mol Ther20051142643415727939
- LinCBlaauboerCJTimonedaMMEngbersen, bioreducible poly(amido amine)s with oligoamine side chains: synthesis, characterization, and structural effects on gene deliveryJ Control Release200812616617418162194
- ParkMRHanKOChoMHNahJWChoiYJChoCSDegradable polyethylenimine-alt-poly (ethylene glycol) copolymers as novel gene carriersJ Control Release200510536738015936108
![Figure 7 Agarose gel electrophoresis of the complexes at various w/w (P123-polyethylenimine [PEI]-R13/DNA) ratios: (A) P123-PEI-R13-h, (B) P123-PEI-R13-m, and (C) P123-PEI-R13-l.](/cms/asset/f18d2ae0-a16e-43c0-b218-09cab01e4ed6/dijn_a_28819_f0007_c.jpg)