Abstract
Background
The sigma-2 receptor is an attractive target for tumor imaging and targeted therapy because it is overexpressed in multiple types of solid tumors, including prostate cancer, breast cancer, and lung cancer. SV119 is a synthetic small molecule that binds to sigma-2 receptors with high affinity and specificity. This study investigates the utility of SV119 in mediating the selective targeting of liposomal vectors in various types of cancer cells.
Methods:
SV119 was covalently linked with polyethylene glycol-dioleyl amido aspartic acid conjugate (PEG-DOA) to generate a novel functional lipid, SV119-PEG-DOA. This lipid was utilized for the preparation of targeted liposomes to enhance their uptake by cancer cells. Liposomes with various SV119 densities (0, 1, 3, and 5 mole%) were prepared and their cellular uptake was investigated in several tumor cell lines. In addition, doxorubicin (DOX) was loaded into the targeted and unmodified liposomes, and the cytotoxic effect on the DU-145 cells was evaluated by MTT assay.
Results:
Liposomes with or without SV119-PEG-DOA both have a mean diameter of approximately 90 nm and a neutral charge. The incorporation of SV119-PEG-DOA significantly increased the cellular uptake of liposomes by the DU-145, PC-3, A549, 201T, and MCF-7 tumor cells, which was shown by fluorescence microscopy and the quantitative measurement of fluorescence intensity. In contrast, the incorporation of SV119 did not increase the uptake of liposomes by the normal BEAS-2B cells. In a time course study, the uptake of SV119 liposomes by DU-145 cells was also significantly higher at each time point compared to the unmodified liposomes. Furthermore, the DOX-loaded SV119 liposomes showed significantly higher cytotoxicity to DU-145 cells compared to the DOX-loaded unmodified liposomes.
Conclusion:
SV119 liposomes were developed for targeted drug delivery to cancer cells. The targeting efficiency and specificity of SV119 liposomes to cancer cells was demonstrated in vitro. The results of this study suggest that SV119-modified liposomes might be a promising drug carrier for tumor-targeted delivery.
Introduction
One major problem with conventional anticancer agents is their severe side effects on normal tissues and organs. To solve this issue, targeted drug delivery to solid tumors was developed as an effective strategy to improve the therapeutic effect and minimize toxicity. Many factors affect the success rate of targeted drug delivery, including the choice of targeting ligands. Many types of ligands have been explored, including antibodies, peptides, aptamers, and small molecule ligands.Citation1 Small molecule ligands have the advantages of minimal immunogenicity, excellent chemical stability, and ease of synthesis.Citation2 Some examples of small molecule ligands that have been developed for tumor targeting include ligands of the folate receptor,Citation3 prostate-specific membrane antigen (PSMA),Citation4 and integrin receptors.Citation5 There has been a recent interest in exploring sigma receptor ligands for tumor imaging and targeted therapy. Sigma receptors are overexpressed in a variety of human tumors including non-small cell lung carcinoma (NSCLC), prostate cancer, melanoma, and breast cancer.Citation6–Citation8 The small molecule ligands of sigma receptors are considered potential tumor imaging agents.Citation6–Citation8 The authors of this study and Dr Huang’s group have reported that sigma receptor ligands, such as anisamide, effectively mediate the selective delivery of liposomes containing doxorubicin (DOX) to prostate cancer cells in vitro and in vivo.Citation9,Citation10 Extensive studies by Dr Huang’s group have demonstrated the targeted delivery of oligodeoxynucleotides and siRNA to lung cancer and melanoma using anisamide-decorated nanoparticles.Citation11–Citation13 Another sigma receptor ligand, haloperidol, was shown to improve the transfection of MCF-7 breast carcinoma cells by more than 10-fold, compared with the control (unmodified) liposomes.Citation14
Neither anisamide nor haloperidol distinguishes between the sigma-1 or sigma-2 receptor subtypes with respect to binding. This study determines whether sigma 2-selective ligands can be employed for the targeted delivery of liposomal vectors. There are several potential advantages associated with sigma 2-selective ligands for tumor-targeted delivery. First, there is a higher density of sigma-2 versus sigma-1 receptors in tumor samples and cultured tumor cells.Citation16–Citation17 Second, ligand binding and photoaffinity labeling studies have demonstrated a lower expression of sigma-2 versus sigma-1 receptors in normal tissues from the central nervous system, gastrointestinal tract, kidney, liver, and heart.Citation18 Third, a correlation between sigma-2 receptor expression and tumor proliferation has been established both in vitro and in vivo,Citation19,Citation20 but no such correlation was observed for the sigma-1 subtype.
SV119 is a small molecule ligand that binds to sigma-2 receptors with high affinity and specificity.Citation21 The tumor-targeting efficiency of SV119 has been demonstrated by conjugating SV119 with apoptosis-inducing peptides, which resulted in the selective killing of tumor cells.Citation22 The present study explores the potential of SV119 to mediate the targeted delivery of liposomes to multiple tumor cell lines. Liposomes are well-established drug carriers for intravenous delivery and several liposomal formulations have been approved for clinical applications.Citation23 Our results show that the incorporation of a SV119 derivatized PEG-lipid significantly increases the cellular uptake of liposomes by several tumor cell lines, but not by normal cells. In addition, DOX-loaded SV119-modified liposomes showed significantly higher cytotoxicity compared to unmodified liposomes loaded with DOX. These findings suggest that SV119 liposomes may have potential for the targeted delivery of anticancer agents to tumors that over-express sigma-2 receptors.
Materials and methods
Materials
L-α-phosphatidylcholine (Soy PC), DSPE-PEG(2000)-OCH3, and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (rhodamine-PE) were purchased from Avanti® Polar Lipids, Inc (Alabaster, AL). Cholesterol (Chol), doxorubicin hydrochloride, FITC-conjugated anti-mouse IgG, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenltetrazolium bromide (MTT), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St Louis, MO, USA). Mouse anti-early endosome antigen 1 (EEA1) monoclonal antibody was purchased from BD Transduction Laboratories. LysoTracker® Green DND-26 was purchased from Life Technologies (Invitrogen®; Carlsbad, CA). A PD-10 desalting column was purchased from GE Healthcare (Little Chalfont, UK). Dulbecco’s phosphate-buffered saline (DPBS) was purchased from Life Technologies (Gibco®) and t-Boc Amine PEG NHS Ester (TBOC-PEG-NHS, MW 3500) was purchased from JenKem Technology USA, Inc (Allen, TX). All other chemicals and organic solvents were purchased from Sigma-Aldrich. 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker FT 400 MHz. The synthesized compounds were also identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Voyager-DE PRO BioSpectrometry Workstation; Applied Biosystems®, Life Technologies). The mass spectrum was obtained in a linear mode with CHCA as the matrix.
Human prostate adenocarcinoma cell line DU-145, human lung carcinoma cell line A549, and human breast cancer MCF-7 cells were obtained from American Type Culture Collection ([ATCC], Manassas, VA) Rockville, MD. BEAS-2B (human bronchial epithelium cell line) was generously gifted by Dr Y Peter Di from the University of Pittsburgh. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; ATCC) and supplemented with 10% FBS (Invitrogen®) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2 in air. All the reagents were obtained from Sigma-Aldrich unless stated otherwise.
Synthesis and chemical characterization of SV119-PEG-DOA conjugates and intermediates
schematically depicts the synthetic procedure for preparing SV119-PEG-DOA. The experimental details are delineated below.
Figure 1 Synthesis scheme for SV119-PEG3500-DOA.
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
Compound 2
NaH (0.8 g, 60% in oil, 20 mmol) was added to a solution of 1 (3.04 g, 10 mmol) and benzyl 2-bromoacetate (2.75 g, 12 mmol) in THF (50 mL).Citation24 The mixture was stirred overnight at an ambient temperature. After ether (75 mL) was added, the organic solution was quenched with water (30 mL) and then washed with a saturated NaCl solution (40 mL), and finally, the organic solution was dried over Na2SO4. After the solvent was removed, the crude product was purified by flash column chromatography with hexane-ethyl acetate (1:1) to obtain compound 2 (4.33 g, 96%) as a pale yellow oil. 1H NMR (CDCl3) δ 7.98 (s, 1H), 7.37–7.21 (m, 6H), 6.77 (m, 2H), 5.17 (s, 2H), 5.00 (m, 1H), 3.83 (s, 3H), 3.42 (m, 2H), 3.27 (m, 2H), 2.33 (m, 2H), 2.30 (s, 3H), 2.13 (m, 1H), 1.98 (m, 2H), 1.51 (m, 3H), 1.17 (m, 2H). ESI-MS m/z 453.5 ([M+H]+).
Compound 3
Chemical name: 2-(3-((2-methoxy-5-methylphenyl) carbamoyl)-9-aza-bicyclo[3.3.1]nonan-9-yl)acetic acid. Pd/C (10%, 500 mg) was added to a solution of 2 (3.62 g, 10 mmol) and stirred under atmospheric H2 pressure in MeOH (50 mL). EtOAc (100 mL) was added after the solvents evaporated under a vacuum. The mixture was washed by a saturated NaCl solution (40 mL), and finally, the organic solution was dried over Na2SO4. EtOAc was then removed to obtain compound 3 (2.82 g, 100%) as a white solid. 1H NMR (CDCl3) δ 10.9 (brs, 1H), 7.96 (s, 1H), 7.15 (s, 1H), 6.79 (m, 2H), 4.96 (m, 1H), 3.83 (s, 3H), 3.32 (m, 2H), 3.04 (m, 2H), 2.35 (m, 2H), 2.29 (s, 3H), 2.08 (m, 1H), 1.64 (m, 3H), 1.50 (m, 4H). ESI-MS m/z 363.4 ([M+H]+).
Compound 4
A mixture of Boc-aspartic acid (2.47 g, 10 mmol), oleyl amine (8.01 g, 30 mmol), DCC (6.18 g, 30 mmol), and DMAP (122 mg, 1 mmol) in anhydrous CH2Cl2 (60 mL) was stirred vigorously at room temperature for 5 hours. After the reaction was completed, the mixture was filtered and evaporated under reduced pressure, and the residue was purified by flash column chromatography (MeOH: CHCl3 1:9) to afford the product (4.40 g, 60%). 1H NMR (CDCl3) δ 7.93 (m, 1H), 5.37 (m, 4H), 4.17 (m, 1H), 3.24 (m, 4H), 1.98 (m, 9H), 1.51 (m, 4H), 1.47 (s, 9H), 1.26 (m, 45H), 0.87 (m, 6H). ESI-MS m/z 719.1 ([M+H]+).
DOA (dioleyl amido aspartic acid)
Trifluoroacetic acid (6 mL) was added to a solution of 4 (4.4 g, 6 mmol) in anhydrous CH2Cl2 (6 mL). After stirring for 2 hours, the reaction mixture was diluted with CH2Cl2 (50 mL) and a saturated aqueous solution of Na2CO3 (20 mL) was added until the gas evolution ceased. The organic layer was then separated and washed with brine. Next, the organic layers were dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure. The residue was purified by flash column chromatography (MeOH:CHCl3 1:9) to afford the product (3.70 g, 95%). 1H NMR (CDCl3) δ 7.53 (m, 1H), 6.33 (m, 1H), 5.37 (m, 4H), 3.67 (m, 1H), 3.22 (m, 4H), 1.98 (m, 9H), 1.50 (m, 4H), 1.26 (m, 45H), 0.87 (m, 6H). ESI-MS m/z 641.0 ([M+Na]+).
Compound 6
A mixture of Boc-NH2-PEG-NHS (350 mg, 0.1 mmol) and DOA (63.1 mg, 0.1 mmol) in anhydrous CH2Cl2 (1 mL) was stirred vigorously at room temperature for 2 hours. The solution was precipitated in cold diethyl ether and the precipitate was washed again by diethyl ether. Anhydrous CH2Cl2 (1 mL) and TFA (1 mL) were added to the precipitate and the mixture was stirred for 2 hours. The reaction mixture was diluted with CH2Cl2 (10 mL) and a saturated aqueous solution of Na2CO3 (2 mL) was then added until the gas evolution ceased. The organic layer was separated and precipitated in cold diethyl ether. The obtained precipitate was washed again by diethyl ether.
Compound 7
A mixture of compound 3 (72 mg, 0.2 mmol), PEG-DOA (300 mg, 0.1 mmol), DCC (41.2 mg, 0.2 mmol), and DMAP (12 mg, 0.1 mmol) in anhydrous CH2Cl2 (1 mL) was stirred vigorously at room temperature for 5 hours. The solution was precipitated in cold diethyl ether and washed again with diethyl ether to obtain compound 7 (245 mg, 73%).
Compound 8
A mixture of compound 1 (304 mg, 1 mmol), succinic anhydride (200 mg, 2 mmol), and DMAP (12 mg, 0.1 mmol) in anhydrous CH2Cl2 (1 mL) was stirred vigorously at room temperature for 5 hours. The residue was purified by flash column chromatography (ethyl acetate) to afford the product (3.70 g, 95%). 1H NMR (CDCl3) δ 10.9 (brs, 1H), 7.96 (s, 1H), 7.15 (s, 1H), 6.79 (m, 2H), 4.96 (m, 1H), 3.83 (s, 3H), 3.04 (m, 2H), 2.72 (m, 2H), 2.35 (m, 2H), 2.29 (s, 3H), 2.24 (m, 2H), 2.08 (m, 1H), 1.64 (m, 3H), 1.50 (m, 4H).
Compound 9
A mixture of compound 8 (75 mg, 0.2 mmol), PEG-DOA (300 mg, 0.1 mmol), DCC (41.2 mg, 0.2 mmol), and DMAP (12 mg, 0.1 mmol) in anhydrous CH2Cl2 (1 mL) was stirred vigorously at room temperature for 5 hours. The solution was precipitated in cold diethyl ether and washed again with diethyl ether to obtain compound 9 (212 mg, 65%).
Preparation of rhodamine-PE labeled liposomes
Rhodamine-PE labeled liposomes with or without SV119 were prepared by thin lipid film hydration followed by probe sonication. Unmodified liposomes were composed of SPC:Chol:DSPE-PEG2000:rhodamine-PE in a 7:3:0.5:0.05 molar ratio. A chloroform solution of the lipid components was mixed and evaporated under a gentle stream of N2 followed by vacuum for at least 4 hours. The dried lipid films were hydrated with DPBS at 4°C overnight with an initial total lipid concentration of 16 mM. After a brief vortex, the suspension was then probe sonicated for 1 hour at a power of 3 watts. The liposomes were then passed through a PD-10 desalting column and used for fluorescence microscopic and quantitative liposome uptake studies. The preparation of the SV119 liposomes followed the same procedures, except the DSPE-PEG2000 was partially substituted by a DOA-PEG(3500)-SV119 conjugate. The size distribution and ζ-potentials of the liposomes were monitored with a dynamic light scattering method using the Malvern Zetasizer (Nano-ZS; Malvern Instruments Ltd, Worcestershire, UK).
Cellular uptake studies
Cells (2 × 104 per well) were seeded onto 48-well plates (Corning Inc, Corning, NY) and allowed to attach overnight. The complete medium was replaced with the serum-free DMEM containing rhodamine-PE labeled liposomes at a final total lipid concentration of 800 μM. Unmodified or SV119 liposomes were incubated with cells at 37°C for 1 hour unless otherwise indicated. After incubation, the cells were washed three times with ice cold PBS and lysed with 200 μl 0.3% Triton X-100 at room temperature for 0.5 hours. The fluorescence intensity of the cell lysate was determined by a PerkinElmer LS 50B luminescence spectrometer (Waltham, MA) (ex = 560 nm; em = 588 nm). The results were expressed as arbitrary fluorescence intensity/mg of protein lysates. All the numbers were subtracted by the autofluorescence of the cells incubated in the serum-free medium without any formulations. The results are reported as the average values of four wells measured from the same plate on the same day.
To evaluate the cellular uptake by microscopy, the cells were fixed with 4% paraformaldehyde in DPBS at 4°C for 20 minutes and washed three times by DPBS. The fluorescence images were obtained using a Nikon fluorescence-phase contrast-optical microscope.
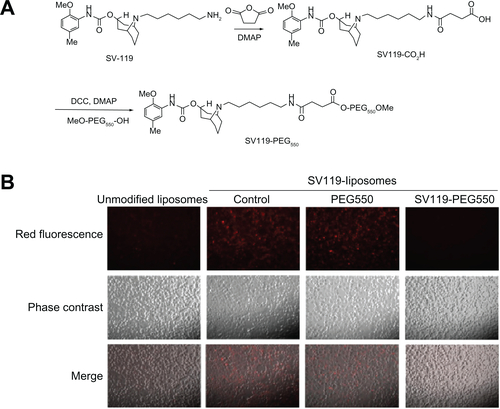
To investigate the effect of free SV119 on the uptake of SV119 liposomes, SV119 was conjugated with PEG550 to increase its water solubility (Figure S1A). The effect of SV119 on the uptake of SV119 liposomes was examined by incubating the DU-145 cells with the liposomes in serum-free media for 1 hour at 37°C in the presence of SV119-PEG550 (200 μM) or PEG550 (200 μM), and a fluorescence microscopy study was carried out as described above.
Preparation of DOX-loaded liposomes
DOX was encapsulated into unmodified and SV119 liposomes using an ammonium sulphate gradient method as previously described.Citation25 The components of the blank liposomes were similar to the liposomes described in the section titled “Preparation of rhodamine-PE labeled liposomes.” The percentage of SV119-PEG3500-DOA in the total lipid of targeted liposomes was 5 mole%. Instead of DPBS, 250 mM ammonium sulfate solution was used to hydrate the lipid membrane. DOX (10 mg/mL) dissolved in 5% glucose was mixed with the liposomes and incubated at 37°C for 20 minutes. The drug/lipid ratio was kept at approximately 100 μg/μmol to achieve an encapsulation efficiency greater than 90%.Citation25 Nonentrapped DOX was removed by filtration through a PD-10 column. After the solubilization of the liposomes in 1% (w/v) Triton X-100, the DOX concentration of the liposomes was determined by measuring the absorbance at 480 nm using free DOX solution as a standard.
Cytotoxicity assay
The viability of cells treated by DOX encapsulated in SV119 liposomes or unmodified liposomes was measured by MTT colorimetric assay. DU-145 cells were trypsinized and resuspended in serum-free DMEM. The formulations were diluted in a serum-free medium and mixed with DU-145 cell suspension, followed by incubation at 37°C for 1 hour. The medium was then removed by centrifugation. The cell pellets were resuspended in a complete medium and plated on a 96-well plate with a density of 5 × 103 cells per well. The cells were cultured at 37°C for another 72 hours followed by an MTT assay. Twenty microliters of MTT solution (5 mg/mL in PBS) were added to each well and the cells were incubated for 4 hours at 37°C. The media were then removed and 150 μL DMSO was added to each well. Absorbance at 550/630 nm was measured using an Ultramark Microplate Imaging System (Bio-Rad Laboratories, Hercules, CA). Six wells were used for each sample, and the data were expressed as the percentage of viable cells compared to the untreated control cells.
Intracellular localization
DU-145 cells grown to 60%–70% confluence in glass bottom dishes (In Vitro Scientific, Sunnyvale, CA) were incubated with rhodamine-PE labeled liposomes (with 3 mole% SV119-PEG-DOA) at 37°C for 3 hours or 16 hours. To detect early endosomes, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% saponin, and stained with anti-EEA1 (1:100), followed by FITC-conjugated anti-mouse IgG (1:100). To label the lysosomes in live cells, LysoTracker Green® (50 nM; Life Technologies) was added to the medium 30 minutes before the incubation period concluded. The stained cells were rapidly washed with ice-cold PBS and observed using an Olympus FV1000 laser scanning confocal microscope (Olympus America Inc, Melville, NY).
Statistical analysis
Statistical analysis was conducted using a two-tailed unpaired Student’s t-test with the SPSS software (version 11.5; IBM Corporation, Armonk, NY). The data were expressed as mean ± SD and differences of P < 0.05 were considered significant.
Results and discussion
Several selective sigma-2 receptor ligands have recently been developed and evaluated in vitro and in vivo. For example, Mach and Wheeler reported the synthesis of a series of N-substituted 9-azabicyclo[3.3.1]nonan-3a-yl phenylcarbamate analogs.Citation26 Several high-affinity sigma-2-selective ligands such as SV119 were identified through structure-activity relationship (SAR) studies. SV119 has a free amino group that is linked to a bridgehead nitrogen atom via a linker group of six methylene units. SAR studies suggest that the amino group is a preferred substituent for assuring a high affinity for σ2 receptors and high σ1 : σ2 selectivity. Nevertheless, various SV119 conjugates were synthesized by chemically modifying this amino group. For example, coupling SV119 with a fluorescence molecule via this amino group led to the development of a sigma-2-selective probe – SW-120 – which was used to effectively assess the sigma-2 receptor expression levels in cultured tumor cells.Citation27 In addition, a recent study by Spitzer et al showed that coupling SV119 with a proapoptotic peptide via this amino group similarly led to the development of a conjugate that demonstrated selective cytotoxicity towards tumor cells that overexpress the sigma-2 receptor.Citation28
In order to develop lipid-based nanoparticles that incorporate SV119 on the surface, we designed and synthesized a SV119-derivatized PEG-lipid. DOA is a double-chain lipid that is synthesized from oleyl amine and aspartic acid and can be inserted into the lipid bilayer of liposomes. The PEG moiety on liposomes provides steric hindrance, which renders the nanoparticles more stable and minimizes nonspecific binding of the liposomes on the cell surface.Citation29 After replacing the amine group with a carboxyl group, a SV119-PEG-DOA conjugate was synthesized by covalent linkage between the SV119 carboxyl group and the PEG terminal amine group. illustrates the synthesis scheme of SV119-PEG3500-DOA.
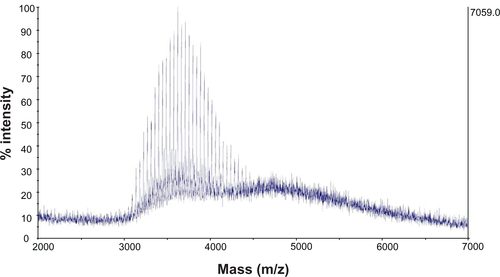
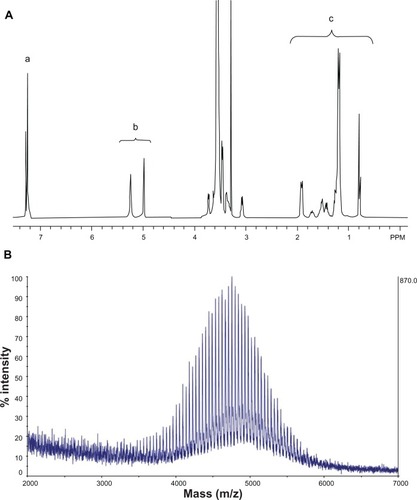
shows the NMR spectrum of synthesized SV119-PEG3500-DOA. The peaks at 7.2–7.3 ppm are attributed to the benzene groups on SV119, the signals at 5.0–5.2 ppm are attributed to the double bond of DOA, the peaks between 3–4 are attributed to the PEG, and the signals at 1–2 ppm are attributed to the carbon chain of DOA. The molecular weight of the synthesized product was determined by MALDI-TOF mass spectrometry. As shown in , the spectra exhibited a bell-shaped distribution of 44 Da-spaced lines centered at 4754.9 Da for SV119-PEG3500-DOA. A comparison between the mass spectrum of SV119-PEG3500-DOA () with PEG3500 (Figure S2) and PEG3500-DOA (Figure S3) suggests the successful synthesis of the SV119-PEG3500-DOA conjugate.
Figure 2 NMR spectrum (A) and MALDI-TOF mass spectrum (B) of SV119-PEG3500-DOA. The peaks in (A) are attributed to the benzene groups on SV119 (a), the double bonds of DOA (b), and the carbon chains of DOA (c).
Abbreviations: DOA, dioleyl amido aspartic acid; MALDI-TOF, matrix-assisted laser desorption/ionisation-time of flight; NMR, nuclear magnetic resonance; PEG, polyethylene glycol.
The synthesized SV119-PEG-DOA was used to formulate targeted liposomes. Liposomes are mainly composed of phospholipids and cholesterol. These molecules are also components of the cells and, thus, are believed to be biocompatible and non-toxic. The present experiment demonstrates that all the liposomal formulations have a mean diameter of approximately 90 nm (), which is optimal for penetrating the blood vessels surrounding the tumor and subsequent endocytosis by the tumor cells.Citation30
Table 1 Size and zeta potential of SV119 liposomes
The targeting efficiency of SV119 liposomes was characterized in vitro. Cellular uptake was studied using DU-145 cells as target cells. DU-145 is a human prostate cancer cell line and these cells are reported to overexpress sigma-2 receptors.Citation8 Human bronchial epithelial cells – BEAS-2B – were chosen to examine the liposomal uptake by normal cells. SV119-PEG3500-DOA was formulated into rhodamine-PE labeled liposomes to partially substitute PEG2000-DSPE, and the total amount of PEG-conjugated lipid in the liposomes was kept at 5 mole%. A longer spacer (PEG3500) was used between SV119 and DOA to minimize the potential steric hindrance imposed by DSPE-PEG2000 and to increase the targeting efficiency.Citation31
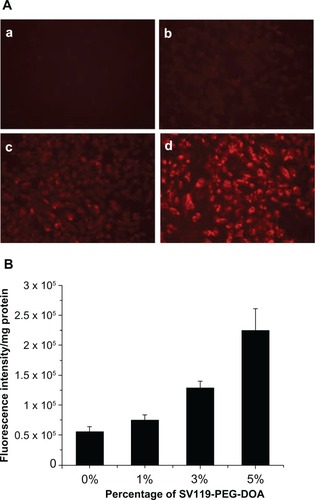
shows the fluorescence images of the DU-145 cells that were treated with rhodamine-PE labeled SV119 liposomes. After incubation with SV119 liposomes or unmodified liposomes for 1 hour, fluorescence was observed on the cell membrane and inside the cells. Increasing the amount of SV119 in the liposomes was associated with a significant increase in the intensity of rhodamine fluorescence. This is in contrast to the unmodified liposomes, which showed minimal binding and cellular uptake with the DU-145 cells (). shows the quantitative measurements of the cell-associated fluorescence 1 hour after the DU145 cells were treated with the SV119 liposomes and the unmodified liposomes. Consistent with the fluorescence imaging studies (), the amount of cell-associated fluorescence for the DU-145 cells increased significantly as the amount of ligand in the liposomes increased. In addition, the binding and uptake of SV119 liposomes was competitively blocked by the presence of the SV119-PEG550 conjugate, but was not inhibited by the same concentration of PEG550 (Figure S1B). These results demonstrate that the enhanced uptake of SV119 liposomes was mediated by SV119.
Figure 3 Enhanced intracellular uptake of SV119 liposomes by DU-145 cells, examined by microscopic study (A) and quantitative analysis (B) (n = 4, mean ± SD). In (A), the percentage of SV119-PEG-DOA in the total lipid was 0% (a), 1% (b), 3% (c), and 5% (d), respectively.
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
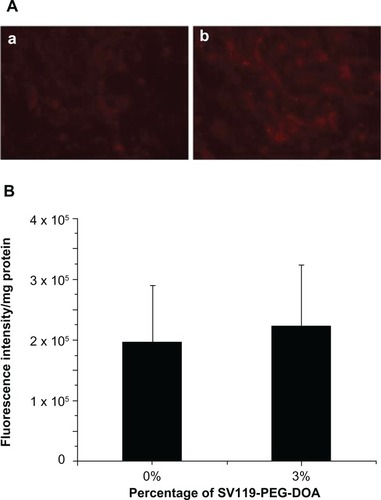
In order to demonstrate the cell-type specificity of the uptake of SV119 liposomes, normal human bronchial epithelial cells (BEAS-2B) were tested as a control. The results showed little binding and cellular uptake of SV119 liposomes and unmodified liposomes by the BEAS-2B cells (). No difference was found between the SV119 liposomes and unmodified liposomes in terms of the intensity of cell-associated fluorescence (). These observations suggest that SV119-mediated liposomal uptake is specific to tumor cells.
Figure 4 Cellular uptake of liposomes by BEAS-2B cells. Microscopic images (A), and quantitative analysis (B) (n = 4, mean ± SD). The percentage of SV119-PEG-DOA in the total lipid was 0% (a), or 3% (b).
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
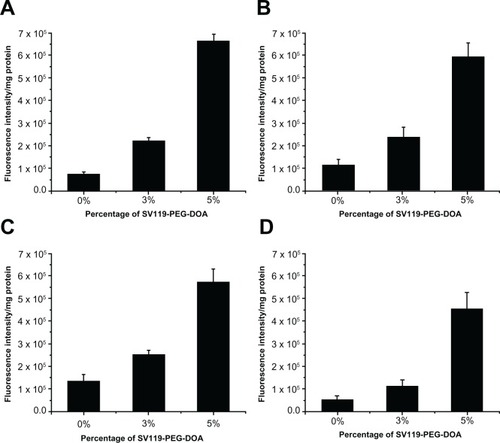
One of the unique features of the sigma-2 receptor is its overexpression in multiple types of cancers. Therefore, following the demonstration of the specific targeting of DU145 cells, we examined the targeting capacity of SV119 liposomes using several other cancer cell lines, including PC-3 (prostate cancer), MCF-7 (breast cancer), A549, and 201T (lung cancer) cells. Similar to the DU-145 cells, the cell-associated fluorescence intensity of the SV119 liposomes was much higher than that of the unmodified liposomes in all the tumor cell lines tested (), which suggests a potential for the SV119 liposomes to target multiple types of tumors.
Figure 5 Uptake of SV119 liposomes by MCF-7 (A), PC-3 (B), 201T (C), and A549 cells (D) (n = 4, mean ± SD).
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
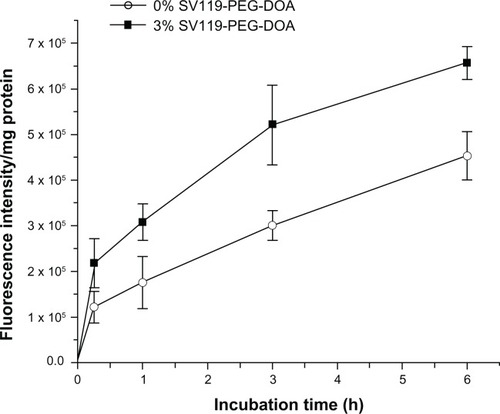
Next, we investigated the time course of liposomal uptake by the DU-145 cells. As shown in , the cell-associated fluorescence intensity increased during the prolonged incubation within 6 hours for the SV119 liposomes and the unmodified liposomes. Note that the cellular uptake of the SV119 liposomes was significantly higher at each time point compared to the unmodified liposomes. This observation further supports our conclusion that the incorporation of SV119 significantly enhances the uptake of liposomes into tumor cells.
Figure 6 Time-course of cellular uptake for SV119 liposomes.
Notes: Rhodamine-PE labeled liposomes with 0% or 3% SV119-PEG-DOA were incubated with DU-145 cells for an indicated period at 37°C. (n = 4, mean ± SD).
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
Like many other sigma-2 receptor ligands,Citation26 SV119 has a tertiary amine group. Therefore, incorporating SV119 onto the surface of liposomes may positively charge the particles, which may increase the nonspecific cellular uptake of the SV119 liposomes. However, the zeta potential analysis showed that the incorporation of up to 5 mol% of lipid-PEG-derivatized SV119 into the liposomes did not significantly alter the surface charge (), which excluded the possibility that the enhanced uptake was mediated by a non-specific charge-charge interaction. Furthermore, to minimize the protonation of the amine nitrogen, a modified SV119 (mSV119) was synthesized by introducing an adjacent carbonyl group, and the mSV119 was similarly conjugated with PEG3500-DOA for incorporation into the liposomes (). This modification did not affect the targeting efficiency of the ligand (). This result may shed new light on the structure-activity relationship, which may aid in the design of new sigma-2 receptor ligands.
Figure 7 Uptake of liposomes decorated with modified SV119 (mSV119) by DU-145 cells. Scheme of synthesis (A) and microscopic images (B) (n = 4, mean ± SD). In (B), the percentage of mSV119-PEG-DOA in the total lipid was 0% (a), 1% (b), 3% (c), and 5% (d), respectively.
Abbreviations: DOA, dioleyl amido aspartic acid; PEG, polyethylene glycol.
After demonstrating the efficiency and specificity of SV119 in mediating the uptake of liposomes by tumor cells, we investigated the utility of the SV119 liposomes for the targeted delivery of anticancer agents. We chose DOX because it is a chemotherapeutic drug widely used in clinical therapy. The DU-145 cells were pre-treated with DOX-loaded SV119 liposomes (SV119-L[D]), DOX-loaded unmodified liposomes (L[D]) and free DOX for 1 hour. The cells were then cultured for an additional 72 hours before the MTT assay was conducted. The results show that the cytotoxic effects of all the treatments were dose-dependent at concentrations of 0.2∼10 μg/mL (). At each of the indicated concentrations, the viability of the cells treated with L[D] showed enhancements of 1.5–3-fold, compared with that of the free DOX, which suggests that the cells were protected by the encapsulation of the drug in the liposomes. In addition, SV119-L[D] exhibited an ∼2-fold higher cytotoxicity to the DU-145 cells than L[D] (), which suggests improved efficacy of the SV119-modified liposomes versus the unmodified liposomes for delivering DOX to the tumor cells. The increased cytotoxicity of SV119-L[D] is consistent with the enhanced uptake of the SV119 liposomes, as shown in . This demonstrates that the SV119 liposome can potentially be used a carrier for the targeted delivery of chemotherapeutics to tumor cells.
Figure 8 Cytotoxic effect of free and liposomal DOX on DU-145 prostate cancer cells. Cells were treated with free DOX, DOX-loaded SV119 liposomes or DOX-loaded unmodified liposomes with DOX concentrations of 0.2, 1, or 10 μg/mL. Cytotoxicity was assessed by MTT assay. Cells receiving no treatment were defined as the maximal cell viability. Values presented are the mean ± SD of six replicates.
Note: *P < 0.05 compared to the control treatments with unmodified liposomes.
Abbreviations: DOX, doxorubicin; L[D], DOX-loaded unmodified liposomes.
![Figure 8 Cytotoxic effect of free and liposomal DOX on DU-145 prostate cancer cells. Cells were treated with free DOX, DOX-loaded SV119 liposomes or DOX-loaded unmodified liposomes with DOX concentrations of 0.2, 1, or 10 μg/mL. Cytotoxicity was assessed by MTT assay. Cells receiving no treatment were defined as the maximal cell viability. Values presented are the mean ± SD of six replicates.Note: *P < 0.05 compared to the control treatments with unmodified liposomes.Abbreviations: DOX, doxorubicin; L[D], DOX-loaded unmodified liposomes.](/cms/asset/2b001240-59fa-40ab-a57b-e3cf49d4724c/dijn_a_31981_f0008_b.jpg)
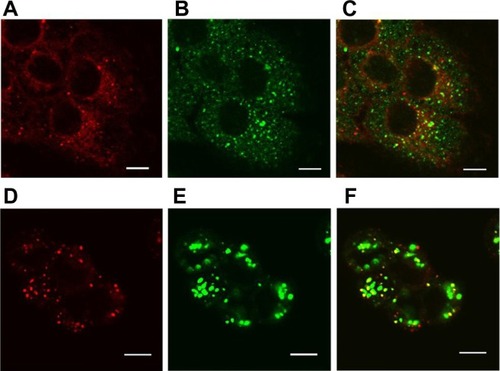
Despite the enhanced cellular uptake of the SV119 liposomes, it is crucial to understand the intracellular fate of the liposomes in order to maximize the bioavailability of the loaded drug. Nanoparticles internalized via the endocytosis pathway are often sequestered in endosomes and lysosomes, which represent an intracellular barrier that limits the accessibility of encapsulated agents to reach their molecular targets. In this study, confocal microscopy was used to examine the intracellular localization of rhodamine-PE labeled SV119 liposomes in DU-145 cells. After 3 hours of incubation, the liposomes showed a peri-nuclei punctuated distribution that is substantially co-localized with early endosomes (). After 13 hours of additional incubation, the liposomes were largely accumulated in the lysosomes, as evidenced by the colocalization with the lysosome markers (LysoTracker®; ). These observations are consistent with the internalization via the endocytosis pathway. Future studies are warranted to improve the formulation and to achieve more efficient release from the endosomes and lysosomes.
Figure 9 Localization of SV119 liposomes in DU-145 cells. Cells were incubated with rhodamine-PE labeled SV119 liposomes for 3 hours (A–C) or 16 hours (D–F) in a serum-free medium, stained with markers of early endosomes (anti-EEA1; A–C) or lysosomes (LysoTracker®; Life Technologies, Carlsbad, CA) (D–F), and imaged using confocal microscopy. Red fluorescence represents rhodamine-PE labeled liposomes (A and D), green fluorescence represents early endosomes B) or lysosomes (E). Yellow color observed in the red + green overlay (C and F) indicates colocalization of red liposomes with the green endosomes (C) or lysosomes (F).
Note: Scale bar = 10 μm.
Conclusion
We have demonstrated the successful synthesis of the functional lipid, SV119-PEG-DOA, as well as its incorporation into a liposome formulation for targeted delivery to cancer cells. As compared with unmodified liposomes, the SV119 liposomes demonstrated enhanced uptake in multiple types of tumor cell lines including DU-145, MCF-7, PC-3, 201T, and A549 cells. In addition, this enhanced uptake was shown to be dependent on ligand density and incubation time, and the uptake of SV119 liposomes was dramatically inhibited by the presence of free SV119. In contrast, the enhanced uptake of SV119 liposomes was not observed in BEAS-2B normal cells, which suggests that enhanced uptake is tumor cell-specific. Furthermore, results of the MTT assay suggest that incorporating SV119 onto the surface of liposomes significantly enhances the delivery and cytotoxicity of liposomal DOX in vitro. At the time of writing this manuscript, Xia and colleagues reported that SV119 mediated the selective delivery of SV119-gold nanocage conjugates to tumor cells.Citation32 Collectively, these data suggest that SV119-conjugated nanoparticles may represent a promising delivery system for the targeted delivery of anticancer therapeutics to tumor cells.
Acknowledgements
This work was supported in part by NIH grants (R21CA128415 and R21CA155983) and a DOD grant (BC09603).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary materials
Synthesis of SV119-PEG550
SV-119 succinate
A solution of SV-119 (2.0 g, 5 mmol) in CH2Cl2 (20 mL) was treated with DMAP (1.2 g, 10 mmol) and succinic anhydride (1 g, 10 mmol). The reaction was reflexed and stirred overnight. After the reaction was completed, the crude product was purified by flash chromatography with silica gel (EtOAc: PE, 3:1) to obtain SV-119 succinate.
SV119-PEG550
A solution of SV-119 succinate (2.5 g, 5 mmol) in CH2Cl2 (10 mL) was treated with DCC (0.82 g, 4 mmol), DMAP (0.12 g, 1 mmol), and MeO-PEG550-OH (2 g, 3.6 mmol). The reaction was stirred at room temperature overnight. After the reaction was completed, 100 mL of Et2O was added and the mixture was filtered to afford the crude product. The crude product was purified by flash chromatography with silica gel (MeOH: CH2Cl2, 1:10) to afford pure SV119-PEG550. 1H NMR δ 7.14 (s, 1H), 6.73–6.80 (m, 2H), 5.14 (m, 1H), 3.85 (s, 3H), 3.60–3.70 (m, 50H), 3.36 (s, 3H), 3.05–3.10 (m, 2H), 2.66–2.71 (m, 2H), 2.55–2.60 (m, 2H), 2.40–2.50 (m, 6H), 2.30 (s, 3H), 2.08–2.24 (m, 1H), 1.82–1.94 (m, 2H), 1.20–1.55 (m, 14H).
Method for testing the effect of SV119 on the uptake of SV119-liposomes
DU-145 cells were treated with SV119-liposomes (with 3% SV119-PEG-DOA) in a serum-free Dulbecco’s modified Eagle’s medium (DMEM; American Type Culture Collection [ATCC], Manassas, VA) for 1 hour in the presence of PEG550 (200 μM) or SV119-PEG550 (200 μM). The cells were treated with unmodified liposomes or SV119-liposomes alone as control groups. Fluorescence images were taken, as described in the section titled “Cellular uptake studies.”
Figure S1 Scheme of synthesis for SV119-PEG550 (A) and effect of SV119 on the uptake of SV119-liposomes by DU-145 cells (B).
Abbreviation: PEG, polyethylene glycol.
References
- WangAZGuFZhangLBiofunctionalized targeted nanoparticles for therapeutic applicationsExpert Opin Biol Ther2008881063107018613759
- WangXLiJWangYA folate receptor-targeting nanoparticle minimizes drug resistance in a human cancer modelACS Nano2012586184619421728341
- LeeRJLowPSFolate-mediated tumor cell targeting of liposome-entrapped doxorubicin in vitroBiochim Biophys Acta1995123321341447865538
- MoffattSPapasakelariouCWiehleSCristianoRSuccessful in vivo tumor targeting of prostate-specific membrane antigen with a highly efficient J591//PEI//DNA molecular conjugateGene Ther200613976177216453011
- ZhangYFWangJCBianDYZhangXZhangQTargeted delivery of RGD-modified liposomes encapsulating both combretastatin A-4 and doxorubicin for tumor therapy: In vitro and in vivo studiesEur J Pharm Biopharm201074346747320064608
- MauriceTSuTPThe pharmacology of sigma-1 receptorsPharmacol Ther2009124219520619619582
- CobosEJEntrenaJMNietoFRCendanCMDel PozoEPharmacology and therapeutic potential of sigma(1) receptor ligandsCurr Neuropharmacol20086434436619587856
- JohnCSVilnerBJGeyerBCMoodyTBowenWDTargeting sigma receptor-binding benzamides as in vivo diagnostic and therapeutic agents for human prostate tumorsCancer Res199959184578458310493511
- JohnCSBowenWDSagaTA malignant-melanoma imaging agent – synthesis, characterization, in-vitro binding and biodistribution of Iodine-125-(2-piperidinylaminoethyl)4-IodobenzamideJ Nucl Med19933412216921758254405
- BanerjeeRTyagiPLiSHuangLAnisamide-targeted stealth liposomes: A potent carrier for targeting doxorubicin to human prostate cancer cellsInt J Cancer2004112469370015382053
- NakagawaOMingXHuangLJulianoRLTargeted intracellular delivery of antisense oligonucleotides via conjugation with small-molecule ligandsJ Am Chem Soc2010132268848884920550198
- LiSDHuangLTargeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cellsMol Pharm20063557958817009857
- LiSDChonoSHuangLEfficient oncogene silencing and metastasis inhibition via systemic delivery of siRNAMol Ther200816594294618388916
- MukherjeeAPrasadTKRaoNMBanerjeeRHaloperidol-associated stealth liposomes: a potent carrier for delivering genes to human breast cancer cellsJ Biol Chem200528016156191562715695518
- VilnerBJJohnCSBowenWDSigma-1 and sigma-2 receptors are expressed in a wide variety of human and rodent tumor-cell linesCancer Res19955524084137812973
- WheelerKTWangLMWallenCASigma-2 receptors as a biomarker of proliferation in solid tumoursBr J Cancer20008261223123210735510
- MatsumotoRRSigma receptors: historical perspective and backgroundMatsumotoRRBowenWDSuTPSigma Receptors: Chemistry, Cell Biology and Clinical ImplicationsNew YorkSpringer2007124
- MachRHSmithCRAlNabulsiIWhirrettBRChildersSRWheelerKTSigma-2 receptors as potential biomarkers of proliferation in breast cancerCancer Res19975711561618988058
- ColabufoNABerardiFContinoMCorrelation between sigma2 receptor protein expression and histopathologic grade in human bladder cancerCancer Lett20062371838816005143
- KashiwagiHMcDunnJESimonPOSelective sigma-2 ligands preferentially bind to pancreatic adenocarcinomas: applications in diagnostic imaging and therapyMol Cancer200764817631687
- SpitzerDSimonPOKashiwagiHUse of multifunctional sigma-2 receptor ligand conjugates to trigger cancer-selective cell death signalingCancer Res201172120120922065721
- Di PaoloDPastorinoFBrignoleCDrug delivery systems: application of liposomal anti-tumor agents to neuroectodermal cancer treatmentTumori200894224625318564613
- VangveravongSXuJZengCMachRHSynthesis of N-substituted 9-azabicyclo[3.3.1] nonan-3α-yl carbamate analogs as σ2 receptor ligandsBioorg Med Chem200614206988699716837201
- EliazRESzokaFCJrLiposome-encapsulated doxorubicin targeted to CD44: a strategy to kill CD44-overexpressing tumor cellsCancer Res20016162592260111289136
- MachRHWheelerKTDevelopment of molecular probes for imaging sigma-2 receptors in vitro and in vivoCent Nerv Syst Agents Med Chem20099323024520021357
- XuJZengCChuWIdentification of the PGRMC1 protein complex as the putative sigma-2 receptor binding siteNat Commun2011238021730960
- SpitzerDSimonPOKashiwagiHUse of multifunctional sigma-2 receptor ligand conjugates to trigger cancer-selective cell death signalingCancer Res201272120120922065721
- LasicDDMartinFJStealth LiposomesBoca RatonCRC Press1995
- GabizonAALiposome circulation time and tumor targeting: implications for cancer-chemotherapyAdv Drug Deliv Rev1995162–3285294
- ZhouWYuanXWilsonAEfficient intracellular delivery of oligonucleotides formulated in folate receptor-targeted lipid vesiclesBioconjug Chem20021361220122512440856
- WangYXuJXiaXSV119-gold nanocage conjugates: a new platform for targeting cancer cells via sigma-2 receptorsNanoscale20124242142422113350