 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
The purpose of this study was to test the efficacy of cyclic Arg-Gly-Asp (RGD) peptide conjugated with polyionic complex nanomicelles as targeted therapy for glioma.
Methods
A stable cyclic RGD polyionic complex nanostructure, ie, a c(RGDfC) polyionic complex micelle, was synthesized and its biocompatibility with cultured neurons was assessed using a cell viability assay. Targeted binding to cultured glioma cells was evaluated by the CdTe quantum dot marking technique and a cell viability assay. The inhibitory effect of the nanomicelles against glioma cells was also evaluated, and their targeted migration into rat brain glioma cells and apoptotic effects were traced by the CdTe quantum dot marking and immunohistochemical staining.
Results
c(RGDfC) polyionic complex micelles did not affect the growth of neurons but bonded selectively to and inhibited proliferation of glioma cells in vitro. When tested in vivo, the micelles migrated into glioma cells, inducing apoptosis in the rat brain.
Conclusion
The c(RGDfC) polyionic complex micelle is an effective targeted therapy against glioma.
Introduction
Glioma is one of the most common primary brain tumors, accounting for about 30%–40% of all intracranial tumors.Citation1 The annual incidence is growing gradually around the world, and has now increased to 2.4–7.0 per 100,000.Citation1,Citation2 Recurrence and mortality rates are still high, even though aggressive therapies have been used, including surgery, radiotherapy, and chemotherapy. The efficacy of antiglioma drugs is limited by their toxicity, as a result of their narrow therapeutic window, which affects therapeutic indices.
Short synthetic peptides containing the tripeptide sequence of Arg-Gly-Asp (RGD) are well known for their integrin-binding activity. RGD have been used recently for tumor-targeted imaging.Citation3–Citation6 This study was undertaken because, although RGD peptide-conjugated nanomaterials are a research focus in the field of antitumor therapies,Citation7 none of the relevant trials so far has resulted in an effective antiglioma drug.
A polyion complex micelle can be formed by electrostatic interaction in an aqueous medium. Polyion complexes have self-assembling core-shell structures which can protect a loaded drug from degradation by enzymes and engulfment by phagocytes. The polyion complex can extend the longevity of a drug in the blood, enhancing permeability and retention. Due to its proven biomedical compatibility, the polyion complex micelle is one of the most frequently used nanodelivery systems in biomedicine.Citation8
In this study, a polyion complex micelle was conjugated with an RGD-containing pentapeptide, ie, c(RGDfC, Cyclo(-Arg-Gly-Asp-D-Phe-Cys), forming an encapsulated poly-(aspartic acid) ion complex, ie, the c(RGDfC) polyionic complex micelle. Experiments were conducted to test the stability of the c(RGDfC) polyionic complex micelle against variations in pH and NaCl concentration, as well as its biocompatibility with neurons and its therapeutic effect against glioma cells. The targeting effect of c(RGDfC) polyion complex micelles was then evaluated by tracing them with CdTe quantum dots.
Materials and methods
Preparation of activated polyion complexes
Maleimide-polyethylene glycol (PEG)-g-N-diethanolamine (PDEA) and maleimide-PEG-g-poly(aspartamide) (PAsp) were gifted for this research by the Department of Macromolecular Science and the Key Laboratory of Macromolecular, Engineering of Polymers, Fudan University. First, 15.8 mg (5 mM) of maleimide-PEG-g-PDEA and 13.8 mg (5 mM) of maleimide-PEG-g-PAsp were dissolved in 10 mL of 0.01 M phosphate-buffered saline (pH 7.4). The maleimide-PEG-g-PAsp solution was slowly dropped into the solution of maleimide-PEG-PDEA under gentle stirring. After 30 minutes, the solution was filtered using a 0.45 μm syringe-type filter before adding an appropriate amount of glutaraldehyde 2.5% solution (Shanghai Biochemical Reagent Company, Shanghai, China), stirring for 4 hours, and refiltering the entire solution again at 25°C. The final product was dried under vacuum for 8 hours. The zeta potential of the nanomicelles was determined using a Zetasizer Nano ZS (Malvern Instruments Ltd, Worcestershire, UK).
Preparation of RGD-conjugated polyion complexes
Because cyclic peptide pentapeptides reportedly had higher binding affinity and selectivity for integrin,Citation9 c(RGDfC) was purchased from Shanghai Sangon Biological Engineering Technology and Services Company (Shanghai, China) for use in this work. c(RGDfC) was suggested with the polyion complex solution for 8 hours at 4°C. The molar ratio of c(RGDfC) to maleimide was 1:3, as indicated in a previous report.Citation10 The reaction mixture was then added to a 1.5 × 20 cm Sepharose CL-4B column and eluted with 0.01 M phosphate-buffered saline (pH 7.4). The turbid c(RGDfC) polyion complex fractions were visually identified and collected, and the polyion complex concentration was determined by turbidimetry using a UV2401 spectrophotometer at 350 nm (Shimadzu, Tokyo, Japan). The zeta potential was assessed using the Zetasizer Nano ZS.
Preparation of c(RGDfC) polyion complex micelles
A cosolvent evaporation method was used to prepare the c(RGDfC) polyionic complex micelles. First, 50 mg of c(RGDfC) polyionic complexes in 10 mL of acetone was added to phosphate-buffered saline (0.01 M, 50 mL, pH 7.4) in a dropwise manner. After 5 hours of stirring the solution at room temperature, a vacuum device was used to ensure complete removal of the organic solvent from the solution.
Transmission electron microscopy and dynamic light scattering
The morphology of the polyionic complex micelles was studied using a transmission electron microscope (Hitachi H-600, Tokyo, Japan, 250 kV). A drop of sample solution was allowed to settle on the copper grid for one minute. Excess solution was removed by wicking with filter paper for one minute. Dynamic light scattering measurements were performed on a Malvern Autosizer 4700, and the laser wavelength (λ) used for the measurements was 514.5 nm.
Neuron culture
Wistar rats were purchased from the Experimental Animal Center of Shanghai Medical School, Fudan University, Shanghai. Primary neuron cultures were prepared from the brains of neonatal Wistar rats sacrificed at postnatal day 1, and cultured in Neurobasal®-A medium containing 2% B27 supplement and glutamine 5 mmol/L (all from Gibco-BRL, Burlington, ON, Canada). The neonatal rats were decapitated and their heads rinsed in phosphate-buffered saline and glucose medium (7.9 mmol/L) solution containing penicillin 40,000 U/L and streptomycin 40,000 U/L (all from Gibco-BRL). The brain was rapidly removed from the skull, placed in phosphate-buffered saline and glucose medium, and dissected to remove the meninges. The hippocampi were placed in a tube containing 1 mL phosphate-buffered saline trypsin-DNase solution (1%, 0.1%, Gibco-BRL) for 15 minutes at room temperature. The forebrains were then rinsed three times with phosphate-buffered saline, and 1 mL Neurobasal-A medium was added. Mechanical dissociation machine was performed to obtain a monocell suspension, which was centrifuged using a Universal 16R (Hettich, Beverly, MA) at 2000 rpm for 10 minutes at 4°C. The supernatant was removed and the pellet was resuspended in 5 mL Neurobasal-A and B27 glutamine medium. The cell number was then counted. Finally, the cells were plated onto 96-well plates (1 × 106 cells/mL) and polylysine-coated glass 6-well plates (1 × 106 cells/mL). The neuron cultures were kept for 7 days in a humidified incubator at 37°C and 5% CO2, (CO2 incubator HERAcell 150, Thermo Electron Corporation, Waltham, MA). The culture medium was renewed every 3 days.
MTT assay for cell viability
For cell viability, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT, Sinopharm Chemical Reagent Company, Shanghai, China), a yellow tetrazole assay, was performed following standard procedures for each well of cells post treatment. Briefly, cells cultured in 96-well plates were treated with 50 μL of MTT per well. After coculture, 150 μL of dimethyl sulfoxide (Sinopharm Chemical Reagent Company) were added per well. After 20 minutes of reaction, the plate was subjected to ocular density (OD) detection using an enzyme-labeled meter (Multiskan 3, Thermo Electron Corporation) at a wave length of 550 nm. Cell viability was expressed as a percentage, and calculated using the following equation:
where CV is cell viability, OD1 is the OD value before treatment, and OD2 is the OD value after treatment.
Human glioma cell culture
U87 human glioma and C6 rat glioma cell lines were gifted for this work by the Shanghai Cancer Institute. The glioma cells were plated into 96-well plates (1 × 104 cells/mL) and polylysine-coated glass 6-well plates (1 × 104 cells/mL), and grown in Dulbecco’s Modified Eagle’s Medium (Gibco-BRL) with 20% fetal bovine serum in 5% CO2 at 37°C for 3 days. Culture medium was renewed every 3 days.
Preparation of quantum dot marked RGD-polyionic complex micelles
To trace the distribution of c(RGDfC) polyionic complex micelles, 0.5 mL of CdTe quantum dots (Quantum Dot Corporation, Hayward, CA) stabilized by thioglycolic acid was added to the copolymer solution. The resulting solution was precipitated repeatedly in ice-cold acetone, and the precipitate was then washed three times with acetone to remove the unloaded CdTe quantum dots.
C6 cell-implanted animal model of glioma
C6 cell lines were maintained in Dulbecco’s Modified Eagle’s Medium, with 15% heat-inactivated fetal calf serum and glutamine 0.2 mM, neomycin 50 g/mL, and streptomycin 100 μg/mL. An animal model of glioma was induced by stereotactic injection of C6 cells following a previously reported procedure,Citation11 with minor modification. Briefly, C6 cells were harvested by trypsinization, pelleted by centrifugation, and resuspended for intracranial implantation. Anesthetized Wistar rats (n = 12 in each group) were placed in a stereotactic head frame, and a small right frontal craniectomy was drilled 2.5 mm from the midline and 1.0 mm anterior to bregma. C6 cells (1 × 106/10 μL) were implanted stereotactically to a depth of 4 mm below the craniectomy using a microinjection syringe. The craniectomy was resealed with bone wax and the scalp closed. For identification of tumor growth, contrast-enhanced computed tomography was conducted using a Somatom Emotion 16 device (Siemens, Shanghai, China) 3 weeks following implantation. All animal experiments were approved by the local animal care committee and were in accordance with international guidelines on the ethical use of animals.
Immunohistochemistry
On day 21 after glioma implantation, quantum dot-marked c(RGDfC) polyionic complex micelles or a control (phosphate-buffered saline) solution was injected into the rats via the tail vein. The rats were decapitated 24 hours later. Their brains were then fixed in 4% paraformaldehyde, frozen-sectioned into 10 μm slices, and stained with FITC-conjugated anticaspase-3 antibody (500×; Chemicon, Shanghai, China) for marking of apoptotic cells, and Hoechst 33258 (200×, Sigma-Aldrich, Shanghai, China) for marking of cell nuclei, following standard procedures. The sections were then subjected to imaging under a fluorescent microscope.
Statistical analysis
The data are presented as the mean ± standard deviation. Statistical significance was determined using one-way analysis of variance followed by a post hoc test with statistical analysis software (SPSS 11.0, Lead Technologies, Chicago, IL).
Results
Micelle morphology
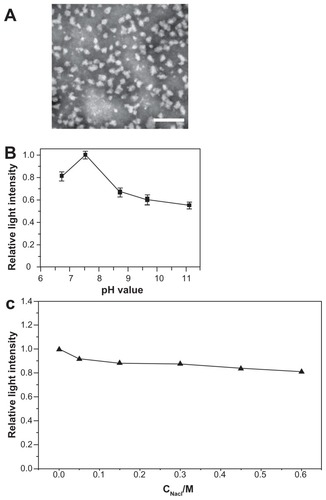
The c(RGDfC) polyionic complex micelles were successfully synthesized and their diameters [averaging 60.1 ± 9.2 nm (counts from 50 independent observations of micelle diameters)] were homogeneous (). The c(RGDfC) polyionic complex micelles demonstrated different shapes. Most of them were dispersed, with rare exceptions, while several micelles aggregated with each other. This phenomenon might result from evaporation of the solvent on the transmission electron microscope grid.
Figure 1 Morphological and physical features of c(RGDfC) polyionic complex micelles. (A) Assemblies of c(RGDfC) polyionic complex micelles in acetone/phosphate-buffered saline (1:5) are shown in a dark-field transmission electron microscopic image (bar 200 nm). The c(RGDfC) polyionic complex micelles show homogeneity in size, with an average diameter of 60.1 ± 9.2 nm. (B) Stability of c(RGDfC) polyionic complex micelles against pH variation. The variations in relative light intensity with increasing pH for c(RGDfC) polyionic complex micelles were measured by dynamic light scattering. The total concentration for each micelle system was 0.5 mg/mL at a temperature of 25°C ± 0.1°C. Relative light intensity showed no abrupt change after pH treatment. (C) Stability of c(RGDfC) polyionic complex micelles against salt. The micelles were subjected to salt treatment at a series of NaCl concentrations of 0, 0.05, 0.15, 0.3, 0.45, and 0.6 M. Relative light intensity was measured by dynamic light scattering (pH 7.4) to show dispersity. The relative light intensity showed no abrupt change after salt treatment.
Abbreviation: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys).
Zeta potential of nanomicelles
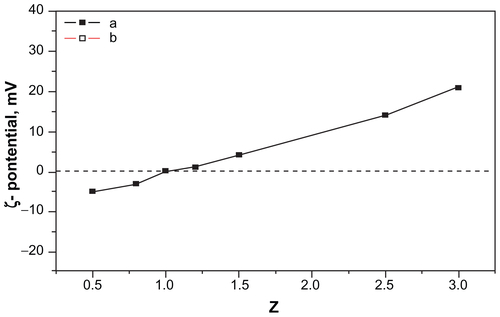
The zeta potential of the c(RGDfC)-polyionic complex micelles is shown in Supplementary Figure 1. When 0 < Z < 1, the zeta potential was negative. When Z = 1, the zeta potential was around zero. When Z > 1, the zeta potential turned positive. The zeta potential increased as a result of increasing PDEA concentration. (Z, Z = C1/C2; C1, concentration of PDEA; C2, concentration of PAsp).
Stability against pH variation and salinity
To quantify micellar stability against pH variation, dynamic light scattering was used to measure the relative light intensity of the c(RGDfC) polyionic complex micelles in solutions with different pH values. Because the micelles were administered intravenously, they would not encounter an acidic environment, so the pH was set at 6–10, covering the physiological pH range from 7.35 to 7.45. The dispersity of the c(RGDfC) polyionic complex micelles changed subtly with pH variation from 6.0 to 11.0 (). To quantifymicellar stability against salt, the micelles were exposed to different NaCl concentrations. Dispersity of the c(RGDfC) polyionic complex micelles was stable against a NaCl concentration range of 0–0.6 mol/L ().
Biocompatibility with neurons
To evaluate the biocompatibility of the c(RGDfC) polyionic complex micelles, 7-day primary cultured neurons in every well of 6-well dishes were treated with either c(RGDfC) polyionic complex micelles 150 μmol/mL or vehicle for 24 hours. The neurons were stained with Hoechst 33258 (a specific marker for the nucleus, 1:100, Invitrogen, Shanghai, China) and TUNEL (an apoptotic cell marker, 1:200, Invitrogen). The stained neurons were then imaged under a fluorescent microscope. No significant differences were found between the apoptotic ratio of the control group (0.63% ± 0.07%, ) and the group treated with c(RGDfC) polyionic complex micelles (0.75% ± 0.05%, ). The apoptotic ratio was expressed as three independent counts of apoptotic cell number/total cell number under a 20 μm × 20 μm microscope.
Figure 2 Biocompatibility of c(RGDfC) polyionic complex micelles with neurons. Neurons stained with Hoechst 33258 for nucleus and TUNEL for apoptosis after coculture with vehicle (A) or c(RGDfC) polyionic complex micelles (B) for 24 hours (bar 40 μm). Seven-day primary cultured neurons in each well of 6-well dishes were treated with c(RGDfC) polyionic complex micelles (150 μmol/mL) for 24 hours, stained with Hoechst 33258 and TUNEL and imaged under fluorescent microscope. (C) Cell viability of neurons following dose-dependent treatments determined by MTT assay. Seven-day primary neurons were given as 0, 50, 150, 300 and μmol/mL c(RGDfC) polyionic complex micelles, c(RGDfC), or polyionic complexes, respectively, for 4 hours. The cells were subjected to MTT assay following treatment. No significant reduction in cell viability was observed in the neurons with the c(RGDfC) polyionic complex, c(RGDfC), or polyionic complex micelles. (D) Viability of the neurons following time-dependent treatments determined by MTT assay. Neurons were cocultured with 150 μmol/mL c(RGDfC) polyionic complex, c(RGDfC), or polyionic complex micelles for a series of times, ie, 0, 2, 24, and 72 hours. The cells were subjected to MTT assay following treatment. No significant reduction of cell viability was observed in neurons treated with the c(RGDfC) polyionic complex, c(RGDfC), or polyionic complex micelles.
Note: All cellular experiments were repeated at least three times as confirmation.
Abbreviation: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys).
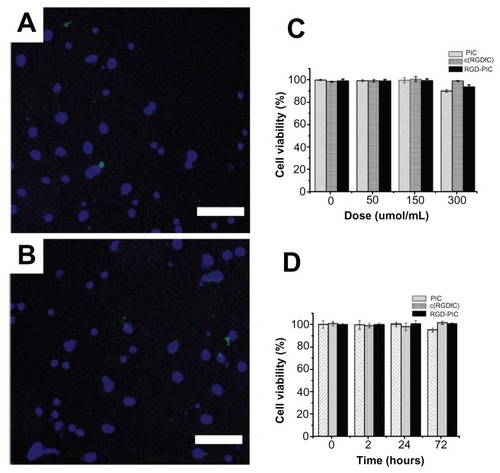
To assess further the biocompatibility of the micelles with neurons, the MTT assay was used to determine cell viability. To perform a dose-dependent assay, 7-day primary neurons were pulsed with c(RGDfC) polyionic complex micelles, c(RGDfC), or polyionic complexes alone at four concentrations (0 μmol/mL, 50 μmol/mL, 150 μmol/mL, and 300 μmol/mL) for 24 hours. The pulsed neurons were then subjected to MTT assay (molarity refers to RGD). The MTT assay showed no significant reduction in cell viability in neurons treated with c(RGDfC) polyionic complexes, c(RGDfC), or polyionic complex micelles ().
To assess the time-dependent relationship between the following treatments and viability of neurons, 7-day primary neurons were pulsed with c(RGDfC) polyionic complex micelles, c(RGDfC), or polyionic complexes in 300 μmol/mL for four durations (hours 0, 2, 24, and 72), respectively, and subjected to MTT assay. The MTT assay showed no significant reduction of viability in neurons treated with the c(RGDfC) polyionic complexes, c(RGDfC), or polyionic complex micelles ().
Inhibition of cultured U87 glioma cells
To investigate for possible inhibitory effects in vitro, U87 cells were treated with c(RGDfC) polyionic complex micelles (150 μmol/mL), polyionic complexes alone (150 μmol/mL), c(RGDfC) alone (150 μmol/mL), or vehicle for 4 hours and subjected to staining with antiglial fibrillary acid protein (red, 1:200, Invitrogen), a specific marker for astrocytes and TUNEL (green, 1:200, Invitrogen), an apoptotic cell marker and imaged under the fluorescent microscope. No significant differences were found between the apoptotic ratio of the vehicle control group (0.77% ± 0.09%, ) and that of the group that received polyionic complexes alone (0.80% ± 0.09%, , P > 0.05). However, significant differences were found between the vehicle control group and RGD alone group (98.19% ± 0.11%, , P < 0.001). Significant differences were also found between the vehicle control group and the group that received c(RGDfC) polyionic complex micelles (99.24% ± 0.13%, , P < 0.001). The apoptotic ratio was expressed as three independent counts of apoptotic cell number/total cell number under a 20 μm × 20 μm microscope.
Figure 3 Inhibition of U87 glioma-derived cells after treatment with c(RGDfC) polyionic complex micelles. Apoptosis of U87 cells post treatment with c(RGDfC) polyionic complex micelles. Three-day U87 cells were treated with (A) phosphate-buffered saline, (B) 150 μmol/mL of polyionic complexes, (C) 150 μmol/mL of c(RGDfC), and (D) 150 μmol/mL of c(RGDfC) polyionic complex micelles for 4 hours and imaged under microscopy (bar 125 μm in each graph), and (E) viability of U87 cells post treatment, determined by MTT assay. To see whether the compound kills glioma cells, 3-day U87 cells cultured in 96-well plates were divided into 12 groups (n = 5), receiving 0, 50, 150, 300 μmol/mL c(RGDfC) polyionic complex micelles, c(RGDfC), or polyionic complexes for 4 hours.
Notes: The cells were subjected to MTT assay post treatment. Significant reduction of cell viability was observed in the group treated with c(RGDfC) polyionic complexes (#P < 0.05) and the group treated with c(RGDfC) (*P < 0.05), while no significant reduction of cell viability was seen in the group treated with polyionic complex micelles. All cellular experiments were repeated at least three times as confirmation.
Abbreviation: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys).
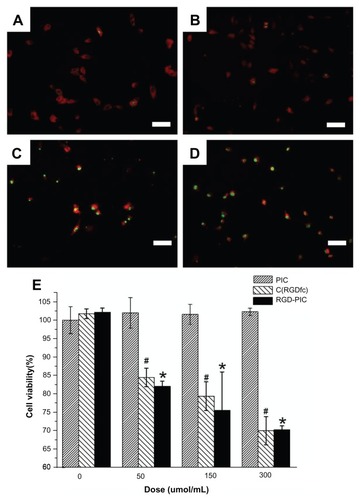
For cell viability, U87 cells were treated with c(RGDfC) polyionic complex micelles, c(RGDfC), or polyionic complexes at four concentrations, ie, 0 μmol/mL, 50 μmol/mL, 150 μmol/mL, and 300 μmol/mL, and then subjected to MTT assay. Significant reductions in cell viability both in the c(RGDfC) polyionic complex-treated group and in the c(RGDfC)-treated group were observed (P < 0.05), while no significant reduction of cell viability was seen in the group treated with polyionic complex micelles ().
Preparation of quantum dot-marked RGD-polyionic complex micelles
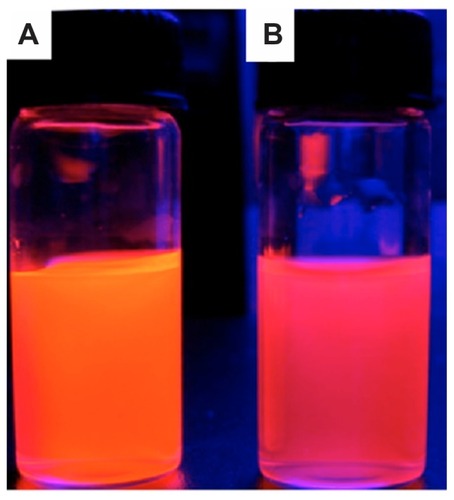
To trace the distribution of c(RGDfC) polyionic complex micelles, the micelles were coated with CdTe quantum dots. The fluorescent shift from orange to reddish was due to electrostatic interaction between the carboxyl groups surrounding the quantum dots and active amine groups within the polyionic complex sphere (). This demonstrated the effective combination and encapsulation of quantum dots within the polyionic complex.
Figure 4 Quantum dot tracing of c(RGDfC) polyionic complex micelles. Quantum dot marked c(RGDfC) polyionic complex micelles in a cup. (A) Quantum dot emitting orange fluorescence. (B) The fluorescent shift from orange to reddish due to the electrostatic interaction between carboxyl groups surrounding quantum dots and active amine groups within polyionic complex spheres (actual size).
Note: This demonstrates the effective combination and encapsulation of quantum dots within a polyionic complex.
Abbreviation: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys).
Glioma inhibition in vivo
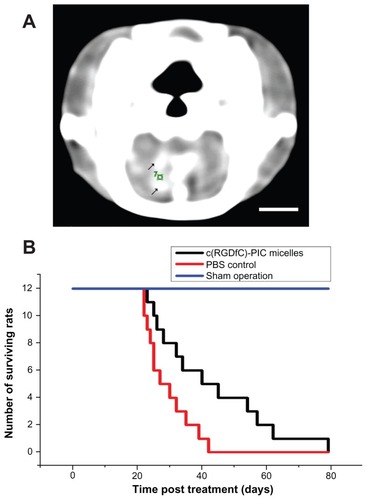
To determine whether the c(RGDfC) polyionic complex micelles could inhibit glioma cells in vivo, Wistar rats were implanted with the C6 glioma cell line and treated with c(RGDfC) polyionic complex micelles or vehicle. A mass with high density found within the contrast-enhanced computed tomographic image of the rat brain () indicated successful implantation of glioma. c(RGDfC) polyionic complex micelles (300 μmol/mL, 1 mL per rat) were given to rats with glioma via the tail vein, setting phosphate-buffered saline (0.01 mol/L, 1 mL per rat) for rats with glioma and c(RGDfC) polyionic complex micelles for sham operation (stereotactic injection of Dulbecco’s Modified Eagle’s Medium without glioma cells) as controls. When the c(RGDfC) polyionic complex micelles were applied to rats with glioma, there was a statistically significant increase in median survival time (). Meanwhile, no deaths occurred in the sham operation group treated with c(RGDfC) polyionic complex micelles, indicating no significant toxicity of the micelles in vivo.
Figure 5 Increased survival time by c(RGDfC) polyionic complex micelles in rat glioma. (A) Contrast-enhanced computer tomography of glioma in rat brain;10 μL medium containing 1 × 106 cells from a C6 malignant glioma cell line cells was directed into the caudate nucleus of the right brain of Wistar rats via stereotactic injection to establish an animal model of glioma. Enhanced computer tomography was conducted 3 weeks later to identify the existence of tumor in rat brain. Because a brain image on computer tomography scan was known to be symmetric, a glioma in the right hemisphere could be recognized by comparison with the left hemisphere. Enhanced computer tomography signals (black arrows) were found within the hemisphere implanted with tumor cells (right). The number “7” was a sectional sequence mark. This figure is the 7th section (bar 3 mm). (B) On day 21, c(RGDfC) polyionic complex micelles (300 μmol/mL, 1 mL/rat) were given to the rats with glioma via their tail veins, setting the phosphate-buffered saline and sham operation as control.
Note: Survival time was plotted and the statistical significance of median survival time was calculated (*P < 0.05).
Abbreviations: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys); PBS, phosphate-buffered saline.
Target effect in vivo
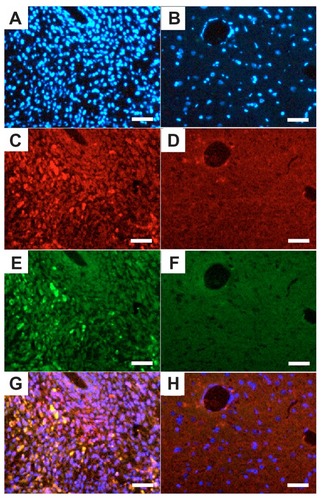
To show whether the c(RGDfC) polyionic complex micelles could bind specifically to glioma cells in vivo, the rats were implanted with a C6 glioma cell line, treated with quantum dot-marked c(RGDfC) polyionic complex micelles, and subjected to standard immunohistochemical staining using caspase-3 (a specific marker for apoptotic cells) and Hoechst 33258 (a specific marker for cell nuclei). The cell density shown by Hoechst 33258 staining (blue) in the tumor area () was much higher than that in the contralateral area (). The c(RGDfC) polyionic complex micelles traced by quantum dots (red) were found within the tumor area (), while little was found within the contralateral area (), indicating a glioma-targeted migration after administration. Apoptosis shown by caspase-3 staining (green) was found within the tumor area () and sparing the contralateral area (), indicating a glioma-selective apoptotic effect. By merging the above fluorescent images, apoptotic staining was found to overlap well with quantum dots, manifesting as yellow spots (). By contrast, this phenomenon was not found within the contralateral area (). Significant differences were found between the apoptotic ratio in the ipsilateral hemisphere (37.42% ± 0.31%) and that of the contralateral hemisphere (0.50% ± 0.12%, P < 0.001). The apoptotic ratio was expressed as three independent counts of (apoptotic cell number)/(total cell number) under a 5 μm × 5 μm microscope. Taken together, these data suggested that in vivo apoptosis of glioma cells was induced by the c(RGDfC) polyionic complex micelles that specifically.
Figure 6 Quantum dot-traced target effect of c(RGDfC) polyionic complex micelles against glioma in an animal model. On day 22, QD marked c(RGDfC) polyionic complex micelles were injected into the rats via their tail veins. The rats were decapitated 24 hours later. The brains were then fixed in 4% paraformaldehyde, frozen-sectioned into 10 μm slices and subjected to standard immunohistochemical staining of caspase-3 (a specific marker for apoptotic cells) and Hoechst 33258 (a specific marker for nucleus). Typical results were shown as representative of six independent experiments. (A) Hoechst 33258 staining (blue) in the glioma area identified by computed tomography. (B) Hoechst 33258 staining (blue) in the contralateral area. (C) Quantum dot-conjugated c(RGDfC) polyionic complex micelles (red) in the glioma area. (D) Quantum dot-conjugated c(RGDfC) polyionic complex micelles (red) in the contralateral area. (E) Caspase-3-positive cells (green) in the glioma area. (F) Caspase-3-positive cells (green) in the contralateral area. (G) Merge of graph A, C, and E. (H) Merge of graph B, D, and F.
Note: Bar 25 μm.
Abbreviation: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys).
Discussion
This study demonstrates the advantages of c(RGDfC) polyionic complex micelles as a targeted therapy against glioma from three perspectives, ie, apoptotic potential, decreased cell viability when applied in vitro, and improved survival rate when used in vivo.
The findings from dynamic light scattering established the stability of the c(RGDfC) polyionic complex micelles against variation in pH and salt exposure. The viability of neurons was shown to be unaffected by c(RGDfC) polyionic complex micelles. This finding provides important evidence for the biocompatibility of a potential drug with activity against brain tumor cells. Biocompatibility was confirmed by the fact that the survival rate of rats was not affected by injection of c(RGDfC) polyionic complex micelles.
RGD is thought to induce apoptosis of tumor cells.Citation12–Citation14 Cilengitide, a cyclic RGD peptide, has been used in clinical trials for treatment of glioblastoma.Citation15, However, RGD peptide-conjugated nanomaterials were mainly used for imaging rather than for treating tumors. RGD peptides can specifically recognize integrin, which is restrictively expressed on cell surfaces of malignant glioma cells.Citation16 RGD peptide conjugated with nanomaterials enabled tracing of glioma in a photoinduced electron transfer imaging system.Citation17–Citation19 We found that RGD and c(RGDfC) polyionic complex micelles, but not vehicle control or polyionic complexes, induced apoptosis of human glioma cells in vitro.
In vivo experiments demonstrated a statistically significant delay in death of rats with glioma treated with c(RGDfC) polyionic complex micelles, compared with those in the phosphate-buffered saline control and sham operation control groups. This phenomenon suggested an inhibitory effect of the c(RGDfC)-polyionic complex micelles on glioma cells. Immunohistochemical staining combined with quantum dot tracing showed glioma-specific migration of the c(RGDfC) polyionic complex micelles after intravenous administration and specific apoptosis of the glioma cells. Most of the c(RGDfC) polyionic complex micelles-bond cells were apoptotic, while the other cells were not apoptotic. This indicates that c(RGDfC) polyionic complex micelles caused the apoptosis of the glioma cells.
The above evidence strongly supports a targeted therapeutic effect of c(RGDfC) polyionic complex micelles against human glioma cells. However, before possible clinical application of c(RGDfC) polyionic complex micelles, further investigations should include in vivo tests in an animal model of glioma. The concentration of micelles used in this study was quite high (ranging from 50 μmol/mL to 300 μmol/mL), which are most likely not to be reached in the brain tumors of patients if used systemically. Further, our experiments did not answer whether the micelles enter the brain through the blood–brain barrier or through craniectomy-induced lesions. In conclusion, we have pushed towards targeted therapy for human glioma by combining the carrier effects of nanoparticles with the targeting and apoptosis-inducing effects of RGD.
Conclusion
This study offers evidence that by conjugating a cyclic RGD peptide with polyionic complex micelles it is possible to achieve targeted therapy for glioma. However, there is still a long way to go from bench to bedside.
Acknowledgments
This work was funded by the Nanotechnology Fund Committee, Science and Technology Commission of the Shanghai Municipality (1052nm05300) and Research Fund for Outstanding Young Teachers in Universities of Shanghai (jdy10017) and Program for SMC-Young Scholar of SJTU. We are particularly grateful to Wei Chen and Changchun Wang from the Department of Macromolecular Science and Key Laboratory of Macromolecular, Engineering of Polymers, Fudan University, Shanghai, without whose instruction and generous help it would have been impossible to synthesize the nanoparticles in this work.
Disclosure
The authors report no conflicts of interest in this work.
Supplementary figure
Figure S1 Zeta potential of the c(RGDfC) polyionic complex micelle solution at pH 10.
Notes: When 0 < Z < 1, the zeta potential was negative. When Z = 1, the zeta potential was around zero. When Z > 1, the zeta potential turned positive. The zeta potential increases as a result of increasing PDEA concentration. Z: Z = C1/C2. C1: concentration of PDEA, C2: concentration of PAsp.
Abbreviations: c(RGDfC), Cyclo(-Arg-Gly-Asp-D-Phe-Cys); PDEA, N-diethanolamine; PAsp, poly(aspartamide).
References
- SchneiderTMawrinCScherlachCSkalejMFirschingRGliomas in adultsDtsch Arztebl Int201010779980721124703
- LarjavaaraSMäntyläRSalminenTIncidence of gliomas by anatomic locationNeuro Oncol2007931932517522333
- ArosioDManzoniLAraldiEMScolasticoCCyclic RGD functionalized gold nanoparticles for tumor targetingBioconjug Chem20112266467221434651
- BraunKWiesslerMPipkornRA cyclic-RGD-bioshuttle functionalized with TMZ by DARinv “Click Chemistry” targeted to αvβ3 integrin for therapyInt J Med Sci2010732633920922134
- LiuZWangJYinPRGD-FasL induces apoptosis in hepatocellular carcinomaCell Mol Immunol2009628529319728930
- ReardonDANeynsBWellerMTonnJCNaborsLBStuppRCilengitide: an RGD pentapeptide ανβ3 and ανβ5 integrin inhibitor in development for glioblastoma and other malignanciesFuture Oncol2011733935421417900
- KimYHJeonJHongSHTumor targeting and imaging using cyclic RGD-PEGylated gold nanoparticle probes with directly conjugated iodine-125Small201172052206021688390
- JoralemonMJMcRaeSEmrickTPEGylated polymers for medicine: from conjugation to self-assembled systemsChem Commun (Camb)2010461377139320162127
- ZhouYChakrabortySLiuSRadiolabeled cyclic RGD peptides as radiotracers for imaging tumors and thrombosis by SPECTTheranostics20111588221547153
- OlivierJCHuertasRLeeHJCalonFPardridgeWMSynthesis of pegylated immunonanoparticlesPharm Res2002191137114312240939
- LiYOwusuALehnertSTreatment of intracranial rat glioma model with implant of radiosensitizer and biomodulator drug combined with external beam radiotherapyInt J Radiat Oncol Biol Phys20045851952714751523
- MatsukiKSashoTNakagawaKRGD peptide-induced cell death of chondrocytes and synovial cellsJ Orthop Sci20081352453219089540
- SmolarczykRCichońTGrajaKHuczJSochanikASzalaSAntitumor effect of RGD-4C-GG-D(KLAKLAK)2 peptide in mouse B16(F10) melanoma modelActa Biochim Pol20065380180517143339
- HuZLuoFPanYArg-Gly-Asp (RGD) peptide conjugated poly(lactic acid)-poly(ethylene oxide) micelle for targeted drug deliveryBiomed Mater Res A200885797807
- GilbertMRKuhnJLambornKRCilengitide in patients with recurrent glioblastoma: the results of NABTC 03-02, a Phase II trial with measures of treatment deliveryJ Neurooncol201210614715321739168
- KimJHZhengLTLeeWHSukKPro-apoptotic role of integrin β3 in glioma cellsJ Neurochem201111749450321332719
- SchnellOKrebsBCarlsenJImaging of integrin alpha(v)beta(3) expression in patients with malignant glioma by [18F] Galacto-RGD positron emission tomographyNeuro Oncol200911686187019401596
- ChenXParkRKhankaldyyanVLongitudinal microPET imaging of brain tumor growth with F-18-labeled RGD peptideMol Imaging Biol2006891516315003
- ZannettiADel VecchioSIommelliFImaging of alpha(v)beta(3) expression by a bifunctional chimeric RGD peptide not cross-reacting with alpha(v)beta(5)Clin Cancer Res2009155224523319671851