Abstract
Background
Nanoparticles undergoing physicochemical changes to release enclosed drugs at acidic pH conditions are promising vehicles for antitumor drug delivery. Among the various drug carriers, high-density lipoprotein (HDL)-like nanoparticles have been shown to be beneficial for cancer chemotherapy, but have not yet been designed to be pH-responsive.
Methods and results
In this study, we developed a pH-responsive HDL-like nanoparticle that selectively releases paclitaxel, a model antitumor drug, at acidic pH. While the well known HDL-like nanoparticle containing phospholipids, phosphatidylcholine, and apolipoprotein A-I, as well as paclitaxel (PTX-PL-NP) was structurally robust at a wide range of pH values (3.8–10.0), the paclitaxel nanoparticle that only contained paclitaxel and apoA-I selectively released paclitaxel into the medium at low pH. The paclitaxel nanoparticle was stable at physiological and basic pH values, and over a wide range of temperatures, which is a required feature for efficient cancer chemotherapy. The homogeneous assembly enabled high paclitaxel loading per nanoparticle, which was 62.2% (w/w). The molar ratio of apolipoprotein A-I and paclitaxel was 1:55, suggesting that a single nanoparticle contained approximately 110 paclitaxel particles in a spherical structure with a 9.2 nm diameter. Among the several reconstitution methods applied, simple dilution following sonication enhanced the reconstitution yield of soluble paclitaxel nanoparticles, which was 0.66. As a result of the pH responsiveness, the anticancer effect of paclitaxel nanoparticles was much more potent than free paclitaxel or PTX-PL-NP.
Conclusion
The anticancer efficacy of both paclitaxel nanoparticles and PTX-PL-NP was dependent on the expression of scavenger receptor class B type I, while the killing efficacy of free paclitaxel was independent of this receptor. We speculate that the pH responsiveness of paclitaxel nanoparticles enabled efficient endosomal escape of paclitaxel before lysosomal break down. This is the first report on pH-responsive nanoparticles that do not contain any synthetic polymer.
Introduction
Over the past decade, various nanoparticles for drug delivery have been developed to target specific cells selectively and to respond to environmental chemical/physical stimuli, such as pH and temperature. Among the various environmental stimuli, pH gradients have been widely employed to design novel tumor-targeting drug delivery systems because the interstitial microenvironment surrounding the solid tumor is acidic, which is due to the hypoxia and lactic acid accumulation that occurs in a rapidly growing tumor.Citation1–Citation3 The lower extracellular pH in most solid tumors than that in the surrounding tissues and blood is now regarded as a phenotype of solid tumor growth and invasiveness. The pH sensitivity of a delivery system is also important for killing cancer cells that are not in a solid tumor. Because the pH of endosomes or lysosomes in the cytoplasm is acidic, pH sensitivity can trigger extensive release of anticancer drugs after endocytosis. This selective release of cytotoxic drugs at low pH decreases the cytotoxicity and promotes the efficacy of chemotherapy.Citation4,Citation5 Consequently, various pH-responsive drug delivery systems have been designed to result in physicochemical changes of the carriers in response to low pH and release the enclosed drugs at the target site.Citation4,Citation6–Citation9
High-density lipoproteins (HDL) consisting of apolipoprotein A-I and lipophilic components, such as phospholipids, cholesterol, and cholesteryl esters, have been used as drug carriers for hydrophobic drugs in cancer chemotherapy.Citation10,Citation11 HDL has a hydrophobic interior core for the transport of cholesterol in vivo, which is favorable for including lipophilic drugs. Furthermore, it is able to target cancer cells, because the apolipoprotein A-I in HDL is recognized by scavenger receptor class B type I (SR-BI), which is overexpressed in most cancer cells.Citation12–Citation14 In contrast with artificial emulsifiers, which have been reported to cause a number of side effects related to immune responses, such as hypersensitivity reactions, nephrotoxicity, and neurotoxicity,Citation15 HDL does not trigger an immune response, because all components of HDL are endogenous molecules that are richly present in the human body. Therefore, many lipophilic drugs have been assembled with apolipoprotein A-I and phospholipids, which result in the formation of soluble HDL-like nanoparticles.Citation10,Citation11 However, despite these beneficial characteristics for cancer chemotherapy, HDL have never been employed to generate a pH-responsive drug delivery system. This is primarily due to the high stability of the HDL-like nanoparticle over a wide range of pH values, which makes it difficult for the enclosed drug to be released upon changes in pH.
The stability of HDL is highly correlated with the α-helical content of apolipoprotein A-I. An increase in the α-helical content of apolipoprotein A-I has been observed when apolipoprotein A-I binds amphipathic phospholipids, indicating that the interaction between phospholipids and apolipoprotein A-I is responsible for the stability of the discoidal structure of HDL.Citation16 However, in the reverse cholesterol pathway, an increase in the nonamphipathic cholesterol content in HDL leads to the conversion of the discoidal structure to a spherical structure and destabilization of apolipoprotein A-I binding, which results in structural disorder at the HDL surface.Citation17 Thus, we hypothesized that exclusion of phospholipids during the assembly of HDL-like nanoparticles may result in sensitivity to environmental changes, such as a pH decrease, because of the expected disordered structure of the HDL in the absence of phospholipids.
All pH-responsive drug carriers developed until now consist of synthetic polymers. In this study, we attempted to generate a pH-responsive HDL-like nanoparticle using only apolipoprotein A-I, which is abundant in the human body. Paclitaxel was assembled with only apolipoprotein A-I in the absence of phospholipids, and was compared with PTX-PL-NP, an HDL-like nanoparticle which contains phospholipids as well as paclitaxel and apolipoprotein A-I. Indeed, the paclitaxel nanoparticles selectively released paclitaxel at low pH, while PTX-PL-NP did not show a pH-dependent release profile. The pH-dependent release of paclitaxel from the paclitaxel nanoparticles resulted in a dramatically enhanced anticancer effect.
Materials and methods
Expression and purification of recombinant apolipoprotein A-I
The expression of codon-optimized human apolipoprotein A-I was performed using the pET protein expression system (Novagen, Madison, WI), with the previously described expression vector pNFXex-apolipoprotein A-I.Citation18 Purification of His-tagged apolipoprotein A-I was performed as elsewhere.Citation19
Assembly of HDL-like paclitaxel nanoparticles
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Avanti Polar Lipids, Alabaster, AL) was prepared as a 25 mM stock solution in chloroform, and was dried under a stream of nitrogen gas to produce a lipid film on the inner wall of a test tube. The glass tube containing the phospholipid film was placed in vacuum overnight to remove the residual solvent. The film of dried POPC was then dissolved in disc formation buffer (10 mM Tris HCl, 100 mM NaCl, pH 7.4) supplemented with 100 mM sodium cholate, resulting in a final lipid to cholate ratio of 1:4, and was dissolved further at 37°C for 2 hours to produce full dissolution. Then, 0.1 mL of 1.64 mM POPC solution was mixed with an equal volume of 1.4 mg/mL (1.64 mM) paclitaxel, which was dissolved in the disc formation buffer containing 100 mM sodium cholate. To induce self-assembly of PTX-PL-NP, the concentration of sodium cholate was lowered below its critical micelle concentration by diluting the mixture solution with 2 mL disc formation buffer containing purified apolipoprotein A-I. In the final mixture, the molar ratio of apolipoprotein A-I to (paclitaxel:POPC) was maintained at 1:75. After the protein/lipid aggregates were removed by centrifugation at 15,000 rpm for 30 minutes, the supernatant was subjected to size exclusion chromatography. The temperature was maintained at 25°C throughout this preparation process. This procedure resulted in a final paclitaxel recovery yield of 50%–60%.
While acceptable amounts of PTX-PL-NP could be obtained by following a widely employed procedure for the HDL-like nanoparticle assembly described above,Citation12,Citation20 paclitaxel nanoparticles not containing phospholipids could not be formed by using the same method. To enhance the assembly yield of paclitaxel nanoparticles, we used several methods, including dialysis, dilution, thermal treatment, and sonication. Of these methods, simple dilution following sonication resulted in a good assembly yield. After size exclusion chromatography purification of the paclitaxel nanoparticles, about 66% of the initially added paclitaxel was found in these nanoparticles, while approximately one third of the paclitaxel had precipitated during the assembly process.
Size exclusion chromatography
The assembled nanoparticles were separated by size exclusion chromatography using a Superdex-200 10/300 GL column on an ÄCTA FPLC apparatus (GE Healthcare, Buckinghamshire, UK). After the column was equilibrated with 10 mM Tris HCl (pH 8.0), a 100 μL sample was injected into the column at a flow rate of 0.5 mL/minute.
Dynamic light scattering and transmission electron microscopy
The size distribution of HDL-like nanoparticles was determined by a dynamic laser light scattering technique using a Dynapro apparatus (Wyatt Technology, Santa Barbara, CA). The scattering angle and temperature were fixed at 90°C and 25°C, respectively. Energy filtered-transmission electron microscopy was performed using a LIBRA 120 electron microscope (Carl Zeiss, Jena, Germany). After deposition of the HDL-like nanoparticles onto a 100 mesh copper grid coated with carbon, the grid was tapped to a filter paper to remove surface water and negatively stained using 2% uranyl acetate. The samples were air-dried before the measurements were performed.
Quantitative assay of paclitaxel
Paclitaxel was quantified using a high-pressure liquid chromatography apparatus equipped with a Capcell Pak C18 MG reversed-phase column (Shiseido, Tokyo, Japan) and an ultraviolet detector. To extract paclitaxel from the paclitaxel nanoparticles or the PTX-PL-NP, 1 mL hexane: n-propanol (5:3) was added to a 1 mL sample and mixed vigorously for 15 minutes. The solvent was removed using nitrogen gas purging after centrifugation and collection of light solution. Dried paclitaxel was dissolved with absolute ethanol and injected into the high pressure liquid chromatography apparatus. Pure ethanol was used as an isocratic mobile phase at a flow rate of 1.0 mL/minute. The concentration of paclitaxel was quantified from a standard curve.
Time-dependent uncoating of apolipoprotein A-I shell from nanoparticles
Release of paclitaxel from the nanoparticles was observed using a static light scattering assay. Static light scattering measures time-averaged intensity of scattered light which is proportional to the size of the macromolecule when the concentration is fixed. Thus, the decrease in light scattering intensity resulting from disassembly of nanoparticles to free paclitaxel and free apolipoprotein A-I, which is accompanied by a size decrease, and revealed by both size exclusion chromatography and dynamic laser light scattering, could be a good measure of paclitaxel release from the paclitaxel nanoparticles. Static light scattering was measured using a Molecular Device SpectraMax M2 fluorometer at an excitation and emission wavelength of 500 nm. After reading the value for 2 hours at the same pH (7.4), the pH values of the samples were lowered or increased to designated pH values (designated to 0), and the light scattering intensity was further recorded for another 6 hours. The plotted data were averaged from three independent experiments.
Circular dichroism analysis
Circular dichroism measurements were carried out using an automated Jasco Model CD spectrometer. The spectra were measured wavelengths ranging from 240 to 190 nm at room temperature, using a 1 mm path length cell. The background circular dichroism spectrum of paclitaxel and/or phospholipid was subtracted from that of the nanoparticles to obtain the apolipoprotein A-I spectrum in the cognate nanoparticles.
Cell lines and culture
MCF-7, SK-Br-3, and MDA-MB-231 breast cancer cell lines were obtained from the American Type Culture Collection. MCF-7 and MDA-MB-231 cell lines were maintained in Dulbecco’s Modified Eagle Medium/high glucose medium supplemented with 10% fetal bovine serum. The SK-BR-3 cell line was cultured in RPMI 1640 with 10 mM HEPES and 10% fetal bovine serum. Cells were incubated with 5% CO2 at 37°C in a humidified incubator.
Cell viability
Cell viability was determined using a modified MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Briefly, cells in exponential growth were harvested by trypsinization and seeded at a concentration of 2 × 104 cells/well into 96-well plates, and incubated overnight to allow for attachment. The medium was then removed, and fresh medium along with graded concentrations of antitumor drugs were added to cultures. Following treatment (8–48 hours), the MTT solution was added to each well at a final concentration of 0.5 mg/mL. After incubation for 3 hours at 37°C in the dark, absorbance was measured at 570 nm using a multiwell plate reader. For each treatment, cell viability was evaluated as a percentage using the following equation: (A570 of drug-treated sample/A570 of untreated sample) × 100. Data presented summarize the mean (± standard deviation) of three independent experiments performed in triplicate.
Results and discussion
Assembly of paclitaxel nanoparticles
No method has been established to assemble HDL-like nanoparticles that are responsive to pH.Citation10,Citation11,Citation20,Citation21 Nearly all HDL-like nanoparticles contain phospholipids, cholesterol, and cholesteryl esters, as well as the lipophilic drug of interest. Because phospholipids such as phosphatidylcholine are important to apolipoprotein A-I binding, which belts the edge of the discoidal nanostructure, we hypothesized that exclusion of phospholipids from the nanoparticle might result in sensitivity to environmental changes, such as pH (). To test this hypothesis, we prepared two forms of paclitaxel-containing nanoparticles using apolipoprotein A-I, ie, PTX-PL-NP that contains both paclitaxel and phosphatidylcholine, and paclitaxel nanoparticles that contains only paclitaxel.
Figure 1 Schematic representation depicting pH-responsive disassembly of paclitaxel nanoparticles, which results in release of paclitaxel under the acid conditions of the tumor surroundings or in endosomes.
Abbreviations: PTX-NP, paclitaxel nanoparticles; apoA-I, apolipoprotein A-I; TEM, transmission electron microscopy.
PTX-PL-NP could easily be generated by following a widely employed procedure with slight modification.Citation21–Citation24 After equal moles of paclitaxel and POPC were mixed together in the presence of 100 mM sodium cholate, the detergent was removed either by dilution or dialysis and apolipoprotein A-I was added at the time of dilution or dialysis. After removing protein and paclitaxel precipitates by centrifugation, the supernatant was subjected to separation by size exclusion chromatography. This procedure resulted in a PTX-PL-NP recovery yield of 50%–60%.
However, in contrast with the formation of PTX-PL-NP, which contained phospholipids,Citation20,Citation21 paclitaxel nanoparticles lacking phospholipids was not formed either by dialysis or dilution (). Even though apolipoprotein A-I is highly soluble in aqueous medium, it unexpectedly coaggregated with paclitaxel upon removal of sodium cholate. Therefore, a method of simple dilution following heating was used. This approach was chosen because it produced the highest assembly yield in the reconstitution of coenzyme Q10 nanoparticles (CoQ10-NP).Citation19 When paclitaxel was assembled with apolipoprotein A-I (initial apolipoprotein A-I to paclitaxel ratio, 1:100) using the same procedure as used for CoQ10-NP, including heating at 60°C for emulsification with sodium cholate, we obtained soluble paclitaxel nanoparticles with an assembly yield of 0.22. However, this assembly yield was much lower than that of CoQ10-NP (0.78). This poor assembly yield is likely attributable to incomplete heat emulsification due to the high melting temperature of paclitaxel (213°C) compared with the relatively low melting temperature of CoQ10 (48°C).
Table 1 Yield of paclitaxel nanoparticles assembled using different methods
Because it has been reported that sonication promotes emulsification with detergents when preparing phospholipid bilayer vesicles including HDL,Citation25,Citation26 sonication was applied instead of heating using 100 mM sodium cholate before the assembly of paclitaxel nanoparticles. The resulting assembly yield of paclitaxel nanoparticles via sonication was 0.66, which was determined after size exclusion chromatography purification of the nanoparticles and was three-fold higher than that of the heat emulsification method (). This result indicates that the assembly yield is likely related to the emulsification yield of paclitaxel with detergent.
The paclitaxel content in the paclitaxel nanoparticles was determined to be 62.2% (). This was much higher than the approximately 10% paclitaxel content in the other nanoparticles, due to the absence of lipophilic adjuvants such as phospholipids, cholesterol, and cholesteryl esters.Citation21 The molar ratio of apolipoprotein A-I to paclitaxel was 1:55, suggesting that about 110 paclitaxel molecules were included in each nanoparticle.
Table 2 Composition of paclitaxel nanoparticles prepared in this study
Size and shape of paclitaxel nanoparticles
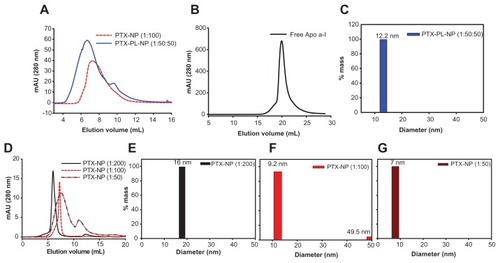
We also prepared several paclitaxel nanoparticles by following the same sonication method but by varying the initial mixing ratio of apolipoprotein A-I to paclitaxel from 1:50 to 1:200. The paclitaxel nanoparticles and PTX-PL-NP were then subjected to size exclusion chromatography and dynamic laser light scattering analysis. In the size exclusion chromatography analysis, the elution volumes of each nanoparticle were in the following order: paclitaxel nanoparticles (initial apolipoprotein A-I to paclitaxel, 1:50) >paclitaxel nanoparticles (initial apolipoprotein A-I to paclitaxel ratio, 1:100) >PTX-PL-NP (initial apolipoprotein A-I to POPC-paclitaxel ratio, 1:50:50) >paclitaxel nanoparticles (initial apolipoprotein A-I to paclitaxel ratio, 1:200, ). This result was consistent with the dynamic laser light scattering analysis. While the PTX-PL-NP was 12.2 nm in diameter, the diameters of the paclitaxel nanoparticles were determined to be 7.0 nm, 9.2 nm, and 16.0 nm for paclitaxel nanoparticles (1:50), paclitaxel nanoparticles (1:100), and paclitaxel nanoparticles (1:200), respectively (). These sizes were similar to the native HDL found in the liver and recombinant HDL particles reconstituted in vitro. This result indicates that the size of the paclitaxel nanoparticles can be controlled by adjusting the initial mixture ratio of apolipoprotein A-I and paclitaxel. All the following experiments were performed using an apolipoprotein A-I to paclitaxel ratio of 1:100.
Figure 2 Size of PTX-NP and PTX-PL-NP. Size exclusion chromatographic analysis of PTX-NP (1:100) and PTX-PL-NP (1:50:50) (A) and free apoA-I (B). (D) Size exclusion chromatographic analysis of the sizes of PTX-NPs assembled by mixing different ratios of PTX and apoA-I, for which the ratio is 1:50–1:200. (C, E–G) Dynamic light scattering assay of PTX-PL-NP (C) and PTX-NP (E–G) with different PTX to apoA-I ratios.
Abbreviations: PTX-NP, paclitaxel nanoparticles; PTX-PL-NP, paclitaxel-phospholipid nanoparticles; apoA-I, apolipoprotein A-I.
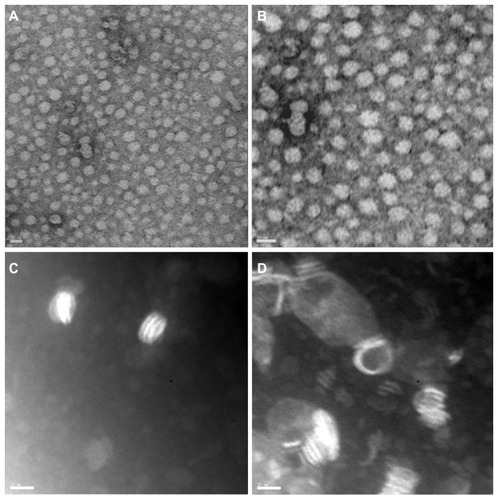
Transmission electron microscopic analysis revealed that the paclitaxel nanoparticle adopted a spherical shape (). It did not show a ring shape with a halo-like circle at the rim, as was observed for the CoQ10 nanoparticle.Citation19 Furthermore, phospholipid nanoparticles which contained only POPC as a lipidic constituent adopted the typical ringlike structure (). This structural difference was likely due to the absence of phospholipids, and we hypothesized that amphipathic molecules, such as phospholipids and CoQ10, thermodynamically stabilized the discoidal structure of the HDL-based nanoparticle. Supporting our observation, in the reverse cholesterol transport pathway, HDL undergoes a structural transition from a discoidal to spherical shape upon absorption of nonamphipathic molecules, such as cholesterol and cholesterol esters. The spherical shape of the nanoparticle has been known to be an essential feature for enhancing drug capacity and stability.Citation21
Figure 3 Transmission electron micrograph images of HDL-like PTX-NP. (A and B) Two representative images of PTX-NP. (C and D) Two representative images of PL-NP.
Note: The scale bars correspond to 20 nm.
Abbreviations: PTX-NP, paclitaxel nanoparticles; PL-NP, phospholipid nanoparticles.
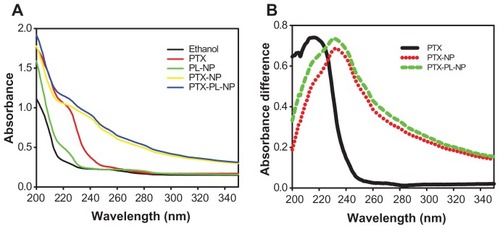
Absorption spectra for the prepared nanoparticles were collected at wavelengths ranging from 200 nm to 350 nm. The absorption spectra for ethanol, paclitaxel dissolved in ethanol, phospholipid nanoparticles, paclitaxel nanoparticles, and PTX-PL-NP are shown in . Representative spectra of only paclitaxel were calculated by subtracting the appropriate reference spectrum. The subtraction spectrum for free paclitaxel () was obtained by subtracting the ethanol spectrum (black line in ) from the paclitaxel spectra dissolved in ethanol (red line in ). The absorption maximum for free paclitaxel appeared at 216 nm. Subtraction spectra for paclitaxel nanoparticles and PTX-PL-NP () were calculated by subtracting the phospholipid nanoparticle spectrum (green line in ) from paclitaxel nanoparticles (yellow line in ) and PTX-PL-NP (blue line in ) spectra, respectively. The absorption maximum of paclitaxel included in the nanoparticles was at 231 nm. This 15 nm red-shift of absorption maximum clearly indicates that the paclitaxel molecules were indeed assembled in nanoparticles.
Figure 4 Absorption spectra of PTX in nanoparticles. (A) Absorption spectra of ethanol, free PTX dissolved in ethanol, PL-NP, PTX-NP, and PTX-PL-NP were scanned in the wavelength range of 200–350 nm. (B) Subtraction spectra for PTX loaded in various forms of nanoparticles. Subtraction spectrum for free PTX was obtained by subtracting ethanol spectrum of (A) from PTX absorption spectrum of (A). The subtraction spectra for PTX included in PTX-NP and PTX-PL-NP were obtained by subtracting the absorption spectrum of PL-NP of (A) from PTX-NP of (A), and PTX-PL-NP of (A), respectively.
Abbreviations: PTX-NP, paclitaxel nanoparticles; PTX-PL-NP, paclitaxel-phospholipid nanoparticles; PL-NP, phospholipid nanoparticles.
pH-responsiveness of paclitaxel nanoparticles
An essential feature of pH-responsive nanoparticles for cancer chemotherapy is selective drug release at low pH, with the drug stably retained inside the nanoparticle at physiological pH.Citation3,Citation27 To measure the pH-dependent behavior of the nanoparticles, a static light scattering assay was employed. Because the size of free apolipoprotein A-I is much smaller than that of the nanoparticle (), uncoating of the apolipoprotein A-I shell and release of paclitaxel from nanoparticles can be easily detected based on a lowered static light scattering. Static light scattering of both nanoparticles was measured for 6 hours at pH 3.8, 5.5, 6.5, 7.4, 8.0, 9.0, and 10.0 (). The paclitaxel nanoparticles showed little change in light scattering intensity above physiological pH (7.4–10.0). However, the light scattering intensity decreased as a function of time at an acidic pH range (6.5–3.8), the rate of which is strongly dependent on pH (). Very interestingly, the PTX-PL-NP was stably maintained over a wide range of pH values from pH 3.8–10.0 (). This result clearly suggests that only the paclitaxel nanoparticle lacking phospholipids responds to pH and dissembles into paclitaxel and free apolipoprotein A-I.
Figure 5 Time and pH-dependent release of paclitaxel from PTX-NP. (A and B) Static light scattering, which is proportional to the amount of soluble nanoparticles, was measured as a function of time at various pH values, where nanoparticles are PTX-NP (A) and PTX-PL-NP (B). Plotted data are the average and standard deviation of three independent experiments. Legends apply to both (A) and (B). Error bars are shown only when there is significant light scattering intensity for brevity. (C) Uncoating of the apoA-I shell from PTX-NP was analyzed by size exclusion chromatography.
Abbreviations: PTX-NP, paclitaxel nanoparticles; PTX-PL-NP, paclitaxel-phospholipid nanoparticles; apoA-I, apolipoprotein A-I; SEC, size exclusion chromatography.
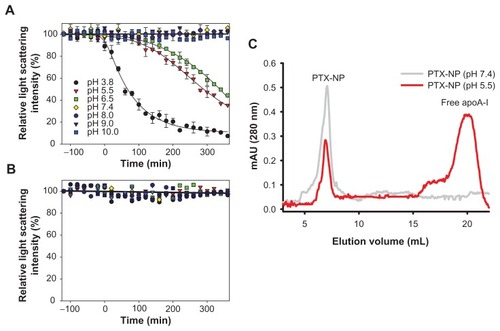
To exclude the possibility that the lowered light scattering intensity of the paclitaxel nanoparticles at low pH was due to aggregation of nanoparticles rather than release of paclitaxel, the solution was analyzed by size exclusion chromatography (). If the apolipoprotein A-I shell was uncoated from the paclitaxel nanoparticles resulting in release of paclitaxel, free apolipoprotein A-I should be present in the solution. Conversely, if paclitaxel nanoparticles just aggregated, there should be no soluble apolipoprotein A-I in the solution. After the paclitaxel nanoparticles had been incubated for 5 hours at pH 7.4 or 5.5, the samples were analyzed by size exclusion chromatography. Indeed, a free apolipoprotein A-I peak appeared after incubation at low pH, while a decrease in the paclitaxel nanoparticle peak was observed. In contrast, the nanostructure of the paclitaxel nanoparticles incubated at physiological pH was stable. This result clearly demonstrates that at low pH, the paclitaxel nanoparticle uncoats its apolipoprotein A-I shell and paclitaxel is selectively released at low pH, which was not observed for the PTX-PL-NP.
The paclitaxel nanoparticle was stable at pH 7–10. In this pH range, paclitaxel nanoparticles did not show significant light scattering intensity changes at 25°C over 5 days. In addition, the paclitaxel nanoparticles were stable over a wide range of temperatures (4°C, 25°C, and 37°C, ), whereas liposomes aggregated rapidly, even at 4°C, which is in agreement with the findings of a previous study.Citation19 These results clearly indicate that the phospholipid-deficient paclitaxel nanoparticles underwent pH-dependent disassembly only in an acidic pH range, suggesting that they are suitable drug carriers for cancer chemotherapy.
Figure 6 Thermostability of PTX-NP.
Notes: Samples were incubated for 120 hours at 4°C, 25°C, and 37°C. As a control, large unilamellar vesicles were incubated for 120 hours at 4°C (open triangle). Light scattering intensity was measured after removing large aggregates by centrifugation (15,000 rpm for 30 minutes).
Abbreviation: PTX-NP, paclitaxel nanoparticles.

Secondary structure of apolipoprotein A-I in nanoparticles
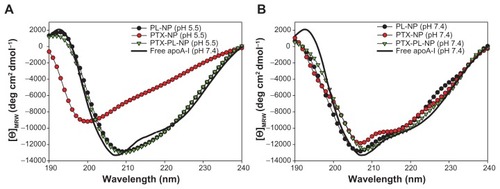
The exclusion of phospholipids from the paclitaxel nanoparticles resulted in pH-responsive release of paclitaxel at an acidic pH range. In order to verify whether this sensitivity at acidic pH was attributable to the structural instability of apolipoprotein A-I due to the absence of phospholipids in the paclitaxel nanoparticles, the secondary structures of apolipoprotein A-I in the nanoparticles were analyzed using far-ultraviolet circular dichroism. The phospholipid nanoparticle consisting of only apolipoprotein A-I and phospholipid, the so-called Nanodisc, was used as a control in this study. The circular dichroism spectrum of apolipoprotein A-I in the phospholipid nanoparticle showed a broad minimum at around 208 nm after subtraction of the phospholipid circular dichroism spectrum (). At pH 7.4, all circular dichroism spectra of apolipoprotein A-I contained in the paclitaxel nanoparticles, PTX-PL-NP, and phospholipid nanoparticles were almost the same as those of free apolipoprotein A-I, which show double minima at 208 nm and 222 nm typical of helical proteins (). In contrast, the spectrum of apolipoprotein A-I in the paclitaxel nanoparticles exhibited a pronounced minimum at around 200 nm at pH 5.5, which indicates a high content of random coils in the protein (). The spectra of apolipoprotein A-I contained in PTX-PL-NP and phospholipid nanoparticles at pH 5.5 were almost the same as those observed at pH 7.4, clearly indicating that phospholipid stabilizes the helical structure of apolipoprotein A-I. This finding suggests that the significant conformational change of apolipoprotein A-I in the paclitaxel nanoparticles was due to the absence of phospholipids, which in turn led to the structural sensitivity of the paclitaxel nanoparticle prone to uncoating of the apolipoprotein A-I shell at acidic pH.
Figure 7 CD analysis of apoA-I in nanoparticles at pH 5.5 (A) and pH 7.4 (B).
Notes: The PL-NP comprising apoA-I and phospholipids was used as a control. Each CD spectrum represents the signal of apoA-I protein, for which the spectrum of the background paclitaxel and/or phospholipids was subtracted from the spectrum of the cognate nanoparticles. The mean residue ellipticity for apoA-I is plotted versus the wavelength.
Abbreviations: CD, circular dichroism; PL-NP, phospholipid nanoparticle; PTX-NP, paclitaxel nanoparticle; PTX-PL-NP, paclitaxel-phospholipid nanoparticles; apoA-I, apolipoprotein A-I.
SR-BI-dependent cytotoxicity of paclitaxel NP
The HER-2 (or ErbB2, neu) gene encodes a 185 kDa transmembrane glycoprotein, and is overexpressed in about 30% of human breast carcinomas.Citation28 HER-2 overexpression in breast cancer cells also results in increased resistance to antitumor drugs, such as paclitaxel.Citation29–Citation31 Furthermore, it is known that apolipoprotein A-I in HDL is recognized by SR-BI, which is expressed by most cancer cells. Based on HER-2-mediated paclitaxel resistance, we tested whether the antitumoral efficacy of paclitaxel could be enhanced when encapsulated into nanoparticles.
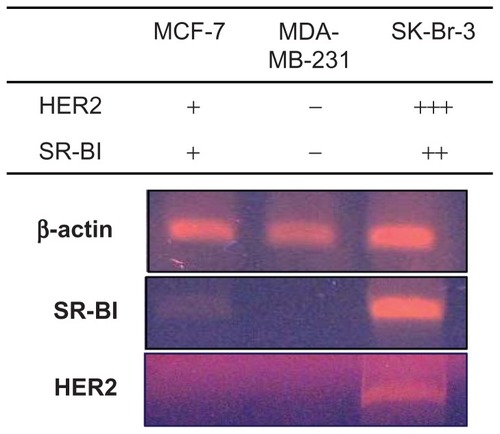
Expression of HER2 and SR-BI in breast cancer cell lines was measured (). Reverse transcription polymerase chain reaction analysis revealed strong expression of HER2 in SK-BR-3 cells, which are known to be HER2-overexpressing cells,Citation32 while MCF-7 breast cancer cells exhibited very low HER2 mRNA levels.Citation33 MDA-MB-231 cells have also been reported to be HER-2 negative.Citation32,Citation34 Expression of SR-BI in MCF-7 cells was mod est, while SR-BI expression in SK-Br-3 cells was high. In contrast, MDA-MB-231 cells did not express SR-BI. Expression of SR-BI and HER2 was found to be mutually correlated in the three cell lines tested.
Figure 8 Reverse transcription polymerase chain reaction analysis of SR-BI and HER2 expression in MCF-7, MDA-MB-231, and SK-Br-3 cells.
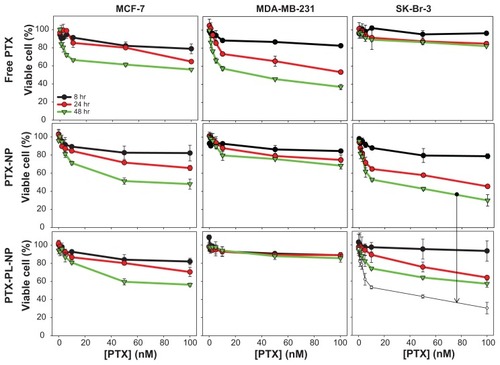
When cells were treated with free paclitaxel, the HER2-overexpressing SK-Br-3 cells were shown to be resistant to paclitaxel, while HER-2 negative control MDA-MD-231 cells were not (), which is consistent with the findings of a previous report.Citation34 The effect of the paclitaxel nanoparticles and PTX-PL-NP on cell death was quite different from the effect of free paclitaxel. The paclitaxel nanoparticles and PTX-PL-NP did not induce cell death in MDA-MD-231 cells, which express neither SR-BI nor HER2. However, MCF-7 and SK-Br-3 cells could be killed well by both nanoparticles. This result suggests that the antitumor activity of paclitaxel nanoparticles and PTXPL-NP is strongly dependent on the presence of the SR-BI receptor. Interestingly, the anticancer effect of the paclitaxel nanoparticles was much stronger than that of PTX-PL-NP when the cancer cells overexpressed HER2 or SR-BI, as in the case of SK-Br-3 cells. We believe this enhanced anticancer effect of paclitaxel nanoparticles was derived from the pH-responsive release of paclitaxel after endocytosis.
Figure 9 Anticancer effect of free PTX, PTX-NP, and PTX-PL-NP. An MTT assay was performed after 8, 24, and 48 hours of treatment with each type of nanoparticle to assess cell viability.
Notes: Legends in upper left panel apply to all panels. The green trace of the middle right panel is also shown again in the lower right panel as a white circle to aid comparison, and is designated by an arrow.
Abbreviations: PTX-NP, paclitaxel nanoparticles; PTX-PL-NP, paclitaxel-phospholipid nanoparticles.
Discussion
As described above, the interstitial microenvironment surrounding the solid tumor is acidic. Thus, a pH-responsive drug carrier may selectively release an anticancer drug around cancer cells while maintaining a low drug concentration near normal cells. Apart from the pH of the tumor environment, the pH-responsiveness of a drug carrier is more important when the nanoparticles containing a drug are endocytosed into the cell. Whether drugs are delivered by cationic lipids, nanoparticles (as in the case of HDL-like nanoparticles), or cell type-specific delivery agents, the intracellular trafficking of drugs begins in early endosomal vesicles. Early endosomes fuse with sorting endosomes, which subsequently transfer their contents to late endosomes. Late endosomal vesicles are acidified (pH 5–6) by membrane-bound proton pump ATPases. Finally, these late endosomes fuse with lysosomes, which are further acidified (pH approximately 4.5). Drugs, whether small chemical compounds like paclitaxel, siRNA, or an antibody, are broken down in the lysosome by various digestive enzymes. To avoid lysosomal degradation, drugs must escape from the endosome into the cytosol, where they can act on their targets. As such, endosomal escape has been a major barrier for efficient drug delivery.Citation35 It is clear that the pH-responsiveness of a drug carrier is a crucial factor for endosomal escape of the drug because of the low pH of endosomes.
Despite the fact that HDL-based nanoparticles have the desired features of antitumor drug carriers, including small size, biodegradability, high drug content, and ability to target cancer cells, they had never been designed for pH-responsiveness to release their enclosed drugs in acidic pH conditions for cancer chemotherapy. Nearly all HDLlike nanoparticles reported to date contain phospholipids, because phospholipids have been considered an indispensable host for a HDL-like nanoparticle, whereby it binds to apolipoprotein A-I and promotes the α-helical conformation required for stable assembly of HDL.Citation36,39 We also failed to assemble paclitaxel nanoparticles in the absence of phospholipids when using the dialysis or dilution method to remove sodium cholate spontaneously. After trying several different reconstitution procedures, we found that simple dilution following sonication allowed for assembly of soluble paclitaxel nanoparticles with a reasonable yield.
The phospholipid-deficient paclitaxel nanoparticles exhibited obvious pH responsiveness selectively at an acidic pH range. In accordance with our hypothesis, the secondary structure of apolipoprotein A-I in the paclitaxel nanoparticles was less stable than that in PTX-PL-NP or phospholipid nanoparticles, rendering the paclitaxel nanoparticles more sensitive to acidic pH, which was clearly due to the absence of phospholipids. Furthermore, exclusion of phospholipids in the preparation of the paclitaxel nanoparticles dramatically increased their paclitaxel content, because the space occupied by phospholipids in other forms of HDL-like nanoparticles was replaced with paclitaxel. When no phospholipids were used, the paclitaxel content of the paclitaxel nanoparticles (1:100) was 62.2% (w/w). Considering that the lipophilic content in native HDL found in the human liver is approximately 45%, this homogeneous incorporation of paclitaxel into the nanoparticle seems to be close to the maximal paclitaxel content.
Conclusion
We generated a pH-responsive nanoparticle containing paclitaxel by using apolipoprotein A-I, which is abundant in the human body. The paclitaxel nanoparticle was stable at neutral and basic pH. It was also stable at temperatures ranging from 4°C to 37°C. The nanoparticle released paclitaxel by selectively uncoating the apolipoprotein A-I shell at low pH. All of these features are required for nanoparticles to be useful in cancer chemotherapy. The pH-dependent release of paclitaxel from the paclitaxel nanoparticles at acidic pH resulted in enhanced antitumor efficacy. The paclitaxel nanoparticles were more potent in killing HER2-overexpressing SK-Br-3 cells, which are known to be resistant to free paclitaxel, than the PTX-PL-NP which is not pH-dependent. The ability of cancer cells to recognize the apolipoprotein A-I of HDL through the SR-BI receptor facilitated uptake of the paclitaxel nanoparticles into the cancer cell via endocytosis. We suggest that the low pH in the endosomes facilitated escape of paclitaxel from the endosome to the cytosol, thereby protecting paclitaxel from degradation in the lysosome, and leading to enhanced anticancer efficacy.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (2011-0006268 and 2011-0004054).
Disclosure
The authors report no conflicts of interest in this work.
References
- HelmlingerGYuanFDellianMJainRKInterstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlationNat Med199731771829018236
- Wike-HooleyJLVan der ZeeJvan RhoonGCVan den BergAPReinholdHSHuman tumour pH changes following hyperthermia and radiation therapyEur J Cancer Clin Oncol1984206196236539698
- ShenYTangHRadoszMVan KirkEMurdochWJpH-responsive nanoparticles for cancer drug deliveryMethods Mol Biol200843718321618369970
- GantaSDevalapallyHShahiwalaAAmijiMA review of stimuli-responsive nanocarriers for drug and gene deliveryJ Control Release200812618720418261822
- OnacaOEneaRHughesDWMeierWStimuli-responsive polymersomes as nanocarriers for drug and gene deliveryMacromol Biosci2009912913919107717
- ConnorJYatvinMBHuangLpH-sensitive liposomes: acid-induced liposome fusionProc Natl Acad Sci U S A198481171517186584905
- BaeYNishiyamaNFukushimaSKoyamaHYasuhiroMKataokaKPreparation and biological characterization of polymeric micelle drug carriers with intracellular pH-triggered drug release property: tumor permeability, controlled subcellular drug distribution, and enhanced in vivo antitumor efficacyBioconjug Chem20051612213015656583
- OhKTKimDYouHHAhnYSLeeESpH-sensitive properties of surface charge-switched multifunctional polymeric micelleInt J Pharm200937613414019394414
- BaeYFukushimaSHaradaAKataokaKDesign of environment-sensitive supramolecular assemblies for intracellular drug delivery: polymeric micelles that are responsive to intracellular pH changeAngew Chem Int Ed Engl2003424640464314533151
- NgKKLovellJFZhengGLipoprotein-inspired nanoparticles for cancer theranosticsAcc Chem Res2011441105111321557543
- RyanRONanobiotechnology applications of reconstituted high density lipoproteinJ Nanobiotechnology201082821122135
- MooberryLKNairMParanjapeSMcConathyWJLackoAGReceptor mediated uptake of paclitaxel from a synthetic high density lipoprotein nanocarrierJ Drug Target201018535819637935
- CaoWNgKKCorbinISynthesis and evaluation of a stable bacteriochlorophyll analog and its incorporation into high-density lipoprotein nanoparticles for tumor imagingBioconjug Chem2009202023203119839633
- PussinenPJKartenBWinterspergerAThe human breast carcinoma cell line HBL-100 acquires exogenous cholesterol from high-density lipoprotein via CLA-1 (CD-36 and LIMPII analogous 1)-mediated selective cholesteryl ester uptakeBiochem J200034955956610880355
- SinglaAKGargAAggarwalDPaclitaxel and its formulationsInt J Pharm200223517919211879753
- DavidsonWSHazlettTMantulinWWJonasAThe role of apolipoprotein AI domains in lipid bindingProc Natl Acad Sci U S A19969313605136108942981
- JayaramanSBenjwalSGantzDLGurskyOEffects of cholesterol on thermal stability of discoidal high density lipoproteinsJ Lipid Res20105132433319700415
- RyanROForteTMOdaMNOptimized bacterial expression of human apolipoprotein A-IProtein Expr Purif2003279810312509990
- ShinJYShinJIKimJSAssembly of Coenzyme Q10 nanostructure resembling nascent discoidal high density lipoprotein particleBiochem Biophys Res Commun200938821722119646960
- LackoAGNairMParanjapeSJohnsoSMcConathyWJHigh density lipoprotein complexes as delivery vehicles for anticancer drugsAnticancer Res2002222045204912174882
- McConathyWJNairMPParanjapeSMooberryLLackoAGEvaluation of synthetic/reconstituted high-density lipoproteins as delivery vehicles for paclitaxelAnticancer Drugs20081918318818176115
- GhoshMSinghATXuWSulchekTGordonLIRyanROCurcumin nanodisks: formulation and characterizationNanomedicine2011716216720817125
- RedmondKANguyenTSRyanROAll-trans-retinoic acid nanodisksInt J Pharm200733924625017412536
- TuftelandMRenGRyanRONanodisks derived from amphotericin B lipid complexJ Pharm Sci2008974425443218271034
- EwertKKAhmadABouxseinNFEvansHMSafinyaCRNon-viral gene delivery with cationic liposome-DNA complexesMethods Mol Biol200843315917518679623
- HuangXCaddellRYuBUltrasound-enhanced microfluidic synthesis of liposomesAnticancer Res20103046346620332455
- GaoWChanJMFarokhzadOCpH-Responsive nanoparticles for drug deliveryMol Pharm201071913192020836539
- SlamonDJClarkGMWongSGLevinWJUllrichAMcGuireWLHuman breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogeneScience19872351771823798106
- HolmesFAWaltersRSTheriaultRLPhase II trial of Taxol, an active drug in the treatment of metastatic breast cancerJ Natl Cancer Inst199183179718051683908
- WahlAFDonaldsonKLFairchildCLoss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosisNat Med1996272798564846
- YuDJingTLiuBOverexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinaseMol Cell199825815919844631
- OrjaloAJohanssonHERuthJLStellaris[trade] fluorescence in situ hybridization (FISH) probes: a powerful tool for mRNA detectionNat Meth8
- LattrichCJuhasz-BoessIOrtmannOTreeckODetection of an elevated HER2 expression in MCF-7 breast cancer cells overexpressing estrogen receptor beta1Oncol Rep20081981181718288420
- TunaMChavez-ReyesATariAMHER2/neu increases the expression of Wilms’ tumor 1 (WT1) protein to stimulate S-phase proliferation and inhibit apoptosis in breast cancer cellsOncogene2005241648165215674342
- DominskaMDykxhoornDMBreaking down the barriers: siRNA delivery and endosome escapeJ Cell Sci20101231183118920356929
- SparksDLLund-KatzSPhillipsMCThe charge and structural stability of apolipoprotein A-I in discoidal and spherical recombinant high density lipoprotein particlesJ Biol Chem199226725839258471464598
- VarkouhiAKScholteMStormGHaismaHJEndosomal escape pathways for delivery of biologicalsJ Control Release201115122022821078351