Abstract
Intracellular processes, including endosomal escape and intracellular release, are efficiency-determining steps in achieving successful gene delivery. It has been found that the presence of acid-labile units in polymers can facilitate endosomal escape and that the presence of reducible units in polymers can lead to intracellular release. In this study, poly(amido amine)s with both bioreducible and acid-labile properties were synthesized to improve gene delivery compared with single-responsive carriers. Transfection and cytotoxicity were evaluated in three cell lines. The complexes of DNA with dual-responsive polymers showed higher gene transfection efficiency than single-responsive polymers and polyethylenimine. At the same time, these polymers were tens of times less cytotoxic than polyethylenimine. Therefore, a polymer that is both reducible and acid-labile is a promising material for efficient and biocompatible gene delivery.
Introduction
The development of a safe and efficient nonviral gene delivery system remains the greatest hurdle to overcome in developing successful gene therapy. Low transfection efficiency of nonviral gene delivery vectors results mainly from multiple extracellular and intracellular barriers.Citation1,Citation2 Among the intracellular barriers, endosomal escape has been considered one of the most significant.Citation3 The endosomal and lysosomal compartments are significantly acidic (pH 4.5–6.5)Citation4 and contain many degrading enzymes. Polyplexes easily degrade in this environment, resulting in low gene transfection. High molecular weight branched polyethylenimine (PEI) shows high transfection efficiency due to its strong buffering capacity over a wide range of pH levels, known as the “proton sponge effect”.Citation5 When PEI-based polyplexes are present in the endosome, protonation of amines leads to endosomal swelling and lysis, thus releasing the polyplexes into the cytoplasm. However, the high cytotoxicity and nondegradability of the branched PEI limit its practical application in clinical therapy.Citation6 Recently, acid-labile PEI was prepared and used in gene delivery with facilitated endosomal escape and biocompatibility.Citation7 The results showed that acid-labile PEI can degrade quickly in a low pH environment, as in the endosome, facilitating efficient endosomal escape. Hydrolysis of the acid-labile group contributes to endosomal disruption by osmotic pressure because of an increased number of small molecules in the endosome. Furthermore, degradation of the polymers reduces the possibility of generating undesirable cytotoxicity.Citation7,Citation8 This acid-labile polymer displays higher gene transfection and lower cytotoxicity, which makes it more suitable for gene delivery.Citation9
The other barrier is the efficient release of nucleic acids into the intracellular environment.Citation10 Once they have escaped from the endosome, polyplexes need to unpack nucleic acids efficiently to release them into the nucleus or a suitable site near the nucleus. A successful strategy to increase intracellular release is to introduce disulfide bonds into the polymer backbone. These disulfide bonds can be reduced by small redox molecules, such as glutathione and thioredoxin, either alone or with the help of redox enzymes.Citation11 The intracellular reduction results in an enhanced release rate, leading to increased availability of free nucleic acids to initiate their biological action.Citation11 Disulfide bonds become an attractive option for gene delivery for two main reasons, ie, their relative extracellular stability and easy intracellular reversibility.Citation12,Citation13 To test this theory, a series of reducible poly(amido amine)s (PAAs) containing various amounts of disulfide bonds was designed.Citation14 The results indicated that the introduction of disulfide bonds into the backbone of polymers enables fast intracellular fragmentation of the polymers, leading to facilitated gene unpacking and enhanced transfection efficiency. In addition, fast degradation of the reducible polymers leads to decreased cytotoxicity.Citation15,Citation16
It has been demonstrated that the presence of reducibleCitation15,Citation17–Citation20 or acid-labileCitation7,Citation8,Citation21–Citation23 units in a polymer can enhance gene delivery separately; however, preparation of a reducible polymer with acid-labile properties has not been reported as yet. We hypothesize that the efficiency of gene delivery will be further enhanced by combining reducible and acid-labile properties in the same polymer. In this study, we synthesized dual redox and acid-responsive polymers. This study suggests that a dual reducible and acid-labile polymer would be a promising gene delivery carrier offering substantially improved gene delivery efficiency and biocompatibility.
Materials and methods
An acid-labile monomer, [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea] (KDA), hexmethylenebisacrylamide (HMBA), Citation7 and N,N′-cystaminebisacrylamide (CBA)Citation24 were synthesized according to previous reports, and the details are described separately in the Supplementary Information section. Ethidium bromide (Alfa Aesar Chemicals, Ward Hill, MA), dithiothreitol (DTT, Alfa Aesar Chemicals), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT, Beijing Solarbio Science and Technology Co., Ltd, China), branched PEI (molecular weight 25,000, Sigma-Aldrich, St Louis, MO) and hexamethylene diamine (HDA, Sinopharm Chemical Reagent Co, Ltd, Shanghai, China) were used as received. Plasmid DNA, gWiz™ high expression luciferase, and gWiz high expression green fluorescent protein were purchased from Aldevron (Fargo, ND). SM105 Polystyrene standards were purchased from Shodex (New York, NY).
Synthesis of dual-responsive PAAs
All PAAs were synthesized by Michael addition of amines (KDA and/or HAD) with acrylamides (CBA and/or HMBA).Citation25 Polymerization was conducted in vials containing a methanol/water mixture (9/1, v/v) at 35°C for a period of 4 days. The reaction conditions and results are shown in . Typically, for the synthesis of DualR1, KDA (243 mg, 1.5 mmol, 1.0 equivalent) was added into a solution of CBA (390 mg, 1.5 mmol, 1.0 equivalent) in a methanol/water mixture (3.0 mL, 9/1, v/v). The reaction was allowed to proceed at 35°C in an argon atmosphere for 4 days. The final polymer was obtained via precipitation in ethyl ether/acetone (1/1, v/v) and drying in a vacuum for 4 hours at room temperature. Characterization of the PAAs is summarized in .
Table 1 Synthesis and characterization of poly(amido amine)s
Characterization of PAAs
1H nuclear magnetic resonance (NMR) spectra were performed on a Bruker AV 300 MHZ spectrometer in d6-dimethylsulfoxide (DMSO). Molecular weights and molecular weight distributions were determined by gel permeation chromatography using a series of three linear Styragel columns. A Waters 1515 pump and Waters 2414 differential refractive index detector were used (set at 35°C). Dimethylformamide was used as an eluent at a flow rate of 1.0 mL per minute, and polystyrene standards were used in the calibration of molecular weights and molecular distributions.
Hydrolysis of DualR1
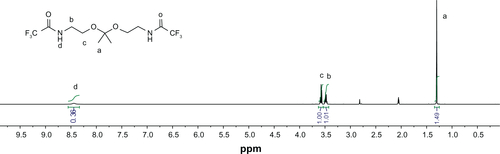
DualR1 was first dissolved in d6-DMSO (100 mg/mL), and the solution was diluted using a deuterated phosphate buffer (pH 5.0 and pH 7.4) to a final concentration of 10 mg/mL. The solutions were transferred to NMR tubes and the spectra were taken at different time intervals. Samples were incubated in a 37°C water bath between spectra acquisitions. The hydrolysis half-life of DualR1 was calculated by comparing the loss of the integral value of the ketal peak (1.34 ppm) with the integral value of the residual DMSO peak from 1H NMR spectroscopy using the Arrhenius equation.Citation26
Ethidium bromide exclusion assay
The ability of the synthesized PAAs to condense plasmid DNA was confirmed by measuring the changes in DNA/ethidium bromide fluorescence in the presence of PAAs at various N:P ratios. In brief, 50 μL DNA solution (20 μg/mL) was mixed with an equal amount of ethidium bromide solution (20 μg/mL) and incubated for 20 minutes. A PAA solution (10 μL) was added to the prepared DNA/ethidium bromide solution at different N:P ratios. Fluorescence was measured using a fluorescence microplate reader (Thermo Scientific Multiskan Flash, San Jose, CA) with λex = 520 nm, λem = 610 nm, and a 5 nm slit width. The fluorescence of DNA/ethidium bromide solution was set to 100% and the background fluorescence was set to 0% using ethidium bromide solution (1 μg/mL) alone.
Agarose gel electrophoresis assay
The ability of PAAs to release DNA from the polyplex in a low pH or reducing environment was evaluated by agarose gel electrophoresis assay. PAA solutions were mixed with 0.4 μg DNA in water (5 μL in total) at an N:P ratio of 10. The polyplexes were vortexed for 10 seconds and incubated at room temperature for 10 minutes. The polyplexes were then further incubated for 4 hours in 100 mM acetate buffer (pH 5.0) or 10 mM HEPES buffer (pH 7.4),Citation21 or 30 minutes with 10 mM DTT at 37°C. The complexes were loaded into a well on a 0.9% agarose gel. Electrophoresis was performed using Tris-acetate running buffer at 90 V for 50 minutes. DNA bands were visualized with ethidium bromide staining using a UVP ED3 imaging system.
MTT assay
The cytotoxicity of the PAAs was evaluated by MTT assay. HepG2, A549, and MDA-MB-468 cells were seeded into a 96-well plate at a density of 9000 per well in 100 μL Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. After 24 hours, polymers in phosphate-buffered solution were added to each well at final concentrations ranging from 5 μg/mL to 100 μg/mL. The medium was removed after 24 hours and replaced with a mixture containing 150 μL of fresh DMEM and 20 μL MTT (5 mg/mL in phosphate-buffered solution). The cells were incubated for 4 hours, and the medium was replaced with 200 μL of DMSO to dissolve the formazan crystal. The relative viability of the cells in each well was measured by ultraviolet absorbance at 493 nm. EC50 was calculated by online software ED50 and IC50 for Graded Response version 1.2 (http://chiryo.phar.nagoya-cu.ac.jp/javastat/Graded50-e.htm).
Luciferase and green fluorescent protein transfection
HepG2, A549, and MDA-MB-468 cells were seeded into a 96-well plate at a density of 8500 cells per well and incubated for 24 hours. The medium was then replaced with 50 μL of fresh fetal bovine serum-free DMEM. To prepare the polyplex, the polymer solution was added into plasmid DNA solutions at the desired N:P ratios and incubated for 20 minutes. Mixtures of 100 μL fresh fetal bovine serum-free DMEM and 10 μL complexes containing 0.16 μg DNA at various N:P ratios were added into each well. After incubating for 4 hours, the medium containing the complexes was replaced with 200 μL DMEM containing 10% fetal bovine serum, and the cells were incubated for another 48 hours. For green fluorescent protein plasmid DNA transfection, the HepG2 cells were analyzed for green fluorescent protein expression with an inverted fluorescence microscope (Olympus BX52). Quantitative transfection efficiency was conducted by flow cytometry at an optimal PAA to DNA ratio determined by fluorescence microscopy. HepG2 cells were seeded into a 24-well plate at a density of 50,000 cells per well and incubated for 24 hours. The medium was then replaced with 200 μL fresh fetal bovine serum-free DMEM. Mixtures of 275 μL fresh fetal bovine serum-free DMEM and 25 μL complexes containing 0.5 μg DNA at their optimal N:P ratios were then added into each well. After incubating for 4 hours, the medium containing the complexes was replaced with 500 μL of DMEM with 10% fetal bovine serum, and the cells were incubated for another 48 hours. The cells were harvested by trypsinization, washed with phosphate-buffered solution twice, and resuspended in 300 μL of phosphate-buffered solution (pH 7.4). Transfection efficiency was evaluated as the percentage of cells expressing green fluorescent protein using FACSCalibur (Becton Dickinson, San Jose, CA). Fluorescence parameters from 1 × 104 cells were acquired and the transfection was carried out in three separate, independent experiments. For the luciferase plasmid DNA expression experiment, the cells were washed with phosphate-buffered solution once. Cell lysate buffer (Promega, Madison, WI, 50 μL per well) was added, and the cells were thoroughly lysed through a onetime freezing-thawing treatment. Luciferase gene expression was quantified using a luciferase reported gene assay kit (Promega), and relative light units were measured with a Varioskan Flash (Thermo Scientific) by automatically adding 100 μL of the luciferase assay buffer to 10 μL of cell lysate in each well. The protein content in the cell lysate was determined by the bicinchoninic acid protein assay kit (Beyotime Institute of Technology, Haimen, China) using a calibration curve constructed with standard bovine serum albumin solution (Beyotime Institute of Technology). Results were expressed in relative light units per milligram of cell protein. All experiments were performed in triplicate to ascertain their reproducibility.
Results and discussion
Synthesis and characterization of PAA
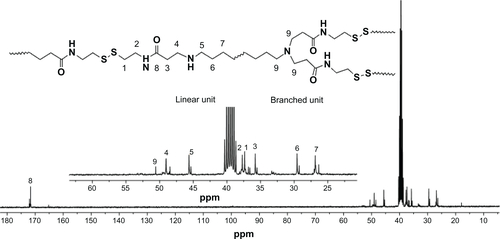
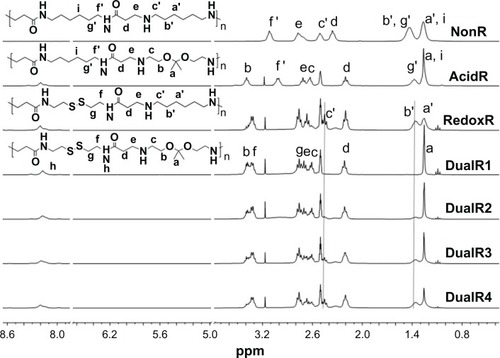
PAAs with different acid degradability were synthesized using a Michael addition reaction of amines (KDA and/or HDA) with acrylamides (CBA and/or HMBA) by varying the molar ratio (). Based on previous findings,Citation27 polymers with an almost linear structure were obtained via Michael addition of an equal molar ratio of KDA and/or HDA with CBA and/or HMBA at a low temperature. Polymerizations were carried out in methanol/water mixture at 35°C in an argon atmosphere for 4 days. The linear structure of the PAAs was confirmed by 13C NMR (Figures S7 and S8). shows the 1H NMR of the PAAs; the disappearance of the characteristic acrylamide peaks between 5 ppm and 6.5 ppm in the final polymer product indicates complete conversion of the acrylamide groups. It was found that the molar ratio of KDA to HDA in the final PAAs was close to the molar ratio in the feed by the integral ratio of methylene protons of HDA at 1.30–1.45 ppm to methylene protons adjacent to the acrylamide groups of CBA at 2.12–2.26 ppm (). Thus, the acid degradability of the PAAs can be easily controlled by varying the content of ketal linkages in the final polymers. The molecular weight and polydispersity index of the polymers were in the range of 16.8–26.8 kDa and 1.36–1.83, respectively ().
Figure 1 Synthetic scheme of dual-responsive poly(amido amine)s.
Abbreviations: CBA, N,N′-cystaminebisacrylamide; HDA, hexamethylene diamine; HMBA, hexmethylenebisacrylamide; KDA, [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea]; NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
![Figure 1 Synthetic scheme of dual-responsive poly(amido amine)s.Abbreviations: CBA, N,N′-cystaminebisacrylamide; HDA, hexamethylene diamine; HMBA, hexmethylenebisacrylamide; KDA, [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea]; NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.](/cms/asset/1827a7e9-533f-4b91-a02d-de957336cb2e/dijn_a_37334_f0001_c.jpg)
Figure 2 1H-nuclear magnetic resonance spectra of DualR1, DualR2, DualR3, DualR4, RedoxR, and AcidR in d6-dimethylsulfoxide, and NonR in D2O.
Abbreviations: NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
Synthesized PAAs have disulfide linkages in their backbone. These linkages are very sensitive to reducing agents, such as DTT and glutathione.Citation15 To confirm reduction of the PAAs, DualR1 was selected and dissolved in dimethylformamide containing 20 mM DTT for 30 minutes, after which the molecular weight was measured by gel permeation chromatography. It was found that DualR1 with a molecular weight of 19.0 kDa was reduced into fragments of <500 Da after incubation with 20 mM DTT for 30 minutes (Figure S9).
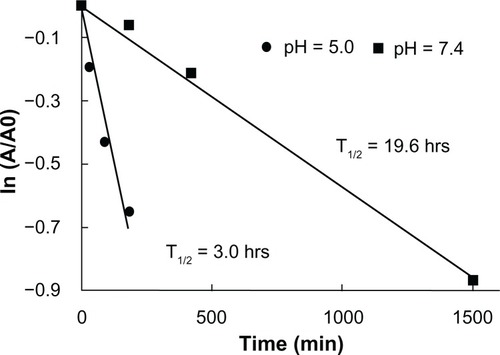
The hydrolysis kinetics of the ketal linkages in the PAAs at pH 5.0 (similar to endosomal pH) and pH of 7.4 (similar to physiological pH) were investigated by incubating DualR1 in deuterated phosphate buffer (pH 5.0 and pH 7.4) at 37°C for various periods of time. The hydrolysis rates and half-lives of the polymers were calculated by monitoring the disappearance of the ketal linkage peaks via NMR spectroscopy and using the Arrhenius equation (). As expected, the PAAs degraded faster at a pH of 5.0 than at a pH of 7.4 because the hydrolysis rate of ketal is proportional to the hydronium ion concentration. The half-lives calculated at a pH of 5.0 and 7.4 were 3.0 hours and 19.6 hours, respectively, indicating the hydrolysis rate of the polymer at a pH of 5.0 is approximately seven times faster than at a pH of 7.4, which is similar to previous reports.Citation21 It has been speculated that in the acidic conditions of the endosome, hydrolysis of ketal bonds would induce endosomal destabilization through increased osmotic force and swollen polyplexes.Citation21
Characterization of polymer/DNA complexes
The ability of the PAAs to condense DNA was studied using the ethidium bromide exclusion assay. As shown in , all PAAs could condense DNA efficiently at an N:P ratio of 5. Among them, DualR1 showed the weakest DNA condensation capability. With the decrease in content of the ketal linkage in the polymer, the ability of DualRs to interact with DNA increases, showing that the hydrophobic nature of relatively bulky dimethyl ketal linkages in polymers interferes with nucleic acid condensation, which can also be confirmed by agarose gel electrophoresis assay ().
Figure 4 DNA condensation efficiency of various DNA/poly(amido amine) polyplexes at different N/P ratios.
Abbreviations: NonR, non-responsive; AcidR, acid responsive; RedoxR, redox responsive; DualR, dual responsive.
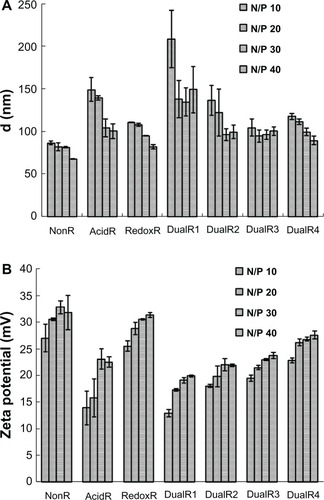
Dynamic light scattering showed that all PAAs, except DualR1, were able to condense DNA into nanosized particles (<200 nm) at and above an N:P ratio of 10 (). The particle size of the DualR1/DNA complexes was in the range of 135–208 nm, which is larger than that of other complexes. The size of the DualR/DNA complexes increased with increasing ketal linkage content in the polymers, which was very consistent with the results of the ethidium bromide exclusion assay. Zeta potential results revealed that all PAAs formed polyplexes with a positive charge at and above an N:P ratio of 10 (). However, the zeta potentials were gradually decreased with increasing ketal linkage content in the polymers, and a possible reason is that the C−O bond in the ketal linkage might shield the positive charges of the polymer/DNA complexes.
Dissociation of nucleic acids from polyplexes
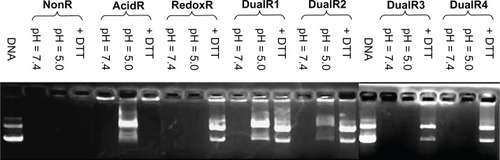
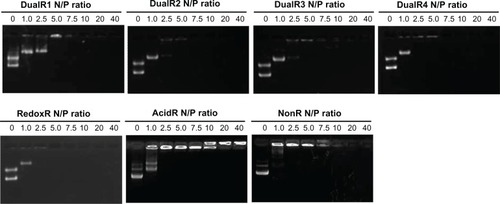
Formation of nucleic acid/PAA polyplexes at an N:P ratio of 10 and the dissociation of nucleic acids from the polyplexes upon hydrolysis or reduction were demonstrated by agarose gel electrophoresis (). No detectable dissociation was observed at a pH of 7.4 even over 4 hours in all polymers. When the polyplexes were incubated in a buffer with a pH of 5.0, DNA was fully released from DualR1 and partly from DualR2. At a pH of 5.0, DualR1 degraded into small cationic residues, which are unable to interact with DNA, so the DNA was released from the polyplexes. However, the DualR3 and DualR4/DNA complexes could not release DNA under the same conditions. These two polymers contain less than 50% ketal linkage, leading the polymer to degrade into fragments during hydrolysis; however, these fragments are still large enough to maintain DNA in its condensed state. For the reducing experiment, complexes with RedoxR, DualR1-4 are expected to be able to be cleaved into small chains and release DNA from the polyplexes because they all contain disulfide bonds. As seen in , when the polyplexes with RedoxR or DualRs were incubated with 10 mM DTT for 30 minutes, all DNA was fully released from the polyplexes.
Although the amount of DNA released from the polyplexes with DualR3 and DualR4 was not significant in an environment of pH 5.0, the low pH actually induced breakage of these two polymers despite the fact that this change was not strong enough to release the DNA. This characteristic may actually benefit transfection efficiency, because the premature release of DNA into an acidic environment, such as that of the endosome/lysosome, can lead to degradation of DNA. The optimum content of ketal bonds in a polymer should be one in which cleavage of the ketal bonds can facilitate endosomal escape without inducing DNA release. This speculation was confirmed in the subsequent transfection study.
In vitro cytotoxicity and transfection
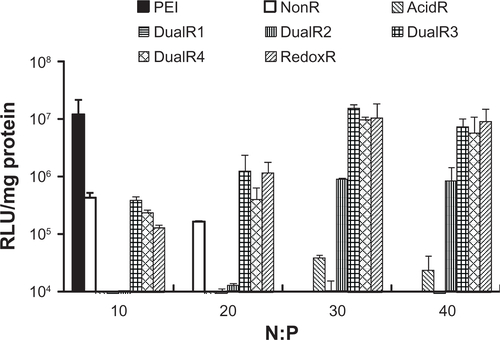
Polycation toxicity is one of the most important issues for development of a successful gene delivery system. The cytotoxicity of PAAs was investigated in HePG2, A549, and MDA-MB-468 cells by MTT assay. The IC50 values are listed in . The higher the IC50, the lower the cytotoxicity. The nonresponsive polymer (NonR) had cytotoxicity similar to that of PEI. As expected, all responsive polymers showed lower cytotoxicity than that of 25 kDa PEI. The reducible bond not only enhances intracellular release, but also reduces cytotoxicity. Similarly, the presence of the acid-labile bond in the polymer not only enhanced endosomal escape but also reduces cytotoxicity. The cytotoxicity of the polymer decreased as the acid-labile bond in the polymer increased. DualR1 demonstrated the highest IC50 in the three different cell lines, indicating that it had the least cytotoxicity. These results demonstrate that dual-responsive polymers have the least cytotoxicity when compared with the nonresponsive, and single-responsive polymers and PEI.
Table 2 Cytotoxicity of poly(amido amine)s in three cell lines
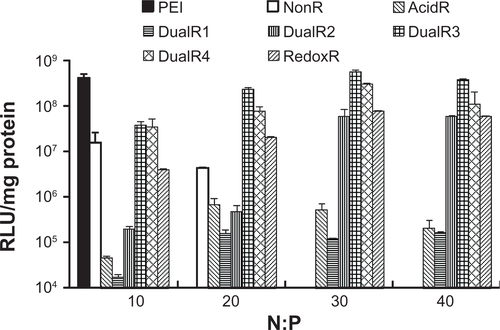
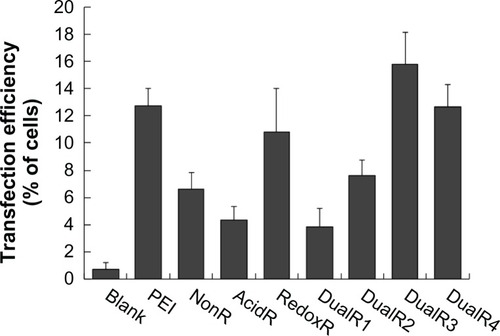
The luciferase transfection results in HePG2 cells () show that DualR3 and DualR4 have a transfection efficiency comparable with that of PEI. The PEI/DNA complex with an N:P ratio of 10 has an optimizing transfection of 6.90 × 108 relative light units per mg protein.Citation28–Citation30 DualR1 has a maximal transfection of 5.28 × 104 relative light units per mg protein when complexed with DNA at an N:P ratio of 40, and RedoxR has a maximal transfection of 2.52 × 108 relative light units per mg protein when complexed with DNA at an N:P ratio of 40. DualR3 and DualR4 have a maximal transfection of 1.23 × 109 and 8.96 × 108 relative light units per mg protein, which is higher than the optimum transfection by PEI. The AcidR (acid-labile) polymer has a low level of cytotoxicity, but it also has a very low level of gene transfection ability. The DualRs, with a maximizing and minimizing content of acid-labile bonds, cannot achieve maximum transfection. The highest gene expression was observed at an optimizing amount of acid-labile content, ie, DualR3. We also confirmed that the DNA complex with DualR3 attained highest transfection in the A549 and MDA-MB-468 cells (Figures S10 and S11) among the tested polymers. This result was further confirmed by fluorescent microscopy () and flow cytometry () in the green fluorescent protein gene expression study. At a different N:P ratio, DualR3 has brighter green fluorescent protein expression than DualR1, DualR2, DualR4, or RedoxR. Among all the tested polymers, DualR3, which has an intermediate amount of acid-labile content, had the highest transfection ability for the luciferase and green fluorescent protein plasmid DNA, and the least cytotoxicity.
Figure 7 Luciferase expression of DNA/poly(amido amine) complexes in HePG2 cells.
Abbreviations: PEI, polyethylenimine; RLU, relative light units.
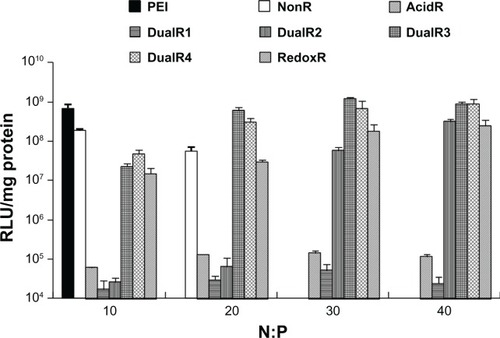
Figure 8 Fluorescence microscopy of green fluorescent protein transfection of DNA/poly(amido amine) polyplexes.
Abbreviation: PEI, polyethylenimine.
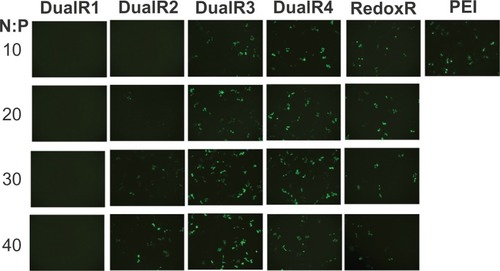
Figure 9 Green fluorescent protein expression of DNA/PAA polyplexes on flow cytometry.
Abbreviations: PAA, poly(amido amine)s; PEI, polyethylenimine.
It is obvious that there should be minimal endosomal escaping ability if there is no acid-labile bond, such as RedoxR. At the same time, if there is an overabundance of acid-labile bonds, such as DualR1, in the polymer, the polyplex will be unstable. As a result, the unstable polyplex prematurely releases plasmid DNA in the endosome and hinders gene expression. In conclusion, maximum gene expression can be acquired in the presence of an optimizing amount of acid-labile bonds. This is because the best time point to release the DNA is the moment after endosomal escape. It is obvious that the amount of acid-labile bonds for optimized transfection is limited in a linear copolymer with reducible and acid-labile bonds. Therefore, our future work will involve synthesizing a brush polymer with the reducible bond on the main chain and the acid-labile bond on the side chain. In this way, hydrolysis of the side chain should not affect the molecular weight of the dual-responsive brush polymer. After endosomal escape, the intracellular environment will ensure full degradation of the main chain of the polymer and release of nucleic acids.
Conclusion
In this study, pH-sensitive and reduction-responsive PAAs were synthesized for gene delivery. To our knowledge, this is the first synthesized polymer with bioreducible and acid-labile properties. Using this dual-responsive polymer, we achieved significant gene transfection ability, which was higher than for nonresponsive or single-responsive polymers. At the same time, the presence of biodegradable bonds in the polymer was associated with less cytotoxicity in comparison with the nondegradable polymers, including PEI. Although the current vector is easy to disassemble in the systemic circulation, the principle described in the current research can be utilized to design novel, potentially serum-resistant gene delivery vectors. Overall, this novel, dual-responsive, gene delivery system represents an innovative approach for gene delivery of plasmid DNA, and its enhanced ability for endosomal escape and cytoplasmic release should be suitable for the delivery of other nucleic acids, such as siRNA.
Acknowledgements
This research received financial support from the National Science Foundation of China (51033005, 20874093, 50973102), the Ministry of Science and Technology of China (2010CB934000), Program for New Century Excellent Talents in University (NCET-11-0882) and the Fundamental Research Funds for the Central Universities (WK2060200004).
Disclosure
The authors report no conflicts of interest in this work.
Supplementary information on synthesis of PAAs
Materials
Pyridinium p-toluene sulfonate (98%), ethyl trifluoroacetate (99%), and 2-methoxypropene (95%) were purchased from Alfa Aesar Chemicals. 2-Aminoethanol (99%) and molecular sieves (5 Å) were purchased from Sinopharm Chemical Reagent Co, Ltd. Cystamine dihydrochloride (98%) was obtained from Changzhou Furong Chemical Co, Ltd. Acryloyl chloride was purified by distillation under reduced pressure. Tetrahydrofuran was dried over molecular sieves (5 Å).
Synthesis of KDA
Synthesis of compound 1 2, 2, 2-Trifluoro-N-(2-hydroxyethyl) acetamide
2-Aminoethanol (8.0 g, 130.96 mmol, 1.0 equivalent), triethylamine (19.88 g, 196.46 mmol, 1.5 equivalent), and ethyl trifluoroacetate (22.32 g, 157.18 mmol, 1.2 equivalent) were added into a flask containing 400 mL methanol under stirring and the reaction was processed at 25°C for 30 hours. The solvent was then evaporated, and the residue was dissolved in 30 mL of water and extracted with 3 × 100 mL ethyl acetate. The crude product was purified by silica gel column chromatography using ethyl acetate as the eluent (see Figures S1 and S2).
Synthesis of compound 2 N, N′-(2, 2′-(Propane-2, 2-diylbis(oxy))bis (ethane-2,1-diyl)) bis(2, 2, 2-trifluoroacetamide)
Pyridinium p-toluene sulfonate (0.61 g, 2.42 mmol, 0.1 equivalent) and compound 1 (9.50 g, 60.51 mmol, 2.5 equivalent) were added into a flask and dissolved in 180 mL of tetrahydrofuran. Molecular sieves (50 g) were added and the mixture was stirred for 10 minutes. 2-Methoxypropene (1.75 g, 24.20 mmol, 1 equivalent) was added, and the mixture was sealed and stirred for 24 hours at 10°C to avoid loss of the highly volatile 2-methoxypropene. The mixture was then stirred at room temperature for another 24 hours. The molecular sieves were removed by filtration, and the solvent was evaporated. The crude product was purified by silica gel chromatography using 8:1 hexane/ethyl acetate, 4:1 hexane/ethyl acetate, and finally 2:1 hexane:ethyl acetate containing 1% (v/v) triethylamine as the eluent (see Figure S3).
Synthesis of KDA
First, compound 2 (4.12 g, 11.64 mmol) was deprotected in 6 M NaOH (30 mL) under stirring for 4 hours. The product was then extracted with 5 × 100 mL CH2Cl2 and the combined organic fraction was evaporated, resulting in an amber-colored oil (see Figure S4).
Synthesis of CBA
Cystamine dihydrochloride (5.8 g, 25 mmol) was dissolved in 50 mL of water in a 250 mL flask. An aqueous solution of NaOH (10 mL, 10 M) and a solution of acryloyl chloride (4.7 g, 50 mmol) in 5 mL of dichloromethane were added dropwise at the same time under stirring at 0°C. The reaction was performed over 3 hours at room temperature until the addition was complete. The product was obtained by filtration and crystallization from ethyl acetate (see Figure S5).
Synthesis of HMBA
1,6-Diaminohexane (2.9 g, 0.025 mol, 1.0 equivalent) was dissolved in 30 mL of water after the solution temperature was decreased down to 0°C–5°C under stirring. An acryloyl chloride dichloromethane solution (4.62 g, 0.05 mol, 5 mL) and an aqueous NaOH solution (4 g, 0.05 mol, 10 mL) were added dropwise simultaneously under stirring over a total time span of 20 minutes. After the addition was complete, the reaction mixture was stirred at room temperature for at least 2 hours. The product was obtain by filtration and washed three times with water (see Figure S6).
Synthesis of branched PAA
As a control, PAA containing branched units was synthesized by Michael addition of HDA with CBA at a higher temperature. HDA (174 mg, 1.5 mmol, 1.0 equivalent) was added into a solution of CBA (390 mg, 1.5 mmol, 1.0 equivalent) in a methanol/water mixture (3.0 mL, 9/1, v/v). The reaction was allowed to proceed at 55°C in an argon atmosphere for 2 days. The final polymer was obtained via precipitation of ethyl ether/acetone (1/1, v/v) and drying under vacuum for 4 hours at room temperature.
Figure S1 Synthetic scheme of [1,1′-(2,2′-(propane-2,2-diylbis(oxy) bis(ethane-2,1-diyl))diurea] (KDA).
![Figure S1 Synthetic scheme of [1,1′-(2,2′-(propane-2,2-diylbis(oxy) bis(ethane-2,1-diyl))diurea] (KDA).](/cms/asset/74267f00-94d4-45ef-918d-ae4598448c88/dijn_a_37334_sf0001_b.jpg)
Figure S4 1H nuclear magnetic resonance spectrum of [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea] (KDA) compound in DCCl3.
![Figure S4 1H nuclear magnetic resonance spectrum of [1,1′-(2,2′-(propane-2,2-diylbis(oxy)) bis(ethane-2,1-diyl))diurea] (KDA) compound in DCCl3.](/cms/asset/6d5472f0-9efa-490a-bf55-fee5c5459fcd/dijn_a_37334_sf0004_c.jpg)
Figure S5 1H nuclear magnetic resonance spectrum of N,N′-cystaminebisacrylamide (CBA) in d6-dimethylsulfoxide.
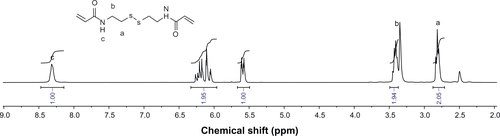
Figure S6 1H nuclear magnetic resonance spectrum of hexmethylenebisacrylamide (HMBA) in d6-dimethylsulfoxide.
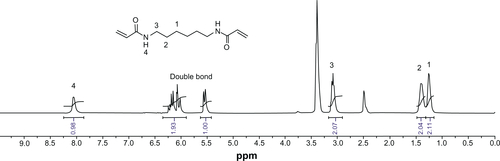
Figure S9 Gel permeation chromatography of DualR1 before (up) and after (down) incubation with 20 mM DTT.
References
- ReadMLLoganASeymourLWBarriers to Gene Delivery Using Synthetic VectorsAdvances in genetics200553PA194616243059
- RuponenMHonkakoskiPRonkkoSPelkonenJTammiMUrttiAExtracellular and intracellular barriers in non-viral gene deliveryJ Control Release125200393221321714636726
- VarkouhiAKScholteMStormGHaismaHJEndosomal escape pathways for delivery of biologicalsJ Control Release5102011151322022821078351
- SchmaljohannDThermo- and pH-responsive polymers in drug deliveryAdvanced Drug Delivery Reviews1230200658151655167017125884
- BoussifOLezoualchFZantaMAA Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - PolyethylenimineP Natl Acad Sci USA811995921672977301
- YuJHQuanJSHuangJAlpha, beta-poly(L-aspartate-graft-PEI): A pseudo-branched PEI as a gene carrier with low toxicity and high transfection efficiencyActa Biomater92009572485249419357003
- ShimMSKwonYJControlled delivery of plasmid DNA and siRNA to intracellular targets using ketalized polyethylenimineBiomacromolecules220089244445518186606
- ShimMSKwonYJDual mode polyspermine with tunable degradability for plasmid DNA and siRNA deliveryBiomaterials6201132164009402021371749
- KnorrVRussVAllmendingerLOgrisMWagnerEAcetal linked oligoethylenimines for use as pH-sensitive gene carriersBioconjugate Chem8200819816251634
- ReadMLBremnerKHOupickyDGreenNKSearlePFSeymourLWVectors based on reducible polycations facilitate intracellular release of nucleic acidsThe Journal of Gene Medicine320035323224512666189
- Soundara ManickamDOupickyDPolyplex gene delivery modulated by redox potential gradientsJ Drug Target9200614851952617050118
- OupickyDDiwadkarVStimuli-responsive gene delivery vectorsCurr Opin Mol Ther820035434535014513676
- MengFHHenninkWEZhongZReduction-sensitive polymers and bioconjugates for biomedical applicationsBiomaterials4200930122180219819200596
- PiestMLinCMateos-TimonedaMANovel poly(amido amine)s with bioreducible disulfide linkages in their diamino-units: Structure effects and in vitro gene transfer propertiesJ Control Release82520081301384518585814
- YouYZManickamDSZhouQHOupickyDReducible poly(2-dime-thylaminoethyl methacrylate): synthesis, cytotoxicity, and gene delivery activityJ Control Release1082007122321722517574292
- Hoon JeongJChristensenLVYockmanJWReducible poly(amido ethylenimine) directed to enhance RNA interferenceBiomaterials4200728101912191717218006
- ChristensenLVChangCWKimWJReducible poly(amido ethylenimine)s designed for triggered intracellular gene deliveryBioconjugate Chemistry Sep-Oct20061751233124016984133
- JeongJHKimSHChristensenLVFeijenJKimSWReducible poly(amido ethylenimine)-based gene delivery system for improved nucleus trafficking of plasmid DNABioconjugate Chemistry217201021229630120078099
- OuyangDShahNZhangHSmithSCParekhHSReducible disulfide-based non-viral gene delivery systemsMini reviews in medicinal chemistry920099101242125019817714
- ManickamDSOupickyDMultiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene deliveryBioconjugate Chemistry Nov-Dec20061761395140317105216
- ShimMSKwonYJAcid-responsive linear polyethylenimine for efficient, specific, and biocompatible siRNA deliveryBioconjugate Chemistry318200920348849919199781
- ShimMSKwonYJAcid-transforming polypeptide micelles for targeted nonviral gene deliveryBiomaterials4201031123404341320122722
- KoIKZiadyALuSKwonYJAcid-degradable cationic methacrylamide polymerized in the presence of plasmid DNA as tunable non-viral gene carrierBiomaterials10200829283872388118585778
- YangWPanCYSynthesis and Fluorescent Properties of Biodegradable Hyperbranched Poly(amido amine)sMacromolecular Rapid Communications1216200930242096210121638500
- van der AaLJVaderPStormGSchiffelersRMEngbersenJFOptimization of poly(amido amine)s as vectors for siRNA deliveryJ Control Release3102011150217718621130817
- JainRStandleySMFréchetJMJSynthesis and Degradation of pH-Sensitive Linear Poly(amidoamine)sMacromolecules 2007/02/012007403452457
- HongCYYouYZWuDCLiuYPanCYThermal control over the topology of cleavable polymers: From linear to hyperbranched structuresJ Am Chem Soc522007129175354 –+.17417853
- SunXMaPCaoXNingLTianYRenCPositive hyaluronan/PEI/DNA complexes as a target-specific intracellular delivery to malignant breast cancerDrug delivery10200916735736219545193
- LiDWangQQTangGPReceptor-mediated gene delive using polyethylenimine (PEI) coupled with polypeptides targeting FGF receptors on cells surfaceJ Zhejiang Univ Sci B200671190691117048306
- HonoreIGrosseSFrisonNFavatierFMonsignyMFajacITranscription of plasmid DNA: influence of plasmid DNA/polyethylenimine complex formationJ Control Release10202005107353754616087268