?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
This study aims to develop a three-dimensional in vitro culture system to genetically engineer mesenchymal stem cells (MSC) to express bone morphogenic protein-2. We employed nanofabrication technologies borrowed from the spinning industry, such as electrospinning, to mass-produce identical building blocks in a variety of shapes and sizes to fabricate electrospun nanofiber sheets comprised of composites of poly (glycolic acid) and collagen. Homogenous nanoparticles of cationic biodegradable natural polymer were formed by simple mixing of an aqueous solution of plasmid DNA encoded bone morphogenic protein-2 with the same volume of cationic polysaccharide, dextran-spermine. Rat bone marrow MSC were cultured on electrospun nanofiber sheets comprised of composites of poly (glycolic acid) and collagen prior to the incorporation of the nanoparticles into the nanofiber sheets. Bone morphogenic protein-2 was significantly detected in MSC cultured on nanofiber sheets incorporated with nanoparticles after 2 days compared with MSC cultured on nanofiber sheets incorporated with naked plasmid DNA. We conclude that the incorporation of nanoparticles into nanofiber sheets is a very promising strategy to genetically engineer MSC and can be used for further applications in regenerative medicine therapy.
Introduction
Scaffolding materials for cell proliferation and differentiation is one of the key technologies for tissue engineering. Porous materials with various three-dimensional (3D) structures have been investigated for the cell scaffold because they have a larger surface area for cell attachment and proliferation than conventional two-dimensional (2D) tissue culture plates and are preferable to assist the formation of 3D cell constructs which may resemble the structure and function of natural tissues. In addition, the 3D scaffold also plays an important role as a suitable substrate for in vitro cell culture to increase the number of cells as high as clinically applicable. Combinational technology of stem cells with 3D in vitro systems is very promising for the achievement of successful tissue regeneration. One of the future approaches with mesenchymal stem cells (MSC) is to genetically engineer the transplanted cells directly in the body. Engineered cells can positively participate in the process of tissue repairing. Several researchers have investigated the potential application of genetically engineered MSC for tissue regeneration.Citation1–Citation3 Gene modification is preferred over the addition of growth factor to the cell as, typically, (1) the half-life of the selected growth factor is short; (2) a single administration is usually not sufficient for a biological effect; (3) the quantities required are prohibitively expensive; and (4) continuous protein production increases the likelihood that a desired outcome will be achieved. In planning gene therapy strategies for tissue engineering two different strategies could be considered: (1) direct gene delivery using viral or nonviral vectors and (2) in vitro cell-mediated gene therapy. In both cases the aim is to deliver a therapeutic gene of a growth factor or cytokine into the target tissue. Better understanding of molecular mechanisms involved in the differentiation of MSC will eventually allow us to properly manipulate MSC both in vivo and in vitro for the regeneration of tissues and organs. Retroviral or adenoviral vectors carrying human bone morphogenic protein (BMP)-2, -4, or -7 have been widely reported in genetically engineering MSC.Citation4–Citation6 However, transfection of MSC by nonviral vectors is considered to be safer although the transfection level is very low compared with viral vectors. We have previously reported on the use of nonviral gene vectors in combination with the principles of tissue engineering to enhance in vitro gene transfection of MSC and in vivo bone regeneration.Citation7–Citation10 However, the previous research was not suitable and easy to handle due to the complicated processes of both the in vitro and in vivo experiments. In this paper, we demonstrate a modified version of our technology with enhanced process feasibility since both nonviral gene vectors and tissue-engineered scaffolds were fabricated through a simple process. We have reported that the dextran-spermine based polycations have been found to be highly effective as nonviral vectors in transfecting many different cells in vitro.Citation11–Citation17 Our recent study also demonstrated that electrospun poly (glycolic acid) (PGA)/collagen nanofibers significantly enhanced cell adhesion compared with PGA/collagen microfibers.Citation18 Based on these results, the present study aims to fabricate a tissue-like 3D transgene cell construct for use as a therapeutic cell-based gene delivery system or as an in vitro model system for testing genetic manipulations to understand the effects of gene expression on tissue development.
Materials and methods
Materials
PGA was purchased from Polysciences Inc (Warrington, PA). Collagen was purchased from Nippon Meat Packers Australia (Sydney, Australia). BMP-2 ELISA Kit was purchased from AN’ALYZA, Human BMP-2 was purchased from Immunoassay System (R&D Systems, Minneapolis, MN). The Alkaline Phosphatase Assay Kit (lot no TJ791) was purchased from Wako Pure Chemical Industries (Osaka, Japan). A polyhedrin promoter of baculovirus (pBacBH2; 1230 bp) was used in this study. The pBacBH2 is the expression vector consisting of coding sequence of hBMP-2 inserted at the downstream into the pBacBH2. The plasmid DNA was amplified in a transformant of Escherichia coli bacteria and isolated from the bacteria by Qiagen Maxi kit-25 (Qiagen, Hilden, Germany). Rhodamine B isothiocyanate (RITC) was obtained from Sigma-Aldrich (St Louis, MO).
Fabrication and characterization of nanoparticles
The synthesis and chemical characterization of dextran-spermine were detailed in our previous publication.Citation17 Incorporation of plasmid DNA into dextran-spermine was performed by simply mixing two materials at various charge ratios (N/P, indicated as the molar ratio of the spermine nitrogen of dextran-spermine to the phosphate molecules of plasmid DNA) in aqueous solution. Briefly, 150 μL of 0.1 M phosphate-buffered saline solution (PBS, pH 7.4) containing 2.5, 5, 10, 25, and 100 μg of dextran-spermine was added to the same volume of PBS containing 10 μg of plasmid DNA. The solution was gently agitated at 37°C for 30 minutes to form dextran-spermine-plasmid DNA nanoparticles. Formation of dextran-spermine-plasmid DNA nanoparticles was confirmed by transmission electron microscopy (TEM). TEM analysis of nanoparticles was performed on a Leo 912 AB Energy Filter TEM machine model (Leo Filter Press Co, Hangzhou, China). Briefly, a drop specimen of nanoparticles was immobilized onto a copper grid for 3–5 minutes, the grid was cleaned using a filter paper, and then stained using 1% polytungsten acid for 3–5 minutes. The particle size and surface charge of DNA nanoparticles were analyzed by using dynamic light scattering (DLS) and electrophoretic light scattering (ELS). The nanoparticles of dextran-spermine and plasmid DNA were prepared by a similar procedure to that described above. All solutions were filtered through a 0.45 μm filter (Millex-HV, Millipore, Billerica, MA) prior to mixing. The mixed plasmid DNA and dextran-spermine solution was placed in a DLS cell and DLS measurement was carried out using a DLS-DPA-60HD instrument (Otsuka Electronic Co, Ltd, Osaka, Japan) equipped with an Ar+ laser at a detection angle of 90° at 37°C for 30 minutes. ELS measurement was carried out on an ELS-7000AS instrument (Otsuka Electronic) at 37°C and electric field strength of 100 V/cm. Light scattering measurement was done three times for every sample.
Fabrication of nanofiber sheets
Solutions for electrospinning were prepared by mixing the PGA and collagen solutions. The PGA solution was prepared by dissolving 67 mg of PGA in 100 mL of 1,1,1,3.3.3 hexafluroro-2-2 propanol under stirred conditions at 80°C for 48 hours. The collagen solution was prepared by dissolving 100 mg collagen in 100 mL of 1,1,1,3,3,3 hexafluroro-2-2 propanol under stirred condition at 4°C for 12 hours. The mixture of PGA and collagen was prepared by mixing two solutions at volume mixing ratios of PGA to collagen of 1/9, 2.5/7.5, 5/5, 7.5/2.5, and 9/1. The mixture solution was then delivered by a programmable pump to the exit hole of the electrode (spinneret with a hole diameter of 0.7 mm). The grounded platform for collecting the nanofibers was a silicon sheet (). The flow rate was set to 10 mL/hour. A high-voltage power supply was used to provide the necessary voltage for the fabrication of fibers. The voltage and distance from spinneret and collector were set at 26 kV and 23 cm, respectively. Our previous results demonstrated that fabricated nanofibers of PGA/collagen at a volume mixing ratio of 5/5 (PGA/collagen weight mixing ratio of 0.4) showed higher cell attachment compared with lower or higher amounts of PGA.Citation18 Based on these results, nanofibers were fabricated at a PGA/collagen volume mixing ratio of 5/5. The electrospinning was continuously operated until a white thick layer of nanofiber sheets (approximately 5 mm) was fabricated. The PGA/collagen nanofiber sheet was removed from the silicon sheet and processed for scanning electron microscopy. The porosity was measured by the liquid substitution method as described in our previous publication.Citation19 Isopropanol with density ρi was used as the displacement liquid at 4°C. A density bottle filled with isopropanol was weighed (W1). A sponge sample of weight (Ws) was immersed into the density bottle and weighed (W2). The sponge saturated with isopropanol was taken out and then the density bottle was weighed (W3). The following parameters of a scaffold including the volume of the sponge pore (Vp), the density of the sponge skeleton (Vs), and the porosity (ɛ) were calculated:Citation20,Citation21
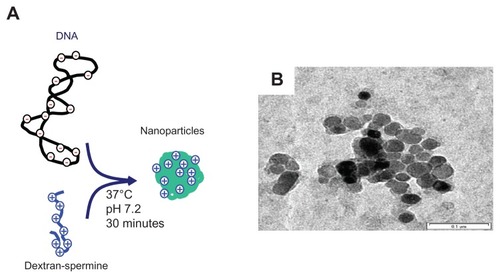
Figure 1 (A) Schematic illustration of nanoparticle formation, and (B) transmission electron microscopy image of dextran-spermine-plasmid DNA nanoparticles.
Note: The concentration of plasmid DNA is 10 μg/mL.
Isolation and culture of MSC
Rats were narcotized by ketamine 10% (40–90 mg/kg, Alfasan, Woerden, Holland) and xylazine 2% (5 mg/kg, Alfasan) and then sacrificed by cervical dislocation. Under sterile conditions, both femurs from each rat were excised. Muscles and the entire connective tissue were detached, and the epiphyses were removed. Femoral bone marrow tissue was extracted from the bones by flushing the medullar cavities of the bones with washing solution using an 18-gauge needle. The washing solution contained high-glucose Dulbecco’s Modified Eagle Medium (Gibco, Gaithersburg, MD), supplemented with 15% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin (source of all components: Gibco). Marrow plug suspension was dispersed by passing through two different needles (21- and 23-gauge), respectively, and centrifuged at 500× g for 5 minutes. The supernatant containing thrombocytes was discarded, and the cell pellet was treated for red blood cell lysing procedures using hypotonic shock. Then the cells were suspended in a growth medium containing high-glucose Dulbecco’s Modified Eagle Medium supplemented with 15% fetal bovine serum, 100 U/mL penicillin, and 100 U/mL streptomycin. Cells in the growth medium were placed on 25-cm2 plastic flasks and incubated at 37°C in a 5% CO2/95% air atmosphere. Non-adherent cells were removed after 4 days by medium exchange and adherent cells were allowed to grow for up to 20 days, with media replacement every 3 days. After 10 days in culture, isolated colonies of MSC became apparent. When the cell cultures reached 80% confluence they were harvested by trypsinization with an aqueous solution of 0.05 wt% trypsin (Gibco) and 1 mM ethylenediaminetetraacetic acids (Merck, Darmstadt, Germany) in 0.1 M PBS (pH 7.4) for 3 minutes at 37°C and reseeded in new flasks at a density of 10–15 × 103 cells/cm2.
Cell seeding into 3D collagen-PGA nanofiber sheets and proliferation assay
MSC were seeded into nanofiber sheets according to an agitated seeding method as this method was effective in seeding cells homogenously into porous 3D cell scaffolds.Citation22 Briefly, 0.5 mL of cell suspension (1 × 106 cells/mL) and the nanofiber sheets were placed in 12-mL tubes on an orbital shaker and agitated at 37°C at 300 rpm for 6 hours. The MSC-seeded nanofiber sheets were thoroughly washed with PBS to exclude non-adherent cells. A static method was used to culture cells for their proliferation. For the static method, the cell-seeded nanofiber sheets were placed into each well of 12-well tissue culture plates. Each nanofiber sheet was incubated in the normal medium for 1–3 weeks at 37°C in a 95% air/5% CO2 atmosphere. To compare the 3D culture with the 2D culture method, we cultured MSC in 12-well tissue culture plates.
The number of cells attached to the nanofiber sheets or tissue culture plate was determined by the fluorometric quantification of cell DNA according to our previous publication.Citation23 Briefly, the cell-seeded nanofiber sheet was washed with PBS and stored at −30°C until assay. After thawing, the cells present in the nanofiber sheet were lysed in a buffer solution (pH 7.4) containing 0.5 mg/mL proteinase K, 0.2 mg/mL sodium dodecylsulfate, and 30 mM saline–sodium citrate at 55°C for 12 hours with occasional mixing. The cell lysate (100 μL) was mixed with saline–sodium citrate buffer (400 μL) in a glass tube. After mixing with a dye solution (500 μL; composition: 30 mM saline–sodium citrate, 1 μg/mL Hoechst 33258 dye), the fluorescent intensity of the mixed solution was measured on a fluorescence spectrometer at Ex 355 nm/Em 460 nm. The calibration curve between the DNA and cell number was prepared by the use of cell suspensions with different cell densities. The DNA assay was performed three times independently for every experimental sample unless mentioned otherwise. The same procedure was applied for MSC cultured in 2D tissue culture plates.
In vitro release test of plasmid DNA
The plasmid DNA-BMP-2 and RITC were mixed in 0.2 M sodium carbonate-buffered solution (pH 9.7) at 4°C for 12 hours, both at concentrations of 1 mg/mL in order to prepare fluorescent-labeled plasmid DNA. The RITC-labeled DNA was separated from the residual RITC by gel filtration of a PD 10 column (Amersham Pharmacia Biotech, Tokyo, Japan), followed by ethanol precipitation to obtain the RITC-labeled plasmid DNA. According to the procedure mentioned above, the cationized dextran was mixed with the RITC-labeled plasmid DNA in PBS at the N/P ratio of 5 to prepare the DNA nanoparticles. The cationized dextran-RITC-labeled plasmid DNA solution in PBS (0.4 mL) was impregnated into the dried scaffolds of the collagen-PGA nanofiber sheets by simple solution dropping, after which they were left for 1 hour at 25°C on a clean bench. The scaffold impregnated with the cationized dextran-RITC-labeled plasmid DNA was placed in 30 mL of the culture medium with gentle stirring at 50 rpm for 1–7 days under the same conditions as the cell culture. As control, RITC-labeled plasmid DNA alone was impregnated into the dried scaffolds of a collagen-PGA nanofiber sheet. At predetermined time intervals, 1 mL of culture medium was sampled and the same volume of medium was added. The fluorescent intensity of the sample solution was measured by a fluorescent spectrophotometer (F-2000 Fluorescent Spectrophotometer, Hitachi, Tokyo, Japan, Ex 570 nm/Em 595 nm) and divided by that initially added to obtain the percentage of plasmid DNA released. Each experiment was carried out independently for six samples.
In vitro experiment to generate genetically engineered MSC
Prior to cell seeding on nanofiber sheets, 0.4 mL of nanoparticle (dextran-spermine-plasmid DNA) solution in PBS was incorporated into the dried nanofiber sheets by simple solution dropping, and then left overnight at 25°C in a clean bench. MSC were seeded into nanofiber sheets as described above. The nanofiber sheets were placed in each well of 24-well culture plates, followed by incubation for 2 days at 37°C in a 95% air/5% CO2 atmosphere. As controls, 0.4 mL of plasmid DNA in PBS solution and culture medium containing MSC (1 × 106 cells/mL) were similarly incorporated into the nanofiber sheets. To measure the level of gene transfection of MSC cultured on nanofiber sheets, the nanofiber sheets collected were washed three times with PBS, cut with scissors, and homogenized in the lysis buffer (0.1 M Tris-HCl, 2 mM ethylenediaminetetraacetic acid, 0.1% Triton X™-100). The sample lysate (2 mL) was centrifuged at 12,000 rpm for 5 minutes at 4°C, and the supernatant was carefully collected and kept in ice. To measure the expression levels of BMP-2 gene, 50 μL of the supernatant was collected and the BMP-2 protein was determined by a human BMP-2 ELISA Kit. Each experiment was carried out three times independently.
Formation of genetically engineered MSC
Two important biological tests were applied to confirm the formation of genetically engineered MSC. The first factor was to check any significant changes in the intracellular alkaline phosphatase (ALP) activity and the second factor was to check bone osteocalcin content (OCN). To check the amount of ALP, the nanofiber sheets collected were washed three times with PBS, cut with scissors, and homogenized in the lysis buffer (0.2% Igepal CA-630, 10 mM Tris-HCL, 1 mM MgCl2, pH 7.5). The sample lysate (2 mL) was centrifuged at 12,000 rpm for 10 minutes at 4°C. The supernatant was assayed for ALP activity, using p-nitrophenyl-phosphate as substrate. To determine the OCN of the cells, the nanofiber sheets collected were washed three times with PBS and then treated with 1 mL of 40 vol% formic acid for 10 days at 4 °C under vortex mixing to decalcify. After the decalcification process, the cell extraction was applied to a SephadexTM G-25 PD-10 column (GE Healthcare Life Sciences, Pewaukee, WI) for gel filtration. The resulting solution was freeze-dried and subjected to an osteocalcin rat enzyme-linked immunosorbent assay.
Statistical analysis
All the data were statistically analyzed to express the mean ± the standard deviation of the mean. Student’s t test was performed and P < 0.05 was accepted to be significant.
Results
Preparation and characterization of DNA nanoparticles and nanofibers
summarizes the DLS and ELS measurement of DNA nanoparticles composed of plasmid DNA and dextran-spermine. DLS measurement revealed that DNA nanoparticles with nanodiameter size were formed when plasmid DNA and cationized dextran were mixed at charge ratio of 5 or higher; their surface charge became positive and the charge tended to increase as the N/P ratio increased. shows the TEM images of the dextran-spermine-plasmid DNA nanoparticles. Morphological characteristics of the collagen-PGA nanofiber sheets revealed that the porosity (%) of fabricated nanofibers was around 79.61 ± 1.2, while the pore size (μm) was 8.0 ± 2.0.
Table 1 Characterization of plasmid DNA, dextran-spermine, and DNA nanoparticles
In vitro release of plasmid DNA
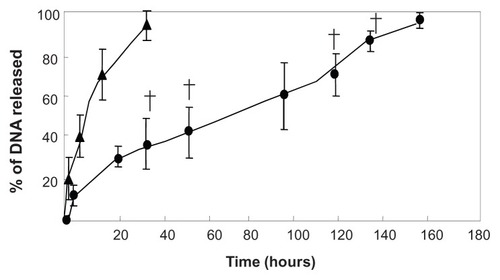
shows the in vitro profiles of plasmid DNA release from collagen-PGA nanofiber sheets in PBS at 37°C. Our results revealed that plasmid DNA was released from nanofiber sheets impregnated with the DNA nanoparticles for longer time periods than those impregnated with the plasmid DNA alone.
Figure 2 In vitro release profiles of plasmid DNA-BMP-2 from DNA nanoparticles incorporated into nanofibers (●). Naked plasmid DNA-BMP-2 incorporated into nanofibers (▴) was used as the control group.
Note: †P < 0.05; significant against the amount of plasmid DNA released from naked plasmid DNA incorporated into nanofibers.
Abbreviation: BMP-2, bone morphogenic protein-2.
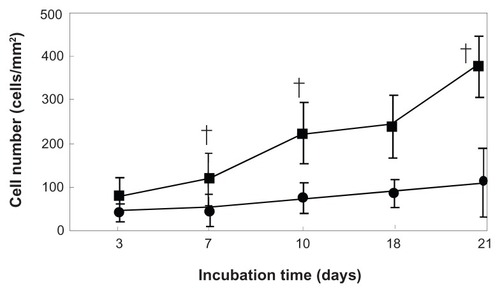
Cell attachment and proliferation in 3D collagen-PGA nanofiber sheets
Our observation confirmed significant differences in the cell numbers from the 3D collagen-PGA nanofiber sheets and those from the 2D static tissue culture plate (). The proliferation of MSC cultured in the 3D collagen-PGA nanofiber sheets showed that the cell number tended to increase significantly with incubation time compared with those cultured by the 2D culture method. Irrespective of the culture method, proliferation profiles tended to increase with incubation time. Among the two methods, the initial cell proliferation was highest for the 3D collagen-PGA nanofiber sheets.
Physical and cellular characterization of nanoparticle incorporated into nanofibers
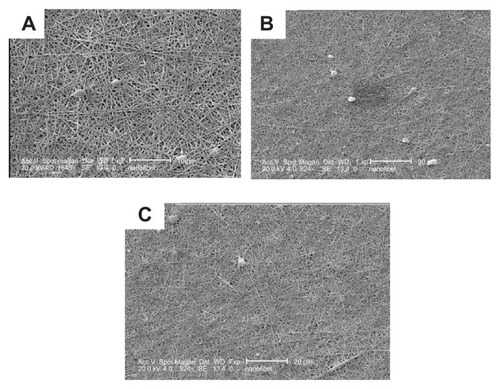
shows a schematic illustration of an electrospinning machine and a scanning electron microscopy image of fabricated PGA/collagen nanofibers. Homogenous nanofibers were fabricated at a PGA/collagen volume mixing ratio of 5/5. shows a scanning electron microscopy image of fabricated PGA/collagen nanofibers 1, 2, and 3 weeks after culturing MSC on the nanofibers already incorporated with nanoparticles, while summarizes the results of the in vitro transfection activity used in this study. Naked plasmid DNA-BMP-2 incorporated into nanofiber sheets did not enhance the level of BMP-2 protein and the BMP-2 protein level was similar to that of the mixture of dextran-plasmid DNA-treated cells. The highest level of BMP-2 was observed for the dose of plasmid DNA, N/P ratio, and the transfection time to be 10 μg/mL, 5 and 2 days after transfection (data are not shown).
Table 2 BMP-2 activity of MSC 2 days after treatment of free plasmid DNA and nanoparticles of dextran-spermine-plasmid DNA
Figure 4 (A) Schematic illustration of electrospinning machine. (B) Cross-sectional scanning electron microscopy photographs of the structural morphology of PGA/collagen nanofibers fabricated by electrospinning.
Abbreviation: PGA, poly (glycolic acid).
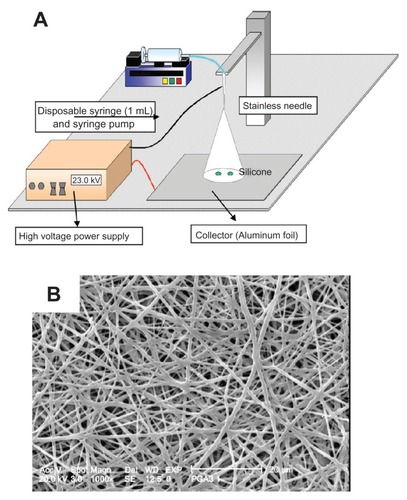
Figure 5 Cross-sectional scanning electron microscopy photographs of the structural morphology of PGA/collagen nanofibers fabricated by electrospinning (A) 1 week, (B) 2 weeks, and (C) 3 weeks after culturing the mesenchymal stem cells on nanofibers incorporated with dextran-spermine-plasmid DNA nanoparticles.
Abbreviation: PGA, poly (glycolic acid).
Biochemical analysis of genetically engineered MSC
indicates the time course of the ALP activity and OCN of MSC cultured on nanofiber sheets that were incorporated with naked plasmid DNA-BMP-2, a mixture of dextran-plasmid DNA, and DNA nanoparticles, 1, 2, and 3 weeks after transfection. The level of ALP activity was significantly enhanced at 1 and 2 weeks after transfection with DNA nanoparticles. The extent of the ALP activity was higher in the nanofiber sheets seeded with transfected MSC. The ALP activity increased with time for the initial 2 weeks, and thereafter leveled off, although the level was still high compared with that of the other groups. Osteocalcin content was continuously increased over the time of study. Nanofiber sheets seeded with genetically engineered MSC exhibited the highest OCN among all the groups.
Table 3 Biochemical analysis of the osteogenic differentiation of genetically engineered mesenchymal stem cells
Discussion
3D in vitro models play an important role in biological evaluations and serve as a template for many biological tests. Natural biodegradable polymers such as collagen have been widely used as 3D models for in vitro and in vivo applications because they have good cell compatibility and processability into various shapes. Synthetic polymers have also been used as 3D models, although cell attachment to the surface of these materials such as PGA is poor compared with natural polymers, but their mechanical properties are appropriated. In the present study, we combined collagen with PGA in order to overcome the poor mechanical properties of collagen on the one hand and the poor cell attachment of PGA on the other, fabricated nanofiber sheets comprised of composites of PGA and collagen through electrospinning technology, and used them as a 3D in vitro model.
It seems that artificial nanoscaled fibers have great potential application in the field of biomaterials and tissue engineering as they are able to mimic the structure and biological function of native extracellular matrices in terms of chemical composition and physical structure as reported by Ma et al.Citation24 Phase separation, electrospinning, and self-assembly have been used in the formation of nanofibers.Citation25,Citation26 Electrospinning is a common and easy technique and it is also extremely cheap and can be used for many different types of polymers. Our recent study demonstrated that fabricated PGA/collagen nanofibers through electrospinning significantly enhanced cell adhesion compared with PGA/collagen microfibers.Citation18
A 3D in vitro model may offer a superior method compared with conventional 2D in vitro gene transfection and can easily be transplanted directly into the defect site. The efficiency of transfection using an expression vector has been shown to be related to cell cycle activity.Citation27 The attachment of cells to the surface of materials is a major factor controlling their structure, function, and cell cycle.Citation28 At first, cells attach to the surface of fibers throughout the polymer scaffold; they then spread and proliferate. Three-dimensional fibrous matrices can provide greater available surface area for cell attachment and spreading than 2D models (ie, tissue culture plate). Moreover, the 3D scaffold surface affects cell adhesion, spreading, and proliferation, and controls the spatial arrangement of cells and their transmission of biochemical and mechanical signals. Cell adhesion, spreading, and cytoskeletal reorganization initiate signaling cascades that govern gene expression.Citation29 It has been demonstrated that the transgene expression of hematopoietic cells was significantly related to the cell adhesion on matrix.Citation30 The higher the cell growth, the better the gene transfection for the cells.Citation29 It has been found that effective transfection of multicellular hepatocyte spheroids required the process of spheroid formation.Citation31 The proposed technique of gene transfer to 3D cell-scaffold constructs is based on the use of a 3D fibrous scaffold to guide cell organization in conjunction with gene manipulation. In comparison with conventional culture, the environment of cells maintained in the 3D culture scaffold is closer to that in vivo, which can affect the gene expression and behavior of cells.Citation32,Citation33
The level of BMP-2 protein was enhanced by mixing plasmid DNA with dextran-spermine nanoparticles and incorporating the nanoparticles into the PGA/collagen nanofiber sheets. It is highly conceivable that the electrostatic interaction between the negatively charged cell membrane and the positively charged nanoparticles is a driving force in attaching nanoparticles to cells, which is an important initial step for gene transfection. When nanoparticles were added into the MSC cultured on tissue culture plates, the level of BMP-2 protein was low (data are not shown). An explanation is that the larger volume of nanoparticles reduced the surface area necessary for the attachment of the nanoparticles to the surface of the cells, resulting in reduced internalization of the nanoparticles into the cells. The sustained release of plasmid DNA from collagen-PGA nanofiber sheets affected the level of gene expression. It may be hypothesized that the controlled release of plasmid DNA by impregnation into nanofiber sheets provides a lower dose of plasmid DNA and a larger surface area both of which enhance the internalization of the DNA nanoparticles into the cells. On the other hand, since plasmid DNA is a large, negatively charged molecule up to 1 μm in length,Citation34 it is impossible to make the plasmid DNA internalize into cells following attachment onto the negatively charged cell membrane. When the plasmid DNA is ionically mixed with a positively charged nonviral vector, it is well recognized that the molecular size decreases by its condensation and form nanoparticles (). It is likely that the condensed DNA-vector nanoparticles which have a positive charge can electrostatically interact with the cell membrane for internalization.
Biochemical analysis of the osteogenic differentiation of MSC on PGA/collagen nanofibers incorporated with nanoparticles indicated significantly increased ALP and OCN levels; both useful markers for osteoblast activity (). ALP activity increased rapidly and saturated at 2 weeks, while the temporal changes in the OCN content increased steadily with time, which corresponded well with the course of the osteogenic differentiation of MSC in the scaffolds. Naked plasmid DNA incorporated into PGA/collagen nanofibers did not increase the level of BMP-2 protein (). Gene therapy is not effective unless the genes administrated come into contact with some cells and generate protein. PGA/collagen nanofiber sheets are a suitable scaffold to enable many cells at the culture site to induce osteogenesis sufficiently by coming into contact with plasmid DNA-BMP-2 and secreting BMP-2 protein. Therefore the administration of plasmid DNA-BMP-2 in combination with dextran-spermine would induce osteogenesis (). It is conceivable that the promoted cell differentiation is due to the enhanced gene expression.
Conclusion
Our results indicated that the level of gene expression in vitro was significantly enhanced by combinational technology of the fabrication of nanoparticles composed of dextran-spermine and plasmid DNA-BMP-2 and incorporation of these nanoparticles into electrospun nanofiber sheets comprised of composites of PGA and collagen. The present findings will provide an attractive combined strategy of tissue engineering principles with gene therapy for tissue regeneration. We chose to use a nanoparticle delivery system based on dextran-spermine to genetically engineer MSC, partly since its nontoxicity, biodegradability, scalability, reproducibility, and high-throughput capability make it potentially beneficial for future clinical trials.
Acknowledgments
This study was funded by the National Science Council of Taiwan through research grant NSC 99-2314-B-010-025-MY3.
Disclosure
The authors report no conflicts of interest in this work.
References
- BlumJSBarryMAMikosAGJansenJAIn vivo evaluation of gene therapy vectors in ex vivo-derived marrow stromal cells for bone regeneration in a rat critical-size calvarial defect modelHum Gene Ther2003141689169514670121
- TurgemanGAslanHGazitZGazitDCell-mediated gene therapy for bone formation and regenerationCurr Opin Mol Ther2002439039712222877
- TsuchidaHHashimotoJCrawfordEManskePLouJEngineered allogeneic mesenchymal stem cells repair femoral segmental defect in ratsJ Orthop Res200321444912507579
- LouJXuFMerkelKManskePGene therapy: adenovirus mediated human bone morphogenetic protein-2 gene transfer induces mesenchymal progenitor cell proliferation and differentiation in vitro and bone formation in vivoJ Orthop Res199917434910073646
- MusgraveDSBoschPGhivizzaniSRobbinsPDEvansCHHuardJAdenovirus-mediated direct gene therapy with bone morphogenetic protein-2 produces boneBone19992454154810375195
- ChengSLLouJWrightNMLaiCFAvioliLVRiewKDIn vitro and in vivo induction of bone formation using a recombinant adenoviral vector carrying the human BMP-2 geneCalcif Tissue Int200168879311310352
- HosseinkhaniHHosseinkhaniMKhademhosseiniAGabrielsonNPPackDWKobayashiHDNA nanoparticles encapsulated in 3-D tissue engineered scaffold enhance osteogenic differentiation of mesenchymal stem cellsJ Biomedical Materials Research Part A2008854760
- HosseinkhaniHYamamotoMInatsuguYEnhanced ectopic bone formation using a combination of plasmid DNA impregnation into 3-D scaffold and bioreactor perfusion cultureBiomaterials2006271387139816139884
- HosseinkhaniHAzzamTKobayashiHCombination of 3-D tissue engineered scaffold and non-viral gene enhance in vitro DNA expression of mesenchymal stem cellsBiomaterials2006274269427816620957
- HosseinkhaniHInatsuguYHiraokaYInoueSShimokawaHTabataYImpregnation of plasmid DNA into three-dimensional scaffolds and medium perfusion enhance in vitro DNA expression of mesenchymal stem cellsTissue Eng2005111459147516259601
- AbediniFIsmailMHosseinkhaniHEffects of CXCR4 siRNAs/dextran-spermine nanoparticles on CXCR4 expression and serum LDH levels in a mouse model of colorectal cancer metastasis to the liverCancer Manag Res2011330130921931504
- AbediniFHosseinkhaniHIsmailMOmarARPei PeiCDombAJIn vitro intracellular trafficking of biodegradable nanoparticles of dextran-spermine in cancer cell linesInt J Nanotechnol20118712723
- AbediniFIsmailMHosseinkhaniHToxicity evaluation of dextran-spermine polycation as a tool for gene therapy in vitroJ Cell Anim Biol20104170176
- AbdullahSYeoYHosseinkhaniHGene transfer into the lung by nanoparticle dextran-spermine/plasmid DNA complexesJ Biomed Biotechnol20102010110
- HosseinkhaniHKushibikiTMatsumotoKNakamuraTTabataYEnhanced suppression of tumor growth using a combination of NK4 plasmid DNA-PEG engrafted cationized dextran complex and ultrasoundCancer Gene Ther20061347948916276347
- HosseinkhaniHTabataYUltrasound enhances in vivo tumor expression of plasmid DNA by PEG-introduced cationized dextranJ Control Release200510854055616219383
- HosseinkhaniHAzzamTTabataYDombAJDextran-spermine polycation: an efficient non-viral vector for in vitro and in vivo gene transfectionGene Ther20041119420314712304
- TianFHosseinkhaniHHosseinkhaniMQuantitative analysis of cell adhesion on aligned micro- and nanofibersJ Biomed Mater Res A20088429129917607759
- MohageriSHosseinkhaniHEbrahimiNGSolimaniMKajbafzadehAMProliferation and differentiation of mesenchymal stem cell on collagen sponge reinforced with polypropylene/polyethylene terephthalate blend fibersTissue Eng Part A2010163821383020673024
- HosseinkhaniHHosseinkhaniMHattoriSMatsuokaRKawaguchiNMicro and nanoscale in vitro 3D culture system for cardiac stem cellsJ Biomed Mater Res A2010941820014298
- HosseinkhaniHHosseinkhaniMKobayashiHProliferation and differentiation of mesenchymal stem cells by using self-assembled of peptide-amphiphile nanofibersBiomed Mat20061815
- HosseinkhaniHHosseinkhaniMTianFKobayashiKTabataYEnhanced angiogenesis through controlled release of basic fibroblast growth factor from peptide amphiphile for tissue regenerationBiomaterials2006275836584416930687
- HosseinkhaniHHosseinkhaniMTianFKobayashiHTabataYBone regeneration on a collagen sponge-self assembled peptide-amphiphile nanofiber hybrid scaffoldTissue Eng2007131917518575
- MaZKotakiMMaZKotakiMInaiRRamakrishnaSPotential of nanofiber matrix as tissue-engineering scaffoldsTissue Eng20051110111615738665
- SmithLAMaPXNano-fibrous scaffolds for tissue engineeringColloids Surf B: Biointerfaces20041012513115556341
- KameokaJVerbridgeSSLiuHCzaplewskiDACraigheadHGFabrication of suspended silica glass nanofibers from polymeric materials using a scanned electrospinning sourceNano Lett2004421052108
- WilkeMFortunatiEVandenBroekMHoogeveenATScholteBJEfficacy of a peptide-based gene delivery system depends on mitotic activityGene Ther19963113311398986440
- IngberDEPrustyDSunZBetenskyHWangNCell shape, cytoskeletal mechanics, and cell cycle control in angiogenesisJ Biomech199528147114788666587
- XieYYangSTKnissDThree-dimensional cell-scaffold constructs promote efficient gene transfect ion: implications for cell-based gene therapyTissue Eng2001758559311694192
- KellerHYunxuCMaritGTransgene expression, but not gene delivery, is improved by adhesion-assisted lipofection of hematopoietic cellsGene Ther1999693193810505119
- WatanabeYAjiokaIAkaikeTGene transfection of multicellular spheroid of hepatocytes on an artificial substrateCytotechnol1998266573
- Mueller-KlieserWThree-dimensional cell cultures: from molecular mechanisms to clinical applicationsAm J Physiol1997273C110911139357753
- HosseinkhaniHInatsuguYInoueSHiraokaYTabataYPerfusion culture enhances the osteogenic differentiation of rat mesenchymal stem cells in collagen sponge reinforced with poly (glycolic acid) fiberTissue Eng2005111476148816259602
- AbdallahBSachsLDemeneixBANon-viral gene transfer: applications in developmental biology and gene therapyBiol Cell199585178882514