Abstract
A highly sensitive and selective method for amplified electrochemical detection for hairpin-stem-loop structured target sequences was developed based on the temperature regulation of DNA hybrids on a sandwich-type electrochemical DNA sensor. Multistep hybridization was applied to promote the hybridization efficiency of each section of sandwich structure. The results showed that both multistep and temperature-controlling hybridization techniques were both especially made to fabricate the sensor for the tendency of internal hybridization of target gene sequences. This strategy provides significantly enhanced hybridization efficiency and sequence specificity of electrochemical detection.
Introduction
The sequence-specific detection of DNA has gained widespread attention in the areas of molecular diagnostics, drug testing and screening, environmental monitoring, and DNA damage analysis over the past decades.Citation1–Citation4 Consequently, a variety of DNA sensors based on DNA hybridization principle was developed to monitor DNA hybridization, including optical, piezoelectric, and electronic methods.Citation5–Citation9 Among them, electrochemical DNA sensors have been rapidly developed and have been widely recognized as a promising solution for medical diagnostics, owing to the fact that electrochemical detectors are simple, sensitive, portable, and low cost.Citation10–Citation13 Already, many researchers have focused on improving the sensitivity and selectivity for the biosensor by designing the special probes or introducing advanced materials.Citation14–Citation17 In our previous work,Citation3,Citation18 we studied the electrochemical biosensor using a sandwich-type biosensor combined with enzyme-amplified technology, and found high sensitivity and selectivity. However, this one-step hybridization strategy was found to be disadvantageous for the special target sequences, which are easy to form secondary structure.
In the hybridization process, oligonucleotide secondary structure and intramolecular hybridization still play an important role in biological recognition processes.Citation19–Citation21 Various secondary structures form when the single-stranded specific target sequence has two or more stretches of complementary bases, including hairpin, which is common, loop-stem, and random-coil (called hairpin-stem-loop structure below).Citation22–Citation24 In recent years, the influence of hairpin-stem-loop structures on kinetics and dynamics mechanism of DNA hybridization have become popular and are widely researched in the solution phase as well as in the solid-liquid interface. Gao et alCitation25 have studied secondary structure effects on DNA hybridization kinetics by comparing the solution phase with the surface interface. The result shows that kinetic rate constants fluctuated with increasing probe and target secondary structure similarly in both solution and surface environments. Specifically, the addition of three intramolecular base pairs in the probe and target structure slows hybridization in solution-phase or on the surface. Surface hybridization rates are also 20- to 40-fold slower than solution-phase rates for identical sequences and conditions. Jo et alCitation26 have also investigated the effect of secondary structure on the hybridization activation energy. In their experiment, a reliable kinetic model was developed to understand how the rate-determining step is influenced by intramolecular secondary structures. They found that hybridization rate constants were determined by different temperatures and salt concentrations. Chen et alCitation27 and Chen et alCitation28 analyzed the origin of the specific recognition of structured DNA and investigated base thermodynamics and dynamics of DNA hybridization/dehybridization at a terminal of a DNA duplex. They proposed that the intermediate states are existent in hairpin folding and unfolding and that the hairpin opens in a concerted manner during the hybridization. Those theoretical studies suggest that hybridization would occur simultaneously for all molecules in an appropriate buffer solution. It implies that, in order to form the duplex, the hairpin-stem-loop should be unjointed in hybridization processes and respective melting temperatures should be comparable for all reactions that were taking place, because the hybridization temperature can impact the stability of double-stranded DNA.Citation27,Citation29,Citation30 In the previous one-step hybridization experiment, we have designed shorter single-stranded probe and target molecules and increasing the incubation temperature. Nevertheless, the result showed that reducing strand length could weaken detection selectivity and increasing temperature could degrade biosensor performance.
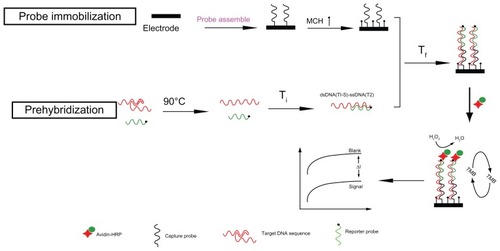
In this work, based on the complex clinical sample and practical application, we establish a simple, novel, low-cost, and temperature-controlled sandwich-like electrical enzyme-linked DNA biosensor to detect those hairpin-stem-loop structures, which were special target sequences, as shown in . In the sandwich-like hybridization process, firstly, the structured target sequences are opened and form single-stranded targets through incubation at 90°C. The single-stranded target sequences were then hybridized with the reporter probes through incubating at the temperature of Ti. This is a competitive process whereby the intramolecular hybridization is displaced by hybridization of the complementary strand. As a result, the stable and partly dsDNA(T1-S)-ssDNA(T2) complexes were formed immediately in solution phase. Following, the dsDNA(T1-S)-ssDNA(T2) complexes were hybridized with the capture probes on the electrode surface under the optimal lower temperature of Tf. Therefore, the stable and complete double helix structure (dsDNA(C-T-S)) can be formed with this multistep temperature-controlling process. Finally, the double helix structure was incubated with streptavidin-horseradish peroxidase (HRP) to amplify the electrochemical current from the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate for the detection of the structured target sequence.
Material and methods
Equipment and reagents
Electrochemical measurements were performed using a CHI1030A electrochemical workstation (Chenhua Co, Shanghai, China). A gold electrode (AuE, Φ = 2 mm) was used as the working electrode. A platinum wire and an Ag/AgCl electrode were used as the counter and reference electrodes, respectively.
The TMB substrate (Neogen K-blue low activity substrate) was from Neogen (Lansing, MI). Streptavidin-HRP was purchased from Beijing Biosynthesis Biotechnology (Beijing, China). Mercaptohexanol (MCH) and bovine serum albumin were from Sigma-Aldrich (St Louis, MO). Phosphate-buffered saline (PBS; 10 mM, pH 7.4) containing 1.0 M NaCl was used as the hybridization solution. The DNA immobilization buffer consisted of 10 mmolL–1 Tris-HCl (pH 7.4), 10 mM ethylene diamine tetraacetic acid (EDTA), and 1.0 M NaCl. All solutions were prepared with double-distilled water.
The synthetic DNA probe and special target sequence of Mycobacterium tuberculosis were purchased from TaKaRa Biotechnology (Dalian, China). The base sequences are shown in .
Table 1 Base sequences of the probes and various target oligonucleotide sequences
Fabrication of ssDNA/MCH/AuE
The AuE was polished with alumina slurry (0.05 μm) on a polishing pad, with ultrasonication in an HNO3 solution (1:1), ethanol and water, successively. The polished AuE was dried with N2 immediately before use and then run for 20 cycles of cyclic voltammetry (CV) in fresh 0.5 M H2SO4 solution to clean the AuE. The electrochemically polished AuE was then rinsed with copious amounts of double-distilled water and dried with N2. The cleaned AuE was then immobilized with 4 μL of capture probe (C) at a 1 μM concentration for 1 hour at room temperature. Subsequently, the capture probe-modified AuE was dipped in 2 mM MCH for 1 hour to obtain the ssDNA/MCH/AuE complex.
Prehybridization of the target DNA
One hundred microliters of hybridization solution containing the target DNA (T) and 50 nM of the reporter probe (S) was heated at 90°C for 5 minutes. The solution was then removed quickly and incubated at 50°C for another 30 minutes to form the partial double helix structure.
The AuE-modified capture probe was dipped in the hybridization solution at 45°C for 40 minutes to form the sandwich-type dsDNA. The AuE was then rinsed carefully with 10 mM PBS buffer and water to remove the remaining DNA, generating the dsDNA/MCH/AuE complex.
Electrochemical detection
The dsDNA/MCH/AuE complex was incubated in a 1 mg/mL bovine serum albumin solution at room temperature for 30 minutes to close the residual nonspecific adsorption. It was then rinsed with PBS buffer and dried with N2. Three microliters of streptavidin-HRP (0.5 U/mL) was dropped on the electrode surface and allowed to react for 15 minutes. The AuE was then stirred and cleaned with PBS buffer containing 0.05% Tween-20 (Biotech, Bio Basic Inc, Toronto, Canada). The formed sandwich biosensor was then subjected to the TMB substrate for electrochemical current–time curves measurement. The initial potential was 0 V; the sampling interval was 0.1 seconds; the sampling time was 100 seconds.
The ssDNA/MCH/AuE and dsDNA/MCH/AuE complex were respectively dipped into 10 mM Tris-HCl and 50 μM Ru[NH3]6+ solution at pH = 8.0 (Tris-HCl buffer) for electrochemical chronocoulometry measurement. The initial potential was −0.5 V and the final potential was 0.2 V.
Results and discussion
Electrochemical responses of detection of DNA hybridization
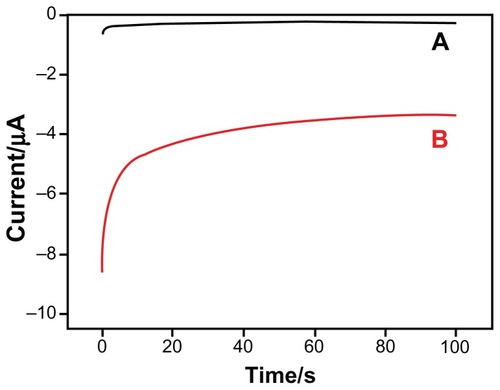
In this work, we compared the electrochemical response of the AuE in different substrate solutions to demonstrate the catalytic activity of HRP in the prepared sandwich-type DNA biosensor. As the current–time curves show in , the current value of the ssDNA-modified AuE (curve A) was lower than 100 nA. When the ssDNA/MCH/AuE complex was hybridized thoroughly with the target DNA sequence to form the sandwich-type DNA model and reacted with streptavidin-HRP (curve B), the current value increased significantly as compared with the ssDNA-modified AuE (curve A) in the TMB substrate solution. In the sandwich-type DNA biosensor model, the avidin-HRP conjugate was immobilized on the electrode surface and TMB was oxidized into a colored compound by H2O2 under the catalysis of HRP, leading to the high electrochemical response. The results illustrate that the constructed sandwich-type DNA biosensor could be used to detect the target DNA sequence successfully.
Figure 2 Current–time curves of the ssDNA-modified electrodes dripped 3 μL streptavidin-HRP in 500 μL TMB substrate before and after hybridization with target sequence. Curve (A) indicates the ssDNA-modified electrode without hybridization; Curve (B) indicates the ssDNA-modified electrode with hybridization.
Abbreviations: HRP, horseradish peroxidase; ssDNA, single-stranded DNA; TMB, 3,3′,5,5′-tetramethylbenzidine.
Formation of the dsDNA(T1-S)-ssDNA(T2) complex through multistep and temperature-controlling hybridization process
Oligonucleotides DNA hybridization/dehybridization is a fundamental process found in biology. The temperature of the hybrid reaction was a key factor that directly influenced the hybridization efficiency. Duplex DNA structure was generally formed under the optimum temperature and separated at the denaturation temperature. The typical method to control the temperature was by heating the mix hybridization solution, which contained the capture probe, reporter probe, and target sequence.
For the specific target sequence with internal hybridized loop, the multistep temperature-controlling process involved both denaturing of the internal hybridized bases of the target, as well as the formation of the target-reporter duplex, where the target-reporter binding was the driving force for the opening of the loop. The response of the biosensor was dependent on the stability of the dsDNA(T1-S)-ssDNA(T2) complex. In order to confirm the improved hybridization efficiency and the formation of dsDNA(T1-S)-ssDNA(T2) complex with this multistep temperature-controlling method, the current–time curves of the prehybridization step under different temperatures were studied. Before that, the target sequences solution was incubated at 90°C for 5 minutes to denature the internal hybridized bases of the target. The results in describe the sensor response for various temperatures of prehybridization. It was observed that the current values were still <1700 nA at temperatures below 49°C, illustrating poor hybridization efficiency. A suddenly significant amplification of the current was then observed to be 3420 nA at the prehybridization temperature of 50 °C. The value was twice that of the traditional one-step hybridization, reflecting good hybridization and the formation of a stable sandwich-type duplex. These results were likely due to the ability of target DNA sequences to easily refold and form partial internal hybrids in the solution phase at such low temperatures, instead of opening, whether denatured at 90°C or not. When the temperature point reached 50°C, the target sequences could be opened and hybridized with the reporter probes to form a stable dsDNA(T1-S)-ssDNA(T2) complex. As shown in , the current value reduced once the temperature was increased further, indicating that the complex was likely to be degenerated and unstable at the higher temperature. It is highly significant that all of the blank experiments showed similar results and the current values are all lower than 180 nA, indicating limited, nonspecific absorption of streptavidin-HRP. The results in clearly show that the temperature controlling of the prehybridization solution was essential for hybridization efficiency and current-response improvement for the multistep method.
Figure 3 Current responses of the different sandwich-type biosensors after hybridization with the complementary target sequence: (A) the prehybridization solution for multistep temperature-controlling method was first heated at 90°C and incubated at different temperatures: 37°C, 45°C, 48°C, 49°C, 50°C, 51°C, 54°C; (B) one-step or multistep temperature-controlling process at 45°C and 50°C, respectively; (C) multistep temperature-controlling process for dsDNA(T1-S)-ssDNA(T2) complex and dsDNA(T2-C)-ssDNA(T1) complex.
Abbreviations: T, target sequence; S, reporter probe; C, capture probe.
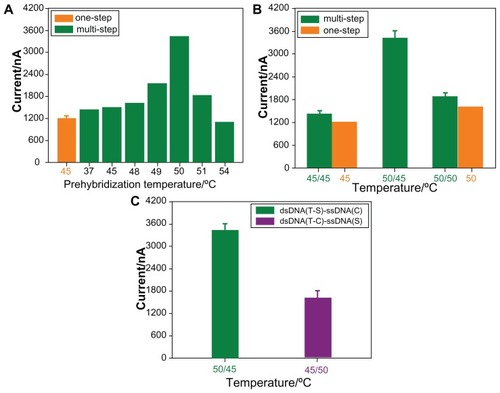
To demonstrate that the temperature of prehybridization and the multistep process both played important roles in the hybridization event, we compared the response of 45°C or 50°C through traditional one-step and novel multistep temperature-controlling methods, respectively. The results are shown in . The current was just 1219 nA for the one-step process (45°C) and 1430 nA for the multistep process (45°C/45°C), demonstrating low hybridization efficiency. A reason for this result was that internal hybridization may have occurred in the target sequence at 45°C, which discourages the hybridization between the capture probes on the AuE and the target sequences. In addition, the responses of the traditional one-step process (50°C) and novel multistep process (50°C/50°C) were 1634 nA and 1885 nA, respectively (). This was because the formed dsDNA(T1-S)-ssDNA(T2) strands were unstable and partially separated at the higher temperature of 50°C. However, significantly enhanced signal of 3420 nA could be observed for the multistep temperature-controlling process (50°C/45°C), indicating high hybridization efficiency can be achieved in this case. These results explained two problems: (1) the traditional one-step temperature control regime was disadvantageous for DNA recognition of hairpin secondary structure. In the case of special DNA structures, such as those that possess an internal double-helical state, the hybridization efficiency was directly reduced and the electrochemical response of the biosensor was weakened; and (2) the multistep temperature-controlling method could improve the current response only if the prehybridization and secondary hybridization temperature were controlled, respectively. The temperature control of each section of dsDNA complex should be considered for the multistep sandwich-type biosensor. Therefore, the special structures were necessary to improve the hybridization efficiency by multistep and temperature-controlling hybrid approaches. Consequently, the hybridization temperature played an important role in the multistep temperature-controlling event.
Furthermore, considering the importance of both the temperature-controlling and multistep processes for hybridization, we carried out another multistep process to make another dsDNA(T2-C)-ssDNA(T1) complex. First, we dipped the capture probe-modified electrode into the target sequence at a temperature of 45°C to form another dsDNA(T2-C)-ssDNA(T1) complex. This complex was then hybridized with the reporter probe at 50°C to form a sandwich-type biosensor. As shown in , the result was compared with the current value of the dsDNA(T1-S)-ssDNA(T2) complex. The current value generated by the sensor formed using this method was as low as 1616 nA. The response was not significantly increased, probably because the structure of the dsDNA(T2-C)-ssDNA(T1) complex was unstable and the target sequence was dissociated from the electrode surface when the secondary temperature was up to 50°C. Significantly, it was suggested that the double-stranded DNA with higher denaturing temperature, such as T-S, should be preferential for the prehybridization process to prevent another double-stranded DNA from denaturing in the following step.
Chronocoulometry was employed to quantify DNA surface density via measuring the redox charges of RuHex.Citation31 We evaluated the hybridization efficiency according to the added number of DNA strands on the electrode surface by electrochemical responses of [Ru(NH3)6]3+ bound to the sandwich-type DNA. As a result, at a concentration of complete target sequence 10 nM, the surface density of dsDNA on the gold surface for multistep temperature-controlling processes was almost twice that of the traditional method (), which demonstrates that the novel multistep method exhibited more excellent hybridization efficiency than the one-step method.
Table 2 The surface density of DNA and the differences of hybrid efficiency between multistep and one-step temperature-controlling hybridization methods
Specificity and sensitivity of the novel sandwich-type DNA biosensor
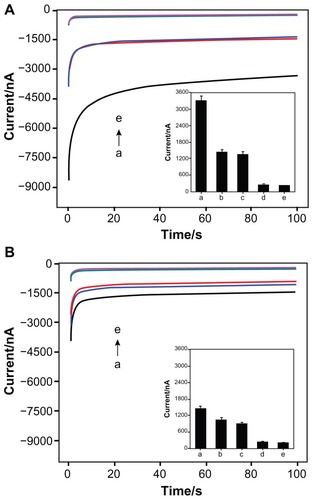
The ability of the DNA biosensor to differentiate the target sequence mismatches was important in the applications. The selectivity of the novel multistep temperature-controlling hybridization method was also investigated. In the experiment, complementary target DNA sequence (curve a), single-base mismatch (curves b and c), noncomplementary (curve d), and the capture probe (curve e) were detected through current–time curves. As shown in , in the present concentration of 10 nM target sequences, the values of the complementary matched DNA sequence (column a) was two times more than the response of two different types of single-base mismatch sequence (column b and c). This optimal difference shows a great ability to distinguish even the complementary target from single-base mismatch sequence. The current value from the noncomplementary DNA (column d) was as low as the blank (column e). Thus, we speculate it was the more strict hybridization temperature control that improved the recognition ability among the bases. The traditional one-step sandwich-type gene biosensor was also compared with the specific gene segment of tubercle bacillus. In , the bar graph from a to d represents the complementary, single-base mismatch (S4 and S5) and noncomplementary target sequences, respectively. However, it was not easy to distinguish the responses between the completely match and single-base mismatch sequences. These results clearly indicated that the novel sandwich-type DNA biosensor could satisfactorily perform the specificity assays, which showed enhanced specificity relative to the traditional sandwich-type DNA biosensor ().
Figure 4 Current–time curves for multistep temperature-controlling process (A) and one-step hybridization (B) with the complementary target (a), one-point mismatch target 1 (b), one-point mismatch target 2 (c), noncomplementary target (d) and the capture probe (e) sequences.
Note: Inserts show the bar graphs of the peak currents when the biosensor was hybridized with the different DNA sequences.
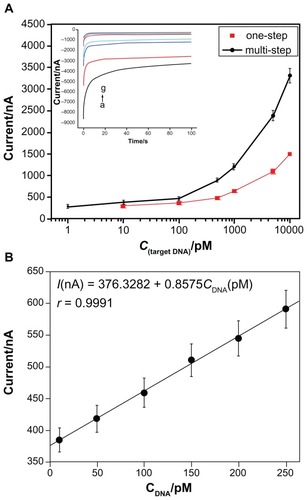
The current–time tests were employed for detection of different concentrations of target DNA sequence to further demonstrate the applicability of the novel sandwich-type DNA biosensor. The sensor was used to test the target DNA sequence at a series of concentrations ranging from 1 pM to 10 nM (, inset). The amperometric responses were found to be nonlinear related to the concentration of the target sequence (). As shown in , the slope of the linear relationship range of the response versus the concentration was 0.85, indicating that the novel biosensor showed higher sensitivity besides the improved selectivity. The detection limits for the novel sensor were observed with target concentration as low as 2.6 × 10−13 M. We assayed five different electrodes of 10 nM target sequence in parallel and the results showed Relative Standard Deviation (RSD) as 7.43%.
Figure 5 (A) The plot of currents for multistep temperature-controlling and one-step hybridization versus target DNA concentration. Insert shows amperometric measurements for the novel sandwich-mode electrochemical DNA biosensor hybridized with the synthetic target DNA at a series of concentrations (from a to g: 10 nM, 5 nM, 1 nM, 500 pM, 100 pM, 10 pM, and 1 pM, respectively). (B) The plot of current versus target DNA concentration (from 10 pM to 250 pM).
Notes: Error bars show the standard deviations of measurements taken from at least three independent experiments. The base solution for hybridization measurement was the TMB substrate.
Abbreviation: TMB, 3,3′,5,5′-tetramethylbenzidine.
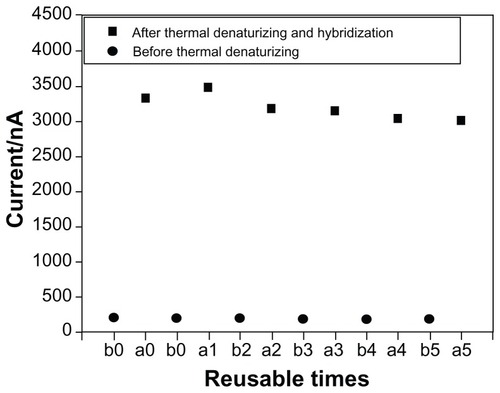
Finally, we tested the reusability of the sensor to support the application of the biosensor. The constructed biosensor was incubated at 90°C for 5 minutes to remove the hybridized target DNA sequence completely via thermal denaturizing. Importantly, the captured probe-modified AuE retained substantial performance even after five regeneration cycles ().
Figure 6 The plot of currents after five cycles of sandwich-type DNA biosensors before and after thermal denaturizing hybridization.
Notes: a0 stands for the captured probe-modified AuE; b0 for hybridization with 10 nM target sequence; b1, b2, b3, b4, b5 for each time of five cycles before thermal denaturizing hybridization, respectively; a1, a2, a3, a4, a5 for each time of five cycles after thermal denaturizing hybridization, respectively.
Conclusion
A sandwich-type DNA biosensor was developed with a modified hybridization process for detection of the hairpin-stem-loop structure sequence. The target sequence was firstly heat-denaturalizated and was then hybridized with the reporter probe to form a dsDNA(T1-S)-ssDNA(T2) complex under controlled temperature conditions. The formed dsDNA(T1-S)-ssDNA(T2) complex was subsequently hybridized with a capture probe immobilized on the electrode to fabricate the sandwich-type DNA biosensor under another controlled temperature. The results were compared to sandwich-type DNA biosensors formed using traditional methods, which showed that the novel multistep temperature-controlling process had higher sensitivity and better specificity. This novel sandwich-type electrochemical DNA biosensor demonstrated improved recognition of special disease-causing DNA sequences and would be especially suitable for the detection of target sequences that could form internal hybrids. It is expected that similarly fabricated sandwich-type DNA biosensors will be of significant use in a clinical setting in the near future.
Acknowledgments
The authors gratefully acknowledge the financial support of the National High Technology and Development of China (863 Project: 2012AA022604), the National Natural Science Foundation of China (20975021, 21175023, 81171668, 21275028), the Fujian Provincial University-Industry Cooperation Science and Technology Major Program (2010Y4003), the Foundation of Fujian Key Laboratory of Hematology (2009J1004), the Scientific Research Major Program of Fujian Medical University (09ZD013), the Natural Science Foundation of Fujian Province of China (2010J06011, 2012J01428) Medical innovation subject of Fujian Province of China (2012-CX-30) and the Science and Technology Project of Fuzhou City of China (2011-S-67-3).
Disclosure
The authors report no conflicts of interest in this work.
References
- MolijnAKleterBQuintWMolecular diagnosis of human papillomavirus (HPV) infectionsJ Clin Virol200532Suppl 14351
- MurrayVNguyenTVChenJKThe use of automated sequencing techniques to investigate the sequence selectivity of DNA damaging agentsChem Biol Drug Des20128011822416919
- WangKSunZLFengMJDesign of a sandwich-mode amperometric biosensor for detection of PML/RAR-fusion gene using locked nucleic acids on gold electrodeBiosens Bioelectron20112662870287621177095
- PalchettiIMasciniMNucleic acid biosensors for environmental pollution monitoringAnalyst2008133784685418575633
- LiuGWanYGauVAn enzyme-based E-DNA sensor for sequence-specific detection of femtomolar DNA targetsJ Am Chem Soc2008130216820682518459781
- LucarelliaFTombelliaSMinunniaMElectrochemical and piezoelectric DNA biosensors for hybridisation detectionAnal Chim Acta2008609213915918261509
- LubinAALaiRYBakerBRSequence-specific, electronic detection of oligonucleotides in blood, soil, and food stuffs with the reagentless, reusable E-DNA sensorAnal Chem200678165671567716906710
- CashKJHeegerAJPlaxcoKWOptimization of a reusable, DNA pseudoknot-based electrochemical sensor for sequence-specific DNA detection in blood serumAnal Chem200981265666119093760
- EndoTKermanKNagataniNLabel-free detection of peptide nucleic acid-DNA hybridization using localized surface plasmon resonance based optical biosensorAnal Chem200577216976698416255598
- LubinAAHuntBVSWhiteRJEffects of probe length, probe geometry, and redox-tag placement on the performance of the electrochemical E-DNA sensorAnal Chem2009812150215819215066
- LiuSJNieHGJiangJHElectrochemical sensor for mercury(II) based on conformational switch mediated by interstrand cooperative coordinationAnal Chem200981145724573019522530
- XiaoYLouXHUzawaTAn electrochemical sensor for single nucleotide polymorphism detection in serum based on a triple-stem DNA probeJ Am Chem Soc200913142153111531619807078
- PinijsuwanSRijiravanichPSomasundrumMSub-femtomolar electrochemical detection of DNA hybridization based on latex/gold nanoparticle-assisted signal amplificationAnal Chem200880176779678418665605
- ZhangJLaoRSongSDesign of an oligonucleotide-incorporated nonfouling surface and its application in electrochemical DNA sensors for highly sensitive and sequence-specific detection of Target DNAAnal Chem200880239029903319551931
- ZhangDDPengYQiHLLabel-free electrochemical DNA biosensor array for simultaneous detection of the HIV-1 and HIV-2 oligonucleotides incorporating different hairpin-DNA probes and redox indicatorBiosens Bioelectron20122551088109419850463
- PeiHLuNWenYLA DNA nanostructure-based biomolecular probe carrier plat form for electrochemical biosensingAdv Mater201022424754475820839255
- ZhangQDPiroBNoëlVApplications of carbon nanotubes to electrochemical DNA sensors: a new strategy to make direct and selective hybridization detection from SWNTsAdv Nat Sci Nanosci Nanotechnol20101418
- WangKChenJHChenJA sandwich-type electrochemical biosensor for detection of BCR/ABL fusion gene using locked nucleic acids on gold electrodeElectroanalysis2009211011591166
- SongGTRenJSRecognition and regulation of unique nucleic acid structures by small moleculesChem Commun2010463972837294
- KimJImCAJungYCSelf-assembled nucleic acid nanoparticles capable of controlled disassembly in response to a single nucleotide mismatchBiomacromolecules20101171705170920527811
- ZhouJBattigMRWangYAptamer-based molecular recognition for biosensor developmentAnal Bioanal Chem201039862471248020644915
- CarrascosaLGMontesSGAvinóASensitive and label-free biosensing of RNA with predicted secondary structures by a triplex affinity capture methodNucleic Acids Res2012408e5622241768
- KuznetsovSVRenCCWoodsonSALoop dependence of the stability and dynamics of nucleic acid hairpinsNucleic Acids Res20083641098111218096625
- HurleyLHSecondary DNA structures as molecular targets for cancer therapeuticsBiochem Soc Trans200129Pt 669269611709056
- GaoYWolfLKGeorgiadisRMSecondary structure effects on DNA hybridization kinetics: a solution versus surface comparisonNucleic Acids Res200634113370337716822858
- JoJJKimMJSonJTSingle-fluorophore monitoring of DNA hybridization for investigating the effect of secondary structure on the nucleation stepBiochem Biophys Res Commun20093851889319422793
- ChenXDZhouYQuPBase-by-base dynamics in DNA hybridization probed by fluorescence correlation spectroscopyJ Am Chem Soc200813050169471695219053418
- ChenCLWangWJWangZInfluence of secondary structure on kinetics and reaction mechanism of DNA hybridizationNucl Acids Res20073592875288417430963
- SchreinerSMHatchALShudyDFImpact of DNA surface interactions on the stability of DNA hybridsAnal Chem201183114288429521561066
- ŠpringerTŠípováHVaisocherováHShielding effect of monovalent and divalent cations on solid-phase DNA hybridization: surface plasmon resonance biosensor studyNucl Acids Res201038207343735120624817
- ZhangJSongSZhangLSequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodesJ Am Chem Soc2006128268575858016802824