 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Agricultural products and by products provide the primary materials for a variety of technological applications in diverse industrial sectors. Agro-industrial wastes, such as cotton and curaua fibers, are used to prepare nanofibers for use in thermoplastic films, where they are combined with polymeric matrices, and in biomedical applications such as tissue engineering, amongst other applications. The development of products containing nanofibers offers a promising alternative for the use of agricultural products, adding value to the chains of production. However, the emergence of new nanotechnological products demands that their risks to human health and the environment be evaluated. This has resulted in the creation of the new area of nanotoxicology, which addresses the toxicological aspects of these materials.
Purpose and methods
Contributing to these developments, the present work involved a genotoxicological study of different nanofibers, employing chromosomal aberration and comet assays, as well as cytogenetic and molecular analyses, to obtain preliminary information concerning nanofiber safety. The methodology consisted of exposure of Allium cepa roots, and animal cell cultures (lymphocytes and fibroblasts), to different types of nanofibers. Negative controls, without nanofibers present in the medium, were used for comparison.
Results
The nanofibers induced different responses according to the cell type used. In plant cells, the most genotoxic nanofibers were those derived from green, white, and brown cotton, and curaua, while genotoxicity in animal cells was observed using nanofibers from brown cotton and curaua. An important finding was that ruby cotton nanofibers did not cause any significant DNA breaks in the cell types employed.
Conclusion
This work demonstrates the feasibility of determining the genotoxic potential of nanofibers derived from plant cellulose to obtain information vital both for the future usage of these materials in agribusiness and for an understanding of their environmental impacts.
Introduction
There is considerable interest in the commercial exploitation of materials derived from cellulose, since it is both renewable and abundant. Cellulose nanofibers (also known as whiskers or nanocrystals) are the crystalline domains of cellulosic fibers that can be isolated using acid hydrolysis. The terminology used to describe these nanometric scale fibers is related to their physical characteristics in terms of rigidity, thickness, and length.Citation1 Although at present these materials have limited industrial applications due to difficulties related to their high hygroscopicity, tendency to agglomerate, and poor interaction with other materials, cellulose whiskers have a variety of potential future uses. These include optical document security, improvement of the mechanical resistance of thin films containing polyelectrolytes in lithium batteries, and reinforcement in the polymeric matrices of environmentally friendly wrapping materials. Additionally, cellulose whiskers could be used in medicine, as well as in agribusiness.Citation2,Citation3
However, before these materials can be considered for these potential uses, investigation of their toxicities is required, considering their safety in terms of both the environment and living beings, as well as possible modifications of production techniques that could reduce any toxicity present. In the health sector, the use of these materials could improve biocompatibility, as required by regulatory agencies.Citation4,Citation5 The aim of nanotoxicological studies is to ensure the safe application of nanomaterials, hence increasing their acceptance and minimizing any future problems that might be associated with their broader usage.
Nanomaterial characteristics such as size, surface charge, and physical structure can vary considerably, even when the same material of origin is used. As a result, the toxicity of the materials can also vary,Citation6 which necessitates investigation of possible toxic effects to safeguard human health and the environment.Citation4,Citation5 Nanotechnological analyses should enable identification of the production methods required to produce nanostructures (such as cellulose nanofibers) that possess optimized characteristics in terms of minimization of toxicity. However, the main difficulty in relation to the toxicological potential of nanomaterials arises from the fact that it is impossible to generalize, since different nanomaterials display distinct toxicological profiles.Citation7,Citation8 In the case of nanofibers, the challenge arises from their tendency to agglomerate and their hygroscopic behavior, which hinders manipulation and analysis and demands the development of new forms of evaluation.
There have been many reports in the literature concerning the genotoxic potential of nanomaterialsCitation9,Citation10 most of which have used comet, micronucleus, and Ames tests. Allium cepa chromosome aberration tests have also been used to determine the toxic potential of these substances.Citation11–Citation18 Many of these procedures have been classically used for risk analysis purposes, including the detection of genotoxic effects following human exposure to various different compounds.Citation19–Citation22 However, many of the standard methods for genotoxic risk analysis need to be modified when the techniques are applied to nanomaterials.Citation10
The aim of this study was to investigate the genotoxicity of cellulosic nanofibers derived from white, brown, ruby, and green cotton, and curaua. These materials have potential uses in health and environmental applications. Information concerning their safety was acquired using the Allium cepa chromosome aberration assay, the comet assay, and molecular analyses.
Materials and methods
Materials
Schiff and Giemsa reagents were obtained from Sigma- Aldrich (St Louis, MO). Supplemented Dulbecco’s modified Eagle’s medium culture medium and colchicine were provided by Cultilab (Campinas, Brazil). The fibroblast cells were donated by the Biomembrane Laboratory of the State University of Campinas (Brazil). All other reagents, including the salts used for buffer preparation (disodium hydrogen phosphate, sodium dihydrogen phosphate, acetic acid, sodium acetate, sodium carbonate, and calcium carbonate), were either of spectroscopic or analytical grade.
Fiber pretreatment
Cotton fibers
Commercial white and colored cotton fibers were supplied by Embrapa Algodão (Campina Grande, Paraiba, Brazil). All the fibers were finely chopped in a knife mill, passed through a 10-mesh sieve, dewaxed with 1:1 (v/v) ethanol/cyclohexane for 12 hours in a Soxhlet apparatus, and then vigorously washed with tap water. The dewaxed samples were dried for 12 hours at 100°C in a recirculating air oven. The color of the fiber was maintained and the dewaxed fibers were used without any type of bleaching.
Curaua fibers
The curaua fibers were supplied by Embrapa Amazonia Ocidental (Belém, Brazil). They were ground in a knife mill to an approximate length of 2 mm then pretreated with aqueous NaOH solution (17.5 wt%) at 70°C for 1 hour under continuous agitation. The fibers were separated by filtration and repeatedly washed with distilled water until the pH reached neutrality. They were then dried at 60°C for either 24 hours or until a constant weight was obtained.
Preparation of nanofibers after fiber pretreatment
Approximately 5.0 g of fibers were dispersed in 100 mL of 6.5 mol/L sulfuric acid at 45°C, and stirred vigorously for 75 minutes. The addition of 500 mL of cold distilled water was then used to stop the reaction. The residual sulfuric acid was partially removed from the resulting suspension by separation using centrifugation at 10,000 rpm for 10–15 minutes. The fibers were then resuspended and dialyzed against tap water using a cellulose membrane until the pH reached 6–7. The suspension was ultrasonicated for 5 minutes then stored in a refrigerator.
Cell description
For the analyses involving lymphocytes, the cells were separated from whole blood using Ficoll-Paque™ PLUS medium (GE Healthcare, Little Chalfont, UK). The blood was provided by donors aged between 18 and 24 years (who freely signed terms of agreement forms) and the project was approved by the Ethics Committee of the University of Sorocaba (protocol #008/08). Blood samples were collected at a suitable location by a qualified professional, using disposable materials throughout the procedure. The lymphocytes were placed in Roswell Park Memorial Institute (RPMI) 1600 culture medium (Cultilab) containing 300 μg/mL of L-glutamine and 200 μg/mL of NaHC3, supplemented with 5% bovine fetal serum, 50 μg/mL of gentamicin sulfate (antibiotic), and 2 μg/mL of amphotericin B (antifungal). The culture was kept at 37°C, under a humidified atmosphere containing 5% CO2.
The 3T3 cell mouse f ibroblasts were provided by the Biomembrane Laboratory of the State University of Campinas (UNICAMP, Brazil). The cells were kept at 37°C, under a humid atmosphere with 5% CO2, in Dulbecco’s modified Eagle’s medium (DMEM) culture medium containing 584 μg/mL of L-glutamine and 370 μg/mL of NaHC3, supplemented with 10% bovine fetal serum, 50 μg/mL of gentamicin sulfate, and 2 μg/mL of amphotericin B. Aliquots, each containing around 106 cells per mL, were stored in a freezer at −80°C in bovine fetal serum with 10% dimethyl sulfoxide. For each new experiment, a fresh aliquot was withdrawn, washed, and plated out.
Toxicity analyses
Cytogenetic analysis
The cytogenetic analyses required the use of adherent cell cultures, since adherence of the nanofibers to the material used for the test necessitated washing after the treatments, and this was only possible using adherent cells. Approximately 5 × 103 cells (3T3 fibroblasts) were cultured on twelve-well plates. Each mixture was incubated for 72 hours at 37°C with 5% CO2. A total of 50 μL of colchicine was added to each culture 4 hours before the end of the incubation period. After 72 hours, the cultures were washed and then treated with trypsin to eliminate the adherence. The unattached cells were transferred to 15 mL tubes and centrifuged at 900 rpm for 10 minutes. The supernatant was discarded and the sediment was subjected to progressive hypotonization, using a solution of 0.075 mol/L KCl at 37°C for 1 hour, with resuspension of the material every 15 minutes. After hypotonization, the product was fixed using a solution of 3:1 (v/v) methanol/acetic acid. Samples were then deposited dropwise onto slides and stained with Giemsa dye for 10 minutes. Analyses were performed using an optical microscope, with an average of 3000 cells counted in each experiment performed in triplicate (approximately 1000 cells per slide).Citation15
The mitotic index (MI) was calculated by dividing the number of cells in division by the total cell count (EquationEquation 1(1) ).
DNA analysis using agarose gel
Since the nanofibers do not allow for the use of the tetrazolium salt reduction test (3-[4,5-Dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide) of cellular viability, due to their interference in colorimetric detection, the DNA of cells exposed to nanofibers was analyzed at a concentration of 1% in agarose gel (1%). This enabled assessment by visualization of breaks in the DNA of the 3T3 cells caused by the nanofibers, indicative of cells in apoptosis.
The effects on DNA of the materials tested were observed using 3T3 cell cultures, in triplicate (). After adhesion, the cells were exposed to each of the nanofiber preparations for a period of 24 hours, using a nanofiber concentration of 1%. Negative controls were incubated in ultrapure water and H2O2 was used as a positive control. The DNA was extracted from all cultures using a cell culture DNA extraction kit (AP-MN-MS-GDNA-250-Axygen; Axygen Biosciences, Union City, CA), according to the method proposed by Sambrook et al.Citation23 The material was submitted to qualitative analysis using 1% agarose gel electrophoresis, run at 5 V/cm. Extractions performed using different methods showed the same results.
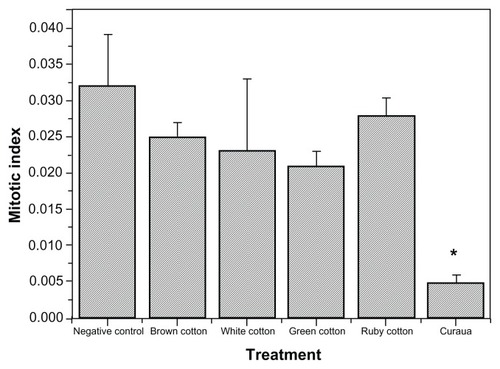
Figure 1 Effects of different types of nanofiber (white, brown, ruby, and green cotton, and curaua) on the mitotic index obtained from cytogenetic analyses using 3T3 cells exposed to the nanofibers at a concentration of 0.1%.
Notes: Values are expressed as the mean of three experiments (n = 3). *Values statistically different to the control (P < 0.05).
Genotoxicity analyses
Allium cepa assay
Allium cepa seeds were germinated in ultrapure water at room temperature. When the roots reached a length of 2 cm, the seedlings were removed from the germination box and placed into contact with the cellulose nanofibers. Ultrapure water was used as a negative control and trifluoraline (0.075 g/L) as a positive control. After 24 hours of contact with the nanofibers, the roots were fixed with Carnoy’s reagent (ethyl alcohol and acetic acid, in a proportion of 3:1 by volume). After fixation, the roots were removed from the reagent, washed, and subjected to acid hydrolysis in a solution of 1 mol/L HCl at 60°C for 9 minutes. They were then washed in distilled water and subjected to a Schiff-base reaction for 2 hours in an amber glass jar, with exclusion of light. After that, the roots were rinsed in distilled water until all excess dye had been removed. The meristematic region of the roots was cut, placed on a slide together with a drop of 2% acetocarmine, and covered with a coverslip, which was used to gently press and spread the cells. The slides were examined under an optical microscope (Zeiss Axiostar Plus, Carl Zeiss Microscopy, LLC, New York, NY), with an average of 500 cells used for each independent assay performed in triplicate (totaling around 1500 cells analyzed).Citation15
The results of the Allium cepa tests were used to calculate the MI (EquationEquation 1(1) ), the relative mitotic index (RMI, EquationEquation 2
(2) ), the chromosomal aberration score (CAS, EquationEquation 3
(3) ), and the relative chromosomal aberration score (RCAS, EquationEquation 4
(4) ). The RMI was obtained by dividing each MI by the value of the MI of the negative control.
Comet assay
Peripheral blood was used for the comet analyses. A casuistic study was undertaken, involving six biotechnology students (three female and three male) from the University of Sorocaba, all of whom were between 18 and 24 years of age. The students were nonsmokers, did not consume alcohol or drugs, and were not on any medications. The lymphocytes were submitted to treatment with the nanofibers for a period of 1 hour. Negative and positive controls employed phosphate buffered saline (PBS) and H2O2 (200 μMol/L), respectively. The comet assay (single cell gel electrophoresis) was performed as described by Tice et alCitation24 and Azqueta et al.Citation22 Each treatment involved the use of 10 μL of lymphocytes in 110 μL of low melting point agarose (0.6%) and the mixture was placed onto microscope slides that had been pre-coated with normal melting point agarose (1.5%). Coverslips were positioned over this material, and the slides were placed in a refrigerator for polymerization. After polymerization, the coverslips were removed, and the slides were treated for 90 minutes with an ice-cold (4°C) lysis solution (2.5 mol/L NaCl, 0.1 mol/L ethylenediaminetetraacetic acid [EDTA], 10 mmol/L tris(hydroxymethyl)aminomethane [Tris], 1% Triton X-100TM, pH 10). All treatments were then incubated in an electrophoresis buffer (0.3 M NaOH, 1 mM EDTA, pH > 13) for 20 minutes, followed by electrophoresis for 20 minutes at 1.3 V/cm. After electrophoresis, the slides were covered with a neutralizing solution (0.4 mol/L Tris, pH 7.5) for 5 minutes, then washed three times in distilled water and allowed to rest overnight at room temperature. Prior to staining, the dry slides were left in a fixing solution (15% w/v trichloroacetic acid, 5% [w/v] zinc sulfate, and 5% glycerol) for 10 minutes, and were then washed three times in distilled water. After these procedures, the slides were allowed to rest at room temperature for 1.5 hours. They were then rehydrated with distilled water and stained for approximately 15 minutes in a silver staining solution consisting of 34 mL of Solution A (0.2% w/v ammonium nitrate, 0.2% w/v silver nitrate, 0.5% w/v tungstosilicic acid, 0.15% v/v formaldehyde, and 5% w/v sodium carbonate) to 66 mL of Solution B (5% sodium carbonate), followed by a bath in distilled water and a bath in stop solution. Lastly, the slides were again washed in distilled water and allowed to dry at room temperature.Citation18 Staining using silver is analogous to fluorescence, where the positive charge of the silver enables it to bind with DNA and DNA fragments, producing the characteristic color.
Throughout the procedures involving cellular material, both natural light and light from fluorescent lamps were avoided to prevent any influence on the results. Analyses were performed using the Zeiss Axovert optical microscope and at least 100 cells were counted on each slide, with 3 slides for each test (around 300 cells). The experiment was performed in triplicate, giving a total of around 900 cells analyzed, on average, for each sample tested. The comet assay analyses were performed by assigning a score of 0 to 4, according to the quantity of DNA in the tail, and the length of the tail: Class 0 corresponded to intact cells, with no damage caused by the exposure; Class 1 corresponded to cells with minimal damage; Class 2 to average damage; Class 3 to severe damage; and Class 4 to cells with maximum damage.Citation19–Citation22,Citation25 For this visual method, the number of cells found for each score was multiplied by the value of the score and the values were summed at the end of the analysis of each slide. Since the score depended on the number of cells observed, an index of tail damage (TD) was created by dividing the score given to the slide by the number of cells analyzed on the slide.Citation26
Data analysis
The cytogenetic and genotoxicity assay results were analyzed statistically by one-way analysis of variance, with the Tukey–Kramer post-hoc test. Statistical significance was defined as P < 0.05.
Results and discussion
Lignocellulosic fibers (such as those of curaua) essentially comprise cellulose, lignin, hemicellulose, and pectins, together with other minor components. The use of cellulose as a support material for polymeric matrices is attractive, since it has low density and abrasivity, and is also nontoxic, inexpensive, biodegradable, and (in principle) renewable.Citation27
The cellulose component of the fibers is therefore of especial interest (since it can be used as a support for polymeric matrices), which means that the lignocellulosic fibers require purification (bleaching) to remove the hemicellulose and lignin fractions. The resulting cotton consists mainly of cellulose and minor components, with lignin acting to maintain the structural integrity of the fiber. As a result, cellulose nanofibers occur naturally but joined in the form of fibrils. To obtain individual cellulosic nanostructures, the fibrils need to be separated. This can be achieved in several ways, such as through high-efficiency shearing, processes employing mechanical refining or sonication as well as degradation by combined enzymatic and mechanical processes, or chemical/mechanical treatments.Citation28
Different particle sizes and degrees of crystallinity can be achieved using the various treatments. In the present work, chemical/mechanical treatment was employed, which consisted of exposing the cellulose to acid attack to remove the amorphous cellulose phase, leaving the crystalline phase. The cellulosic nanostructures resulting from the acid treatment are known as “whiskers.” The use of relatively small amounts of this type of highly crystalline material can greatly improve polymer mechanical strength and an additional benefit is that the material is also environmentally friendly.
Nanocellulose has been investigated with the aim of obtaining polymeric nanocomposites with superior strength and barrier properties. These are characteristics that are important in the packaging industry. In medicine, cellulose nanomaterials have potential applications as indicators, tissue scaffolds, hydrogel supports, drug delivery systems, sensors, and valves.Citation28
This work investigated the genotoxic effects of cellulose nanofibers originating from different sources (white cotton, brown cotton, ruby cotton, green cotton, and curaua). The coloration of the cotton used to prepare the nanofibers is natural, and obtained using genetic breeding techniques, hence avoiding the need for synthetic dyes that could have adverse effects in humans and the environment. The color results from the presence of natural pigments, including flavonoid compounds such as flavonone, flavonol, and anthocyanidin,Citation29,Citation30 with the final color determined by genetic factors and unknown environmental factors.Citation29,Citation31
The synthesis and characterization of the nanofibers has been described previously.Citation32,Citation33 Their main properties (average length, diameter, and zeta potential, respectively) were as follows: white cotton: 135 ± 50 nm, 14 ± 4 nm, −31 mV; brown cotton: 140 ± 45 nm, 11 ± 3 nm, −26 mV; green cotton: 180 ± 45 nm, 13 ± 2 nm, −23 mV; ruby cotton: 130 ± 25 nm, 10 ± 4 nm, −25 mV; and curaua: 80–170 nm, 6–10 nm, −30 mV.Citation20,Citation28
The lengths of the nanofibers were around 150 nm, while differences were observed in the diameters. The diameters of the cotton nanofibers were in the region of 12 nm, while those of the curaua nanofibers were smaller, in the 6–10 nm range. The zeta potentials of all the nanofibers were close to −25 mV. Correlation between these parameters and the results of genotoxicity tests can be used to better understand the interaction between cells and nanostructured systems, which is affected by particle characteristics including size, surface area, surface charge, functional groups, ligands, hydrophobicity, and hydrophilicity.Citation34
An initial analysis of possible cytotoxic effects of the different cellulose nanofibers (at a concentration of 0.1%), cytogenic studies and DNA analysis using agarose gel were performed with cultures of Balb/c mouse 3T3 fibroblast cells. The cytogenetic analyses () showed that significant alterations in the MI only occurred in the tests using the curaua nanofibers. One method of assessing cytotoxicity is by analysis of changes in the MI value,Citation35 where a significant reduction relative to the control could indicate a potentially lethal effect on the organism.Citation36 The curaua nanofibers therefore presented toxic effects to the fibroblast cells. No statistically significant alterations were observed for the remaining nanofibers, indicating that the nanofibers did not induce cellular stress.
Nonetheless, even though the nanofibers may not cause cell death, this does not necessarily mean that effects cannot occur following interaction with the genetic material. Damage to the DNA, even when reparable, is potentially dangerous since errors in the repair can occur, especially at higher levels of exposure. The 3T3 cells were exposed to 1% concentrations of the nanofibers for 24 hours, after which the DNA was extracted using an extraction kit, and analyzed by electrophoresis on 1% agarose gel.
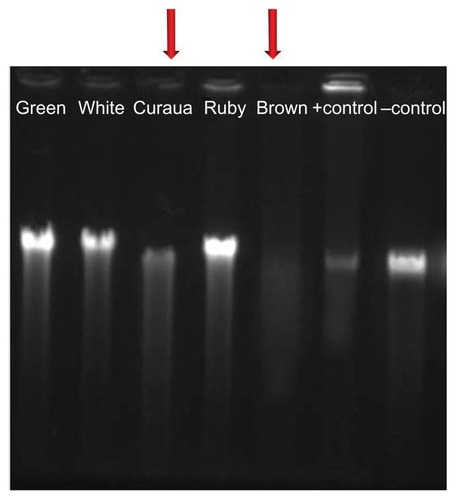
The results () showed that the brown cotton and curaua nanofibers caused many breaks in the genetic material, resulting in poor quality of the DNA extraction. This test was performed immediately after the treatment, so that there was no time for any possible repair of the DNA. It could be observed that even in the absence of any alteration in the MI value () the brown cotton nanofibers caused changes (breaks) in the DNA, while the curaua nanofibers caused alterations in the MI as well as breaks in the DNA. These findings suggest that cellular interaction was different for the two materials.Citation37
Figure 2 Agarose gel electrophoresis results for the effects of the different types of nanofiber (white, brown, ruby, and green cotton, and curaua), at a concentration of 1%, on DNA extracted from 3T3 cells.
Note: The arrows indicate the nanofibers that caused the greatest DNA damage.
Previous work by our research group investigated the genotoxicity of different polymeric nanoparticles composed of chitosan and poly(methacrylic acid), with determination of chromosome damage using the Allium cepa testCitation15,Citation38 and cytogenetic analyses employing human lymphocyte cell cultures.Citation15 These tests were shown to be valid for the genotoxicological analysis of nanostructured materials.Citation11–Citation13,Citation15–Citation18
In the present work, the Allium cepa assay was performed by placing germinated roots in contact with the different types of nanofiber for 24 hours, at nanofiber concentrations of 0.01, 0.1, and 1%. After the treatment, slides were prepared and counts were made of the numbers of cells in division (prophase, metaphase, anaphase, and telophase) as well as the numbers of damaged cells (). The results of the Allium cepa tests were used to calculate the MI (EquationEquation 1(1) ), the RMI (EquationEquation 2
(2) ), the CAS (EquationEquation 3
(3) ), and the RCAS (EquationEquation 4
(4) ), as shown in and . The results showed that there was no significant difference in the RMI (), indicating that the nanofibers did not present cytotoxicity to this cell type.
Table 1 Summary of the effects caused by different types of nanofiber (white, brown, ruby, and green cotton, and curaua), obtained in the Allium cepa tests
Figure 3 Examples of normal Allium cepa cells (A: interphase; B: prophase; C: metaphase; D: anaphase; E: telophase) and chromosomal abnormalities found in Allium cepa cells exposed to nanofibers (F–J and L–R).
Notes: Anaphase bridges are visible in images F, L–N and P–R. Images G and J show anaphase fragments, and images I, O, R, and S show c-metaphases.
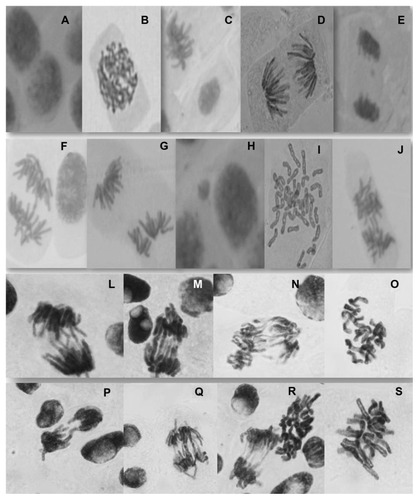
Figure 4 Effects of different types of nanofiber (white, brown, ruby, and green cotton, and curaua) on (A) the relative mitotic index and (B) the relative chromosomal aberration score after exposure of Allium cepa cells.
Notes: *Significant changes compared with the control (analysis of variance, P < 0.05). Values are expressed as the mean of three experiments (n = 3).
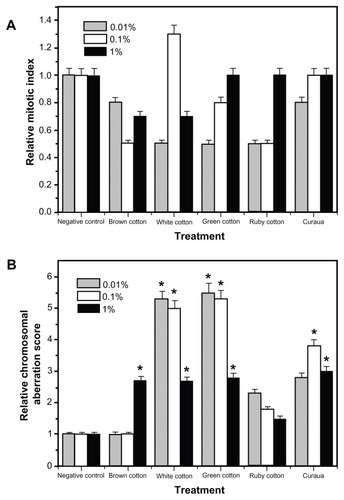
Use of the white and green cotton nanofibers produced significant DNA breaks, as shown by the different CAS and RCAS values ( and ), relative to the control, at all concentrations (0.01, 0.1, and 1%), while, in the case of the curaua nanofibers, DNA breaks were only observed at concentrations of 0.1% and 1%. For the brown cotton nanofibers, there was only a significant difference at the highest concentration tested. An important finding was that the white cotton, green cotton, and curaua nanofibers produced the highest chromosomal aberration index values at the lowest fiber concentrations, supporting the notion that aggregate formation at higher concentrations reduced the contact surface as well as the degree of interaction with the Allium cepa cells, resulting in fewer cellular DNA breaks. The brown cotton nanofibers (at concentrations of 0.01% and 0.1%) and ruby cotton nanofibers (at concentrations of 0.01, 0.1, and 1%) caused practically no significant DNA breaks, relative to the control, indicating that these materials were less toxic to the Allium cepa cells.
Given these results, the comet test was performed using nanofiber concentrations of 0.1%. The comet test is a test of genotoxicity that is able to quantitatively detect damage to DNA induced by alkylating, intercalating, and oxidizing agents.Citation19–Citation22 The alkaline version of this test allows the detection of breaks in single and double strands, alkali-labile sites, and crosslinks.Citation39 The neutral version, initially proposed by Ostling and Johanson,Citation40 produces tails that are denser and shorter, and is only able to detect lesions involving simple breaks, which are of less interest since the breaks can be repaired rapidly, and are not considered to be mutagenic lesions.Citation19–Citation22,Citation25
The comet test was performed using peripheral blood. Lymphocytes were separated from whole blood using Ficoll-Paque PLUS medium, and exposed to the different types of nanofiber, with incubation at concentrations of 0.1% for 1-hour periods. Analysis of the results considered the sizes of the tails produced by the chromosome breaks, to which were attributed different scores, with values of 0 to 4 corresponding to the tail intensity, as shown in .
Figure 5 Representative images (at ×40 magnification) illustrating the tail intensity for the fibroblast comet test scores.
Notes: (A) score = 0 (negative control); (B) score = 1 (nanofibers); (C) score = 2 (nanofibers); (D) score = 3 (nanofibers); (E) score = 4 (nanofibers and positive control).
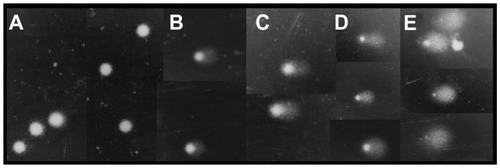
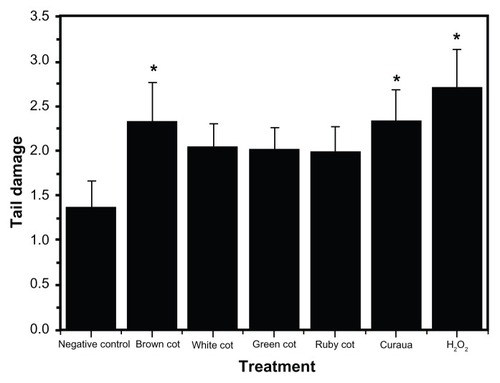
The comet tests using lymphocytes showed that the various nanofibers (at a concentration of 0.1%) induced different DNA breaks in the cells, as shown by the damage scores (). All the nanofibers tested showed the capacity to induce DNA breaks in the genetic material; however, significant alterations (P < 0.05) were only observed for the brown cotton and curaua nanofibers, with the latter inducing the greatest changes.Citation26 The differences were probably associated with the mechanisms of entry into the cells, intracellular signaling, and interaction with biological macromolecules.Citation34,Citation41–Citation43 The results also suggested that, compared with plant cells, animal cells may possess an apparatus that is better able to repair DNA damage.
Figure 6 Results of tail damage obtained for the lymphocyte cells after treatments with the nanofibers (white, brown, ruby, and green cotton, and curaua) at a concentration of 0.1%.
Notes: *Significant alterations compared with the control (analysis of variance, P < 0.05). Values are expressed as the mean of three experiments (n = 3).
Studies of the genotoxicity of nanofibers are still in their infancy, so although DNA changes were observed in the present work, the mechanism of action remains unknown. Investigations of other types of nanomaterials have shown that there are several ways in which the materials can penetrate cells;Citation37 however, there are no reports concerning cellulose nanofibers.
There are many mechanisms of cellular signaling, including those responsible for the transmission of responses that can even involve cell apoptosis. Further work will be needed to determine the mechanisms of action by which cellulose nanofibers are able to damage DNA.
There were no important differences in the size, zeta potential, and diameter of the nanofibers that might be able to explain the greater effects of the brown cotton and curaua nanofibers. Possible explanations could be differences in the forms of the nanofibers, as observed previously using microscopy,Citation32,Citation33 or differences in functional groups and other characteristics arising from the nanofiber synthesis process.Citation34
It was evident that higher indices of genotoxicity were obtained in the Allium cepa tests, compared with the alterations observed in the fibroblast and lymphocyte cells. This difference was probably due to the type of signaling and the cellular response caused by the nanofibers in each of the different cellular materials. Another interesting finding was that the lowest alteration values were obtained using the ruby cotton nanofibers, which, under the conditions employed, were therefore the least genotoxic of the nanofibers investigated.
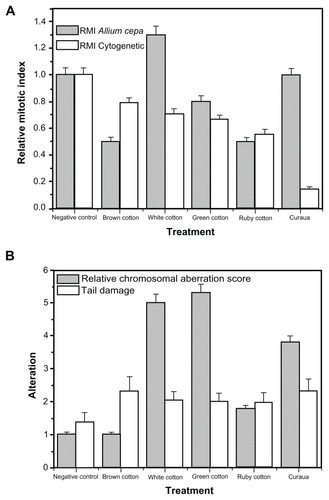
It is clear that the various types of nanofiber caused different genetic alterations in the cellular materials used in this study, with the brown cotton and curaua nanofibers presenting the greatest toxicity to fibroblast cells and human lymphocytes. Considering only the 0.1% nanofiber concentration level, the RMI showed greater alterations in animal cells compared with plant cells (), while the plant cells showed higher damage index (RCAS) values than the animal cells (using comet test TD analyses) (). When the same effects on animal cells of the treatments using 0.1% nanofiber concentrations were compared with the effects on plant cells, the RCAS and TD score showed generally similar trends in terms of genotoxicity.
Figure 7 Effects on animal and plant cells caused by the cellulose nanofibers (white, brown, ruby, and green cotton, and curaua) at a concentration of 0.1%. (A) Values of the relative mitotic index (RMI); (B) values of the relative chromosomal aberration score (Allium cepa test) and the tail damage score (comet assay).
Note: All values are expressed as the mean of three experiments.
Considering the issue of different processes of cellular signaling, mentioned above, an important point is that cellulose nanofibers show a strong tendency to agglomerate, especially after drying and in highly concentrated aqueous solutions. This is due to strong inter- and intramolecular hydrogen bonding. The nanofibers are highly hydrophilic and in nonpolar media tend to form aggregates. Agglomerated nanofibers could provoke effects in cells different to those caused by individual fibers, which could help to explain the different responses observed.
Conclusion
The future use of cellulosic nanofibers first requires a better understanding of their effects, including possible environmental impacts when used in agribusiness applications. Cellular responses can differ according to the organism concerned, the nanofiber concentration, and the exposure medium. The results demonstrated that nanofibers derived from cotton (white, green, and brown) and curaua could cause alterations in plant cells, while the brown cotton and curaua nanofibers were genotoxic in animal cells (human lymphocytes and mouse fibroblasts). The animal cells therefore appeared to be less liable to genetic alterations and possessed more effective repair mechanisms. An important observation was that neither plant nor animal cells showed any genotoxic alterations when exposed to ruby cotton nanofibers, under the conditions employed, indicating that ruby cotton might be the material of choice for nanofiber production.
Aggregation of nanofibers appeared to reduce toxicity, and was greater in culture media (used for the experiments involving animal cells) than in water (used for the plant cell tests). Further studies will be needed to better understand the influence of aggregation on nanofiber toxicity. It is therefore important to consider the environment in which the nanofibers will be used, since in some applications fiber aggregates are used to reinforce polymeric matrices, where the fibers can improve thermal stability, mechanical resistance, and permeability to liquids and gases, even at low fiber concentrations. In their intact state, these matrices are unlikely to release nanofibers; however, decomposition processes could result in slow release of fibers to the wider environment.
Since nanofibers have considerable potential for future commercial applications, additional work is in progress to obtain further information concerning their toxicological characteristics as well as their possible impacts on human health and the environment.
Acknowledgments
This research was supported by the Brazilian agencies FAPESP, CNPq, CAPES, Fundunesp, Finep, and Embrapa.
Disclosure
The authors report no conflicts of interest in this work.
References
- de Souza LimaMMBorsaliRRodlike cellulose microcrystals: structure, properties, and applicationsMacromol Rapid Commun2004257771787
- EichhornSJBaillieCAZafeiropoulosNReview: Current international research into cellulosic fibres and compositesJ Mater Sci200136921072131
- TeixeiraEMLottiCCorrêaACTeodoroKBMarconciniJMMattosoLHThermoplastic corn starch reinforced with cotton cellulose nanofibersJ Appl Polym Sci2011120424282433
- ChauCFWuSHYenGCThe development of regulations for food nanotechnologyTrends Food Sci Technol2007185269280
- BouwmeesterHDekkersSNoordamMYReview of health safety aspects of nanotechnologies in food productionRegul Toxicol Pharmacol2009531526219027049
- NafeeNSchneiderMSchaeferUFLehrCMRelevance of the colloidal stability of chitosan/PLGA nanoparticles on their cytotoxicity profileInt J Pharm2009381213013919450671
- HerzogECaseyALyngFMChambersGByrneHJDavorenMA new approach to the toxicity testing of carbon-based nanomaterials – the clonogenic assayToxicol Lett20071741–3496017920791
- PisanicTR2ndBlackwellJDShubayevVIFiňonesRRJinSNanotoxicity of iron oxide nanoparticle internalization in growing neuronsBiomaterials200728162572258117320946
- NgCTLiJJBayBHYungLYLCurrent studies into the genotoxic effects of nanomaterialsJ Nucleic Acids20102010 pii:947859
- LandsiedelRKappMDSchulzMWienchKOescFGenotoxicity investigations on nanomaterials: methods, preparation and characterization of test material, potential artifacts and limitations – many questions, some answersMutat Res20096812–324125819041420
- KumariMMukherjeeAChandrasekaranNGenotoxicity of silver nanoparticles in Allium cepaSci Total Environ2009407195243524619616276
- PandaKKAcharyVMKrishnaveniRIn vitro biosynthesis and genotoxicity bioassay of silver nanoparticles using plantsToxicol In Vitro20112551097110521419840
- FachinettoJMOuriqueAFLubiniGTedescoSBSilvaACBeckRCTretinoin-loaded polymeric nanocapsules: evaluation of the potential to improve the antiproliferative activities on Allium cepa root-tip compared to the free drugLatin American Journal of Pharmacy2008275668673
- GhoshMBandyopadhyayMMukherjeeAGenotoxicity of titanium dioxide (TiO2) nanoparticles at two trophic levels: plant and human lymphocytesChemosphere201081101253126220884039
- De LimaRFeitosaLPereiraAEEvaluation of the genotoxicity of chitosan nanoparticles for use in food packaging filmsJ Food Sci20107568996
- KlančnikKDrobneDValantJDolenc KoceJUse of a modified Allium test with nanoTiO2Ecotoxicol Environ Saf20107418592
- GhodakeGSeoYDLeeDSHazardous phytotoxic nature of cobalt and zinc oxide nanoparticles assessed using Allium cepaJ Hazard Mater2011186195295521122986
- ParidaUKBindhaniBKNayakPGreen synthesis and characterization of gold nanoparticles using onion (Allium cepa) extractWorld Journal of Nano Science and Engineering2011149398
- DusinskáMCollinsAKazimírováAGenotoxic effects of asbestos in humansMutat Res20045531–29110215288536
- DusinskaMCollinsARThe comet assay in human biomonitoring: gene–environment interactionsMutagenesis200823319120518326867
- CollinsAROscozAABrunborgGThe comet assay: topical issuesMutagenesis200823314315118283046
- AzquetaAGutzkowKBBrunborgGCollinsARTowards a more reliable comet assay: optimising agarose concentration, unwinding time and electrophoresis conditionsMutat Res20117241–2414521645630
- SambrookJFritschEFManiatisTMolecular Cloning: A Laboratory Manual2nd edNew York, NYCold Spring Harbor Laboratory Press1989
- TiceRRAgurellEAndersonDSingle cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testingEnviron Mol Mutagen200035320622110737956
- CollinsARDobsonVLDusinskáMKennedyGStetinaRThe comet assay: what can really tell us?Mutat Res199737521831939202728
- LovellDPOmoriTStatistical issues in the use of the comet assayMutagenesis200823317118218385511
- KaliaSDufresneACherianBMCellulose-based bio- and nanocomposites: a reviewInt J Polym Sci20112011 Article ID 837875
- HubbeMARojasOJLuciaLASainMCellulosic nanocomposites: a reviewBio Resources200833929980
- XiaoYHZhangZSYinMHCotton flavonoid structural genes related to the pigmentation in brown fibersBiochem Biophys Res Commun20073581737817482578
- HuaSWangXYuanSCharacterization of pigmentation and cellulose synthesis in colored cotton fibersCrop Sci200747415401546
- de CarvalhoLPdos SantosJWRespostas correlacionadas do algodoeiro com a seleção para a coloração da fibra [Correlated responses to selection of cotton for dyeing fiber]Pesq Agropec Bras2003387983 Portuguese
- CorrêaACTeixeiraEMPessanLAMattosoLHCCellulose nanofibers from curaua fibersCellulose201017611831192
- TeixeiraEMCorrêaACManzoliALeiteFLOliveiraCRMattosoLHCellulose nanofibers from white and naturally colored cotton fibersCellulose2010173595606
- NelAEMädlerLVelegolDUnderstanding biophysicochemical interactions at the nano-bio interfaceNat Mater20098754355719525947
- OdeigahPGCNurudeenOAmundOOGenotoxicity of oil field wastewater in NigeriaHereditas19971262161167
- AntosiewiczDAnalysis of the cell cycle in root meristem of Allium cepa under the influence of ledakrinFolia Histochem Cytobiol1990281–279952097185
- SinghNManshianBJenkinsGJNanoGenotoxicology: the DNA damaging potential of engineered nanomaterialsBiomaterials20093023–243891391419427031
- RankJNielsenMHA modified Allium test as a tool in the screening of genotoxicity of complex mixturesHereditas199311814953
- SinghNPMcCoyMTTiceRRSchneiderELA simple technique for quantitation of low levels of DNA damage in individual cellsExp Cell Res198817511841913345800
- OstlingOJohansonKJMicroelectrophoretic study of radiation- induced DNA damages in individual mammalian cellsBiochem Biophys Res Commun198412312912986477583
- DufresneAComparing the mechanical properties of high performances polymer nanocomposites from biological sourcesJ Nanosci Nanotechnol20066232233016573028
- DobrovolskaiaMAMcNeilSEImmunological properties of engineered nanomaterialsNat Nanotechnol20072846947818654343
- CliftMJFosterEJVanheckeDInvestigating the interaction of cellulose nanofibers derived from cotton with a sophisticated 3D human lung cell cocultureBiomacromolecules201112103666367321846085