?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Infectious diseases cause a huge burden on healthcare systems worldwide. Pathogenic bacteria establish infection by developing antibiotic resistance and modulating the host’s immune system, whereas opportunistic pathogens like Pseudomonas aeruginosa adapt to adverse conditions owing to their ability to form biofilms. In the present study, silver nanoparticles were biofunctionalized with polymyxin B, an antibacterial peptide using a facile method. The biofunctionalized nanoparticles (polymyxin B-capped silver nanoparticles, PBSNPs) were assessed for antibacterial activity against multiple drug-resistant clinical strain Vibrio fluvialis and nosocomial pathogen P. aeruginosa. The results of antibacterial assay revealed that PBSNPs had an approximately 3-fold higher effect than the citrate-capped nanoparticles (CSNPs). Morphological damage to the cell membrane was followed by scanning electron microscopy, testifying PBSNPs to be more potent in controlling the bacterial growth as compared with CSNPs. The bactericidal effect of PBSNPs was further confirmed by Live/Dead staining assays. Apart from the antibacterial activity, the biofunctionalized nanoparticles were found to resist biofilm formation. Electroplating of PBSNPs onto stainless steel surgical blades retained the antibacterial activity against P. aeruginosa. Further, the affinity of polymyxin for endotoxin was exploited for its removal using PBSNPs. It was found that the prepared nanoparticles removed 97% of the endotoxin from the solution. Such multifarious uses of metal nanoparticles are an attractive means of enhancing the potency of antimicrobial agents to control infections.
Introduction
Infectious agents and their toxins are a constant burden on healthcare systems worldwide. Infections associated with surgical devices and medical implants pose persistent and severe problems that account for high morbidity in hospital settings. According to Center for Disease Control and Prevention (CDC), surgical site infections account for ~22% of hospital-acquired infections.Citation1 Initial bacterial adhesion to biomedical device surfaces is a crucial step for pathogenesis. As the biofilm matures, it becomes refractory to both host defense mechanism and antibacterial agents.Citation2 These bacteria also release toxins that cause complications like sepsis and septic shock syndromes.Citation3 Therefore, several prevention strategies are being developed that include coating of antimicrobial molecules either by modifying the physicochemical properties of the surface or direct coating on the surfaces of medical devices. These methods are employed with the aim of either inhibiting the adhesion of the planktonic cells or killing the bacterial cells that have adhered.Citation4 Owing to resistance in microorganisms toward antimicrobial agents, abolition of established biofilms often requires a high concentration of antimicrobial agents. The situation is further complicated by the development of multiple antibiotic resistance among bacteria associated with nosocomial infections.Citation5,Citation6 Advances in the field of nanotechnology have motivated scientists to develop innovative strategies to utilize nanomaterials that not only effectively control bacterial infections but can also mitigate their toxins.
One such strategy is the use of silver nanoparticles (SNPs) as antimicrobial coatings on the surface of medical devices. Among the antimicrobial metal nanoparticles, SNPs have a clear advantage as they kill target bacterial cells by disrupting the bacterial cell wall, resulting in leakage of cytoplasmic contents and inactivation of respiratory enzymes and proteins responsible for RNA and DNA replication.Citation7 Owing to these multiple modes of action of SNPs, there is a low chance of bacteria developing resistance, with minimal damage to mammalian cells. Since SNPs interact with the bacterial surface by random collision, a high concentration of SNPs is required to exert an antimicrobial effect.Citation8 Toxicity issues associated with silver demand that silver be used at as low a concentration as possible. To overcome this problem, SNPs have been synthesized using various biological moieties including antibiotics, enzymes, polysaccharides, and oligopeptides so as to enhance the antibacterial potential.Citation9–Citation11 In recent years, antimicrobial peptides (AMPs) have emerged as excellent antimicrobial agents to address multidrug resistance in bacteria.Citation12,Citation13 AMPs act like a “molecular knife” and facilitate the damage to bacterial cell membrane even at low concentrations.Citation14 Additionally, AMPs can stabilize SNPs by exerting a polyvalent effect as they form layers on the surface of SNPs and thus prevent agglomeration.Citation15,Citation16
The purpose of this study was to develop an innovative strategy to enhance the antibacterial potency of SNPs and test these functionalized SNPs for various medical applications. For this, 1) we have chosen polymyxin B (an AMP) to synthesize biofunctionalized nanoparticles by a simple and efficient method. Polymyxin B is a well-established cationic, nonribosomal peptide known to exert antibacterial activity by interacting with the lipid A component of lipopolysaccharide (LPS) of the outer membrane of Gram-negative bacteria.Citation17 2) We assessed the polymyxin B-capped silver nanoparticles (PBSNPs) for their antibacterial and antibiofilm activity against multidrug-resistant clinical isolate of Vibrio fluvialis and Pseudomonas aeruginosa PAO1. 3) Further, we coated stainless steel surgical blades with PBSNPs and tested their ability to inhibit the growth of nosocomial pathogen P. aeruginosa. 4) Finally, we tested the endotoxin removal efficiency of PBSNPs from solution.
Materials and methods
Chemicals, reagents, and bacterial strains
Polymyxin B and proteinase K were obtained from Sigma Aldrich (St Louis, MO, USA). Silver nitrate (AgNO3), sodium borohydrate (NaBH4), crystal violet (CV), Luria Bertani (LB) broth, Mueller Hinton (MH) broth, and molecular biology grade methanol were obtained from EMD Millipore (Billerica, MA, USA). The P. aeruginosa PAO1 strain resistant to gentamycin, ciprofloxacin, tobramycin, oflaxacin, nalidixic acid, kanamycin, tetracycline, novobiocin, and vancomycin was provided by Prof Lori Burrows of McMaster University, Canada. Vibrio fluvialis L-15318, a multiple drug-resistant clinical strain, was isolated from Infectious Diseases Hospital, Kolkata, India. The strain shows resistance to ampicillin, kanamycin, rifampicin, norfloxacin, cephalothin, oxacillin, and vancomycin, and was a kind gift from Prof Amit Ghosh of the National Institute of Cholera and Enteric Diseases, Kolkata, India.
Facile synthesis and characterization of PBSNPs
Synthesis of PBSNPs was based on a method described earlier with some modifications.Citation18 Briefly, SNPs were prepared by the addition of freshly prepared silver nitrate (2 mM) and NaBH4 (0.6 mM) to a polymyxin B solution in methanol. The resultant mixture was incubated for 0–30-minute time periods at 30°C under illumination at 40 W yellow light (141.3 lux). The optimum polymyxin B concentration for this reaction was determined by a set of batch experiments where the concentration of the peptide was varied between 0 and 100 μg/mL. The prepared PBSNPs were dialyzed against miliQ water for 12 hours using a 10 kDa cut-off membrane to remove unbound polymyxin B and free silver ions. The amount of polymyxin B on biofunctionalized nanoparticles and their stability in terms of polymyxin B leaching was estimated by using the fluorescent ortho-phthaldialdehyde (OPA) assay.Citation19 The unbound polymyxin B was removed from PBSNPs by passing the sample through a 10 kDa molecular weight cut-off spin-filter (Vivaspin, Vivaproducts, Inc., Littleton, MA, USA). Twenty-five microliters of filtrate was mixed with 250 μL of OPA solution (1 mg/mL OPA dissolved with 2 μL/mL β-mercaptoethanol in 0.2 M borate buffer, pH 9.4) and incubated for 1 minute. The OPA was excited at 350 nm, and emission was measured at 460 nm. The amount of unbound polymyxin B in solution was calculated on the basis of a standard curve plotted for polymyxin B. For stability analysis, PBSNPs were kept for 12 hours at 37°C and samples were withdrawn at different time intervals and checked for leaching of polymyxin B in the solution. Further, the stability of PBSNPs stored at 4°C was also determined by measuring the antibacterial activity over a period of 1 month. Citrate- capped silver nanoparticles (CSNPs) were synthesized using a previously published method and were used as control in the experiments.Citation20 PBSNPs and CSNPs were characterized by using UV–visible spectroscopy, transmission electron microscopy (TEM), zeta potential, dynamic light scattering (DLS), Fourier transform infrared (FTIR), and circular dichroism (CD) spectroscopy. Surface plasmon resonance (SPR) spectra of PBSNPs and CSNPs were recorded using the UV–visible absorbance spectrometer (SpectraMax Plus spectrophotometer, Molecular Devices LLC, Sunnyvale, CA, USA). The stability of nanoparticles was determined by measuring zeta potential using a Zetasizer (Malvern Instruments, Malvern, UK). The morphology and size of nanoparticles were observed using TEM (JEOL JEM-2100F, JEOL, Tokyo, Japan). For TEM analysis, samples were prepared on carbon-coated copper grids and examined with an accelerating voltage of 200 keV. Concentration of silver ions (Ag+) in CSNPs and PBSNPs was determined by Atomic Absorption Spectrophotometer (AAS), (GBC Scientific Equipment, Hampshire, IL, USA) using silver nitrate (AgNO3) standards. The presence of polymyxin B on the surface of SNPs and the effect of attachment on the secondary structure of peptide were measured with FTIR (Thermo Nicolet NEXUS 670 FTIR; GMI, Ramsey, MN, USA) and CD spectroscopy (Chirascan™ CD Spectrometer; Applied Photophysics Ltd, Leatherhead, UK), respectively. CD measurements were carried out at 25°C using a cylindrical fused quartz cell of 0.2 mm path length. Free polymyxin B and citrate-capped nanoparticles were used as control.
Determination of minimum inhibitory concentration (MIC), fractional inhibitory concentration (FIC), and characterization of bacterial morphology by scanning electron microscopy (SEM)
The susceptibility of the microbial cells to CSNPs and PBSNPs was determined by microbroth dilution assays of the Clinical and Laboratory Standards Institute in MH broth for both Gram-negative bacterial strains.Citation21,Citation22 The cellular growth was observed at OD600 after 10 hours. Growth above 10% of the OD at 600 nm (OD600) of the positive control (lacking any antimicrobial agent) was considered uninhibited growth. MIC is the lowest concentration of PBSNPs that inhibited the visible growth of microorganisms. CSNPs with the same concentration were taken as a control. The assays were repeated twice with three replicates in each plate. To determine whether CSNPs (compound A) and polymyxin B (compound B) worked synergistically or additively, the FIC index was used. The FIC index was calculated as follows:
FIC index values lower than 0.5 indicate synergistic activity, values in between 0.5 and 2.0 indicate additive effects, whereas values above 2.0 indicate antagonistic effects.Citation23 For SEM analysis, bacterial cells treated with MIC concentration of CSNPs and PBSNPs were collected by centrifugation at 10,000 rpm for 10 minutes. The cells were primarily fixed with 2% glutaraldehyde followed by gradual dehydration with ethanol gradient (25%, 50%, 70%, 80%, 90%, and 100%). The prepared samples were coated with a layer of gold and observed under SEM (Ultra Plus Field Emission; Carl Zeiss Meditec AG, Jena, Germany).
Biofilm inhibition assay
P. aeruginosa PAO1 biofilm formation was achieved by modifying the previously described assays.Citation24,Citation25 Briefly, P. aeruginosa PAO1 was grown overnight in LB medium at 37°C with shaking at 250 rpm. Fresh LB medium was seeded with 1% of overnight culture and incubated at 37°C till an OD600 of 1.0 was attained. To investigate the bactericidal effect of CSNPs and PBSNPs on planktonic cells of P. aeruginosa PAO1, different concentrations of CSNPs (2.4–25,000 Ag0 ng/mL) and PBSNPs (4.3–4,500 Ag0 ng/mL) were suspended in M63 minimal medium in a 96-well plate. The above culture was diluted 1:100 in M63 minimal medium and added to varying concentrations of CSNPs and PBSNPs to reach a concentration of 108 CFU/mL. Media subcultured with diluted cells without SNPs was taken as positive control, and the plate was incubated at 37°C for 6, 12, and 24 hours under static condition. M63 minimal medium alone was used as the negative control. After incubation, the planktonic cells were removed, and the wells were gently washed three times with 300 μL of sterile 0.9% saline. The biofilm was stained with 300 μL of 0.1% aqueous CV solution for 15 minutes at 30°C. The stained film was washed thrice with 0.9% saline, air-dried, and solubilized with 300 μL of 30% acetic acid. The solubilized CV from each well was transferred to a new 96-well plate, and the inhibition of the biofilm was quantified by measuring the OD550.Citation26 The above experiments were conducted in triplicates, and the results were represented using two-way ANOVA statistics of GraphPad Prism (GraphPad Software, San Diego, CA, USA) for variation in independent experiments.
Live/Dead staining of the bacteria in the biofilm
To study the effect of PBSNPs on biofilm formation, P. aeruginosa PAO1 was grown to an OD600 of 1.0 and diluted 1:100 in M63 minimal medium. Aliquots of 1 mL (108 CFU/mL) of diluted culture were transferred to a 6-well plate with a sterile 13 mm coverslip along with MIC and sub-MIC of PBSNPs in a final volume of 2 mL. Control samples were prepared on a sterile glass coverslip with P. aeruginosa PAO1 cells (positive) and M63 minimal media (negative) without any SNPs.Citation27,Citation28 The wells were washed twice with 3 mL of sterile 0.9% saline. The coverslips were stained with a 1.5 mL mixture of 0.8 μM SYTO9 green fluorescent dye and 10 μM propidium iodide (PI) red fluorescent dye of Live/Dead staining kit (LIVE/DEAD® BacLight™ Bacterial Viability Kit, Thermo Fisher Scientific, Waltham, MA, USA) and observed under a fluorescence microscope (Axio Scope A1; Carl Zeiss Meditec AG) at 1,000× magnification.
Viability analysis of PBSNPs-treated P. aeruginosa PAO1
To examine the bactericidal effect of PBSNPs on planktonic cells, P. aeruginosa PAO1was subcultured in fresh LB broth and incubated at 37°C. After an OD600 of 0.4 was attained, 2 mL of culture was centrifuged at 6,000 rpm for 5 minutes. The cells were washed twice with sterile 0.9% saline and finally resuspended in 2 mL of sterile 0.9% saline. To carry out the Live/Dead staining of strain PAO1, 100 μL (108 CFU/mL) of resuspended cells were treated with MIC (4.5 Ag0 μg/mL) of sonicated PBSNPs, whereas untreated cells were taken as control. Three different samples, untreated (stained and unstained) and MIC-treated (stained) PAO1 cells, were prepared. After incubation of 2 hours, samples were centrifuged at 10,000 rpm for 10 minutes at 4°C, supernatant was discarded, and the cell pellet was resupended in 100 μL of 0.9% saline. Untreated and MIC-treated samples were stained with a 1 μL of 1:1 mixture of solution A and B of Live/Dead staining kit. Untreated, unstained sample was used as the control. The samples were analyzed on a flow cytometer (BD FACS Verse™ BD Biosciences, San Jose, CA, USA) to discriminate between live and dead cells.Citation29,Citation30 The above stained samples were also visualized using fluorescent microscopy at 1,000× magnification.
Electrophoretic deposition of polymyxin and CSNPs
Electrophoretic deposition of PBSNPs and CSNPs was performed on stainless steel surgical blades (No 15 Glassvan; DNP Enterprise, Gujarat, India) using procedure described earlier.Citation31 To observe the PBSNP coating on surgical blades, atomic force microscopy (NTEGRA; NT-MDT, Moscow, Russia) was performed. Antibacterial activity of PBSNPs, CSNPs, and uncoated surgical blades (negative control) were tested on an MH agar plate spread with 100 μL of 0.2 OD600 P. aeruginosa PAO1 cells and incubated overnight at 37°C. To ascertain the contribution of polymyxin B toward antibacterial activity, PBSNPs-coated surgical blade was treated with proteinase K (5 mg/mL) for 1 hour at 37°C followed by incubation with
P. aeruginosa cells on agar assay, as mentioned above.
Removal of endotoxin by PBSNPs
Endotoxin (Escherichia coli) removal efficiency of PBSNPs was assessed using endpoint endotoxin assay kit (ToxinSensor™ Chromogenic Limulus Amebocyte Lysate Endotoxin Assay Kit; GenScript USA Inc., Piscataway, NJ, USA). The Limulus Amebocyte Lysate (GenScript USA Inc.) (0.05 EU/mL) was incubated with 100 μL of PBSNPs and CSNPs (with the same silver concentration, 4.5 μg/mL) separately for 30 minutes at 37°C, followed by centrifugation at 14,000 rpm for 15 minutes. The presence of endotoxin was determined in the supernatant as per manufacturer’s instructions. CSNPs and standard endotoxin provided with the kit were taken as controls. Endotoxin removal ability of PBSNPs and CSNPs was compared by measuring the absorbance of chromogenic mixture at OD545. Concentration of the remaining endotoxin in the supernatant was estimated from the standard curve.
Results and discussion
Characterization of biofunctionalized nanoparticles and evaluation of the antibacterial activity
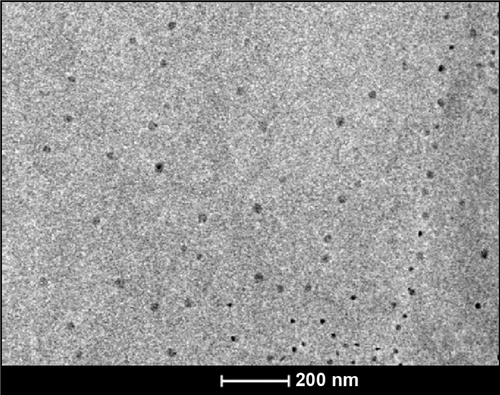
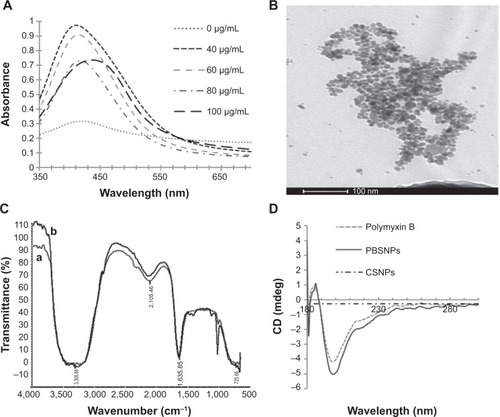
To optimize the synthesis of PBSNPs, a batch of experiments were performed with varying concentrations of polymyxin B (0–100 μg/mL) and a fixed concentration of silver nitrate (340 μg/mL) with different incubation times (0–30 minutes). The appearance of a yellow color and a sharp plasmon resonance absorption band at 414 nm () in the UV–vis spectrum of the preparation is indicative of nanorange monodispersed nanoparticles.Citation32 TEM analysis revealed that the PBSNPs were monodispersed and spherical in shape with a mean diameter of 15 nm (). However, an increase in nanoparticle size (~130 nm; in the presence of 0.1% tween-20) with polymyxin B capping as estimated by DLS was observed, which is likely due to biomolecule layering on the nanoparticles. These results are in agreement with the findings that the hydrodynamic size of the nanobioconjugates (through DLS) is often more than what was observed under TEM.Citation33 For the rest of the experiments, a solution consisting of 340 μg/mL of silver nitrate and 60 μg/mL of polymyxin B in methanol and an incubation time of 30 minutes was taken for nanoparticle synthesis. For CSNPs, a characteristic SPR peak was observed at 415 nm (data not shown). TEM data suggested that the prepared nanoparticles were monodispersed and spherical in shape with a size range of 10–20 nm (). The zeta potential of PBSNPs and CSNPs was recorded to be 18.1±6 and −23±15 mV, respectively, indicating the stability of the prepared nanoparticles. The FTIR spectrum of PBSNPs was similar to the spectrum of free polymyxin B (). Any change in the secondary and tertiary protein structures alters hydrogen bonding between the CO and NH groups in the peptide backbone, which results in alterations of the primary, secondary, or tertiary amine bands between 1,200 and 1,700/cm.Citation34 No such significant shifts were observed in this IR region of PBSNPs and free polymyxin B. In both cases, there was a peak at 1,635.85/cm typical of the amide group, indicating no major alterations in the functional groups of polymyxin B after capping on SNPs. CD spectrum further confirmed that the polymyxin B preserved its secondary structure after capping on the nanoparticles.Citation35 A negative peak typical of helix structures was detected at 200 nm (peptide π–π* transition) in the spectra of both the free polymyxin B in water and the polymyxin B-capped nanoparticles ().Citation36 No such peaks were observed in the case of CSNPs, which were used as control.
Figure 1 Synthesis and characterization of PBSNPs.
Notes: (A) UV–visible absorption spectra of PBSNPs prepared using different concentrations of polymyxin B. (B) Size and morphology analysis of PBSNPs by TEM, (C) FTIR spectra summarizing the presence of functional groups on (a) pure polymyxin B, (b) PBSNPs, (D) CD spectra for polymyxin B, PBSNPs, and CSNPs.
Abbreviations: PBSNPs, polymyxin B-capped silver nanoparticles; TEM, transmission electron microscopy; FTIR, Fourier transform infrared; CD, circular dichroism; CSNPs, citrate-capped silver nanoparticles.
It has been shown earlier that combined antibacterial action of SNPs and polymyxin B in solution exerts a synergistic effect on bacterial pathogens.Citation23 A simple method to immobilize polymyxin B on SNPs will be helpful to use these biofunctionalized nanoparticles as coatings for medical devices. Several studies have reported the conjugation of biomolecules via simple adsorption, covalent attachment, or electrostatic binding on the surface of nanoparticles for developing antibacterial surfaces.Citation9,Citation31 Recently, Soonhyang et alCitation19 reported antibacterial property of polymyxin B that was electrostatically conjugated to gold nanoparticles in a complex two-step process. In the present study, the conditions optimized for capping SNPs with polymyxin B are facile, thus leading to a stable (no evidence of significant release of polymyxin B in solution as measured by OPA assay) and biologically relevant interaction of polymyxin B with nanoparticles. Moreover, the antibacterial property of the PBSNPs did not alter significantly over a period of 1 month.
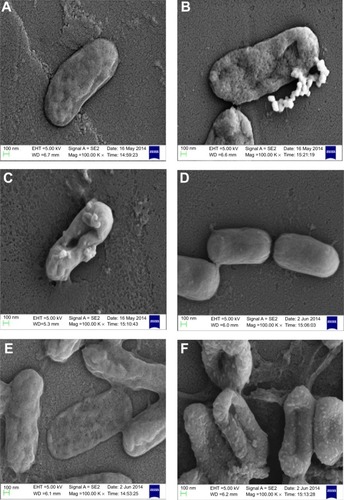
Owing to the presence of an extra outer membrane that prevents antibiotics from crossing the double layer, Gram-negative pathogens are known to be resistant to a variety of antibiotics. To overcome this problem, Gram-negative antibacterial drugs are being discovered that may be effective even without crossing the inner layer.Citation37 A dearth of new antibiotics and the development of antibiotic resistance has led researchers to explore antimicrobial metals such as silver in innovative ways. The biofunctionalized SNPs used in this study clearly showed a decrease in MIC as compared with CSNPs for their antibacterial activity against both antibiotic-resistant Gram-negative bacteria (). The FIC index was used to investigate the synergistic, additive, or antagonistic effect between CSNPs and polymyxin B. The results showed that the FIC indices were 1.485 for P. aeruginosa PAO1 and 0.922 for V. fluvialis, indicating that capping of polymyxin B to the SNPs has produced the additive antimicrobial effect on both pathogens (). Further, the morphological effect of MIC of CSNPs and PBSNPs on both the cells were studied using SEM. The results confirm extensive damage to the cell membrane in the presence of PBSNPs () as compared with CSNPs () and untreated cells (). The plausible explanation for the damage is that highly cationic polymyxin B peptides first get attached to the bacterial surface and make small and transient ion-permeable holes that disturb the functions of the bacterial membrane. Subsequently, the combined action of SNPs and polymyxin B led to a complete rupture of the bacterial membrane, resulting in leakage of the cell contents and finally cell death.Citation38 These results indicate that the enhanced antibacterial activity of PBSNPs was due to the presence of polymyxin B molecules on the surface of SNPs. The interaction of these conjugates provided ample opportunity for both agents to act on the bacteria simultaneously, thus enhancing its inhibitory effect.Citation23,Citation39 SEM images of PBSNPs, CSNPs, and untreated cells at different magnification for V. fluvialis (–) and P. aeruginosa PAO1 (–) are shown in the supplementary material.
Table 1 MIC of polymyxin B, CSNPs, and PBSNPs
Figure 2 Scanning electron micrograph.
Notes: (A) and (D) show control cells (without SNPs). (B) and (E) show CSNPs-treated cells. (C) and (F) PBSNPs-treated cells. (A–C) Vibrio fluvialis L-15318 and (D–F) Pseudomonas aeruginosa PAO1.
Abbreviations: SNPs, silver nanoparticles; CSNPs, citrate-capped silver nanoparticles; PBSNPs, polymyxin B-capped silver nanoparticles.
Effect of biofunctionalized nanoparticles on biofilm formation
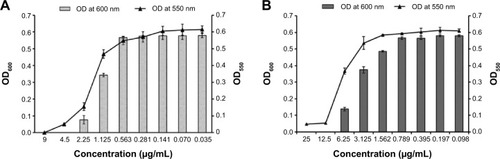
Adhesion of planktonic cells on the surface of biomedical implants and devices is a primary and indispensable step for the initiation of biofilm formation.Citation40,Citation41 This observation prompted us to test the ability of these functionalized nanoparticles for their antibiofilm activity against P. aeruginosa. shows that upon exposure of PAO1 cells to MIC of PBSNPs (4.5 μg Ag0/mL, ) and CSNPs (12.5 μg Ag0/mL, ), a complete inhibition of bacterial growth (OD600) occurred, with the disappearance of biofilm formation (OD550) after 6 hours of incubation. These results reveal that PBSNPs display ~3-fold higher antibiofilm activity as compared with CSNPs, suggesting a paramount role being played by the capping agent polymyxin B. Concurrent inhibition of planktonic cell growth and biofilm formation with increasing concentration of PBSNPs suggests that this formulation is refractory to the establishment of bacterial biofilm likely due to inhibition of planktonic cells.
Figure 3 Inhibition of biofilm formation of Pseudomonas aeruginosa PAO1 cells.
Notes: (A) P. aeruginosa PAO1 cells treated with PBSNPs, (B) P. aeruginosa PAO1 cells treated with CSNPs. The line graph indicates the biofilm biomass at 550 nm (OD550), and the bar graph represents the bacterial growth at 600 nm (OD600).
Abbreviations: CSNPs, citrate-capped silver nanoparticles; OD, optical density; PBSNPs, polymyxin B-capped silver nanoparticles.
Live/Dead staining of bacteria in biofilm
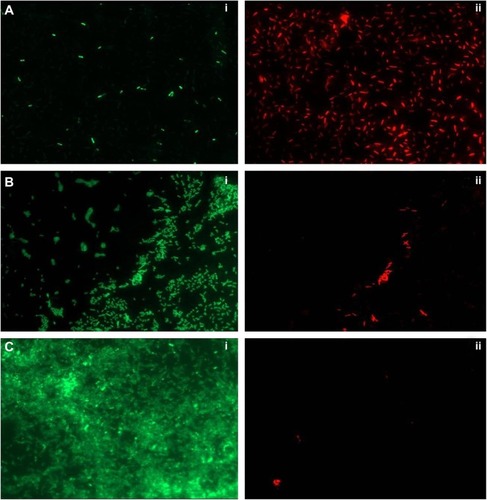
To ascertain the antibiofilm activity of PBSNPs, P. aeruginosa PAO1 biofilm was assayed on the glass coverslip with MIC (4.5 μg Ag0/mL) and sub-MIC concentration (2.25 μg Ag0/mL) of PBSNPs under stationary conditions for 6 hours. The viability of the attached cells on the coverslip was evaluated by a mixture of SYTO9 (stains the total nucleic acid content of the cells, thus staining dead as well as live cells) and PI dye (unable to penetrate the intact cytoplasmic membrane of healthy cells, thus staining only the dead cells).Citation29,Citation42 Although cells were able to adhere to the coverslip in MIC (4.5 μg Ag0/mL) and sub-MIC (2.25 μg Ag0/mL) treatments, the relative cell density was reduced as compared with the untreated cells, thus demonstrating the inhibitory effect of PBSNPs. (A) shows the effect of MIC-treated cells that resulted in the death of most of the bacterial cells. The ratio of (i) live to (ii) dead cells was significantly higher in the case of sub-MIC-treated cells (B) as compared with the MIC-treated cells (A), while a confluent layer of live cells on the coverslip was observed in the case of untreated cells (C). The inhibitory effect of silver nanoparticle treatment on planktonic cells of P. aeruginosa PAO1 was also observed in a recent study.Citation27 The killing efficiency of PBSNPs in our study was found to be 2.5 times more effective than the reported values.Citation27 This highlights the importance of biofunctionalization of nanomaterials at surfaces for controlling infections.
Figure 4 Live/Dead staining of biofilm on glass coverslip.
Notes: (A) PBSNPs MIC-treated Pseudomonas aeruginosa PAO1 cells, (B) PBSNPs sub-MIC-treated P. aeruginosa PAO1 cells, and (C) untreated P. aeruginosa PAO1 cells. A–C: (i) represents green cells as live, and (ii) red cells as dead.
Abbreviations: PBSNPs, polymyxin B-capped silver nanoparticles; MIC, minimum inhibitory concentration.
Viability analysis of PBSNPs-treated P. aeruginosa PAO1 using flow cytometer
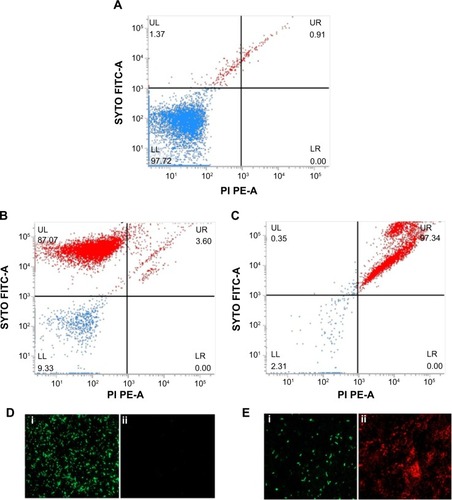
Viability analysis by standard plate assay may give a false impression of the bactericidal nature of an antibacterial agent by reducing the cultivability of cells, while they remain dormant and recover later after a certain period of time.Citation43 To confirm the killing efficacy of PBSNPs against PAO1 planktonic cells, MIC-treated cells were stained with Live/Dead stains and analyzed by flow cytometery, providing us with a real-time assessment of bacterial viability. Citation29,Citation30 shows the flow cytometry analysis of untreated (A) unstained cells, (B) stained cells as control samples, and MIC-treated (C) stained cells as test sample. On the basis of the staining profile, the cells were divided into quadrants highlighting their viability status. In , 97.72% of untreated cells were gated in the LL region of the scatter plot, signifying the unstained nature of cells. shows that untreated but stained cells displayed an increase in green fluorescence signal, representing 87.72% of the cells in the UL quadrant as viable. However, in the case of MIC-treated PAO1 cells, 97.34% of the cells were nonviable and displayed a clear shift in both green and red fluorescence signals. These cells were observed in the UR quadrant (signifying the killing efficiency of PBSNPs toward PAO1 cells) since dead cells take up both the stains (). The flow cytometry data are in agreement with the fluorescence microscopy data for both untreated () and treated () stained cells. Live/Dead staining results in combination with flow cytometry and microscopy data strongly support the enhanced bactericidal potency of PBSNPs against the nosocomial pathogen P. aeruginosa.
Figure 5 Live/Dead staining of planktonic Pseudomonas aeruginosa PAO1 cells by flow cytometer and FM.
Notes: Flow cytometer scatter plot for (A) untreated and unstained P. aeruginosa cells, (B) untreated SYTO9/PI stained, and (C) PBSNPs MIC-treated P. aeruginosa PAO1 cells stained with SYTO9/PI stains was analyzed; LL shows the unstained cells, UL region represents live cells, and dead cells are seen in the UR region. FM of cells prepared (D) untreated stained cells and (E) PBSNPs MIC-treated stained cells (i) represents live cells stained as green, (ii) dead cells stained as red.
Abbreviations: FITC, fluorescein isothiocyanate; PE, phycoerythrin; FM, fluorescence microscopy; PBSNPs, polymyxin B-capped silver nanoparticles; MIC, minimum inhibitory concentration; LL, lower left; LR, lower right; PI, propidium iodide; UL, upper left; UR, upper right.
Antibacterial potency of polymyxin B-capped nanoparticles coating
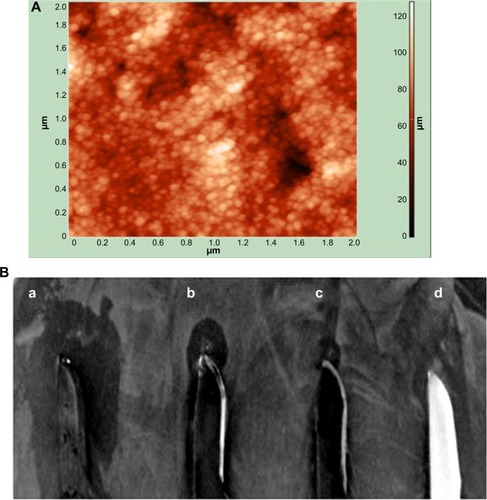
Antibacterial surfaces of materials used for medical devices can complement the fight against the pathogens that propagate in the sessile stage. Since PBSNPs inhibited one of the biofilm-associated pathogen P. aeruginosa PAO1 effectively, we tested whether the functionalized nanoparticles could be adapted for coating on the surgical devices and whether they could retain their inhibitory property following the composite coating. For this, PBSNPs were electroplated on stainless steel surgical blades to scrutinize their antibacterial potency. Coating of PBSNPs onto the surgical blade was observed by AFM study (). The antibacterial activity assayed by the agar-diffusion test () revealed that (a) PBSNPs-coated blades showed enhanced antibacterial activity as compared with blades coated with (b) CSNPs alone (smaller inhibition zone), (c) PBSNPs-coated stainless steel blade when treated with proteinase K (as broad spectrum serine-protease) exhibited significantly reduced antimicrobial activity, and (d) uncoated stainless steel blades (no inhibition zone). These results confirmed that the enhancement in antibacterial activity of PBSNPs-coated surgical blades was due to polymyxin B.
Figure 6 Characterization of coating and antibacterial activity of CSNPs and PBSNPs against Pseudomonas aeruginosa PAOI.
Notes: (A) AFM image of PBSNPs coated on surgical blade, (B) antimicrobial assay displaying the antimicrobial activity of (a) blade coated with PBSNPs, (b) blade coated with CSNPs, (c) blade coated with PBSNPs followed by proteinase K treatment, and (d) uncoated blade against Pseudomonas aeruginosa PAO1. Zone of inhibition is observed surrounding the blade.
Abbreviations: AFM, atomic force microscopy; PBSNPs, polymyxin B-capped silver nanoparticles; CSNPs, citrate-capped silver nanoparticles.
Removal of endotoxin by PBSNPs
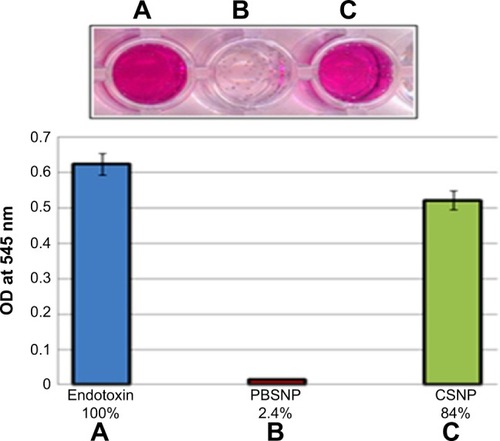
We expanded the utility of such biofunctionalized SNPs further in medical applications. Polymyxin B is known to interact with the LPS part of lipid A of Gram-negative bacterial cell walls. LPS is also known as endotoxin, which is shed by Gram-negative bacteria and is notorious for causing sepsis and septic shock.Citation44 Preservation of the biological activity of polymyxin B upon capping on SNPs prompted us to investigate the ability of such biofunctionalized nanoparticles to remove endotoxins from solutions using a conventional chromogenic Limulus Amebocyte Lysate endotoxin assay (). The PBSNPs efficiently removed endotoxin from the test samples (~97%) as compared with control (~16%). The efficient removal of endotoxin could be attributed to the polymyxin molecules present on SNPs owing to their enhanced surface-to-volume ratio as compared with the conventional column matrix.Citation45 Herein, we have exploited PBSNPs as an efficient agent for endotoxin removal, which could be developed further for removal of endotoxins from other biological fluids while keeping the infections under control.
Figure 7 Endotoxin removal capability of PBSNPs.
Notes: (A) Control endotoxin (no agent added); removal by (B) PBSNPs, (C) CSNPs. Original picture of the wells (top inset) along with graphical representation (bottom). Percentage on X-axis represents the residual endotoxin after the treatment.
Abbreviations: OD, optical density; PBSNPs, polymyxin B-capped silver nano-particles; CSNPs, citrate-capped silver nanoparticles.
Conclusion
Owing to multiple modes of action against bacterial pathogens, the inability of bacteria to develop resistance against silver is well established. In the present study, we have increased the potency of silver by synthesizing biofunctionalized SNPs (PBSNPs) using a facile method and have characterized them using various biophysical and analytical techniques. Owing to a simple and straightforward method of synthesis, on functionalization, the AMP was found to retain its antibacterial property, which was a problem as reported in a recent study.Citation44 The capping of polymyxin B on the SNPs enhanced the antimicrobial effectiveness of the nanoparticles. The extant knowledge about the synergistic mode of action of SNPs along with polymyxin B is well established. In this study, we have immobilized this antibacterial peptide on SNPs and have thus proposed an application of that knowledge in hospital settings. The composite antimicrobial coating of PBSNPs on surgical blades via electrophoretic deposition proved effective in inhibiting the biofilm-forming multiple antibiotic-resistant bacterial strains. Biofilm formation on solid surfaces is a survival strategy of bacterial populations. These biofilms have been found to be associated with recalcitrant bacterial infections in medical devices and patients with compromised immune system.Citation46 The development of efficient antibacterial coating has been recognized as a major challenge in mitigating biofilm formation and controlling the infectious agents in hospital settings. Polymyxin B-capped nanoparticles reported in this study offer an opportunity to develop an effective strategy to fight infectious agents in general and hospital-acquired infections in particular. The usefulness of PBSNPs has also been extended to endotoxin removal from solutions, which widens the applications of these surface-modified nanoparticles. Future challenges to this approach will be to study the stability and physical layering of the AMP coating on medical devices. Also, the performance of the antimicrobial coating under different pathological conditions, including the ability to resist biofilm formation and performance under necrotic tissue with peptidases, needs detailed investigations before this approach can be tested at the clinical stage.
Acknowledgments
This work is supported by the Government of India and Department of Biotechnology Grant No-BT-MED-TF (3) – 2012, National Agricultural Innovation Project (NAIP) grant number C4-30032 of the Indian Council of Agricultural Research (ICAR) to NKN and RP.
Supplementary materials
Figure S2 Scanning electron micrograph of PBSNPs-treated Vibrio fluvialis L-15318 at different magnifications (A) 20,000× and (B) 50,000×.
Abbreviation: PBSNPs, polymyxin B-capped silver nanoparticles.
Figure S3 Scanning electron micrograph of CSNPs-treated Vibrio fluvialis L-15318 at different magnifications (A) 5,000×, (B) 20,000×, and (C) 50,000×.
Abbreviation: CSNPs, citrate-capped silver nanoparticles.
Figure S4 Scanning electron micrograph of untreated Vibrio fluvialis L-15318 at different magnifications (A) 5,000×, (B) 10,000×, and (C) 50,000×.
Figure S5 Scanning electron micrograph of PBSNPs-treated Pseudomonas aeruginosa PAO1 at different magnifications (A) 5,000×, (B) 10,000×, and (C) 50,000×.
Abbreviation: PBSNPs, polymyxin B-capped silver nanoparticles.
Figure S6 Scanning electron micrograph of CSNPs-treated Pseudomonas aeruginosa PAO1 at different magnifications (A) 5,000×, (B) 10,000×, and (C) 50,000×.
Abbreviation: CSNPs, citrate-capped silver nanoparticles.
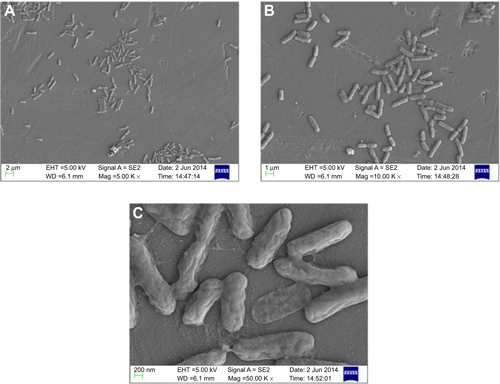
Figure S7 Scanning electron micrograph of untreated Pseudomonas aeruginosa PAO1 at different magnifications (A) 5,000×, (B) 10,000×, and (C) 50,000×.
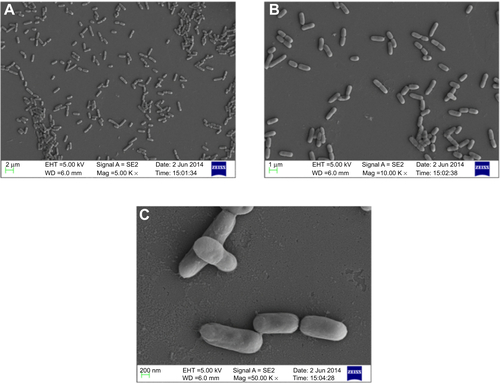
Table S1 Synergy or additive effect between CSNPs and polymyxin B as presented in FIC index
Disclosure
The authors declare that they alone are responsible for the content and writing of this article. The authors report no conflicts of interest in this work.
References
- MagillSSEdwardsJRBambergWMultistate point-prevalence survey of health care-associated infectionsN Engl J Med20143701198120824670166
- GristinaAGImplant failure and the immuno-incompetent fibro-inflammatory zoneClin Orthop Relat Res19942981061188118964
- MunfordRSSevere sepsis and septic shock: the role of gram-negative bacteremiaAnnu Rev Pathol2006146749618039123
- HetrickEMSchoenfischMHReducing implant-related infections: active release strategiesChem Soc Rev200635978078916936926
- GuerreroGAmalricJMutinPHSottoALavigneJPInhibition of bacterial adhesion and prevention of biofilm formation: use of organic self-assembled monolayers on inorganic surfacesPathol Biol (Paris)200957364318824310
- DefezCFabbro-PerayPCazabanMBoudemagheTSottoADauresJPAdditional direct medical costs of nosocomial infections: estimation from a cohort of patients in a French university hospitalJ Hosp Infect20086813013618201796
- MeiLLuZZhangWBioconjugated nanoparticles for attachment and penetration into pathogenic bacteriaBiomaterials20133438103281033724090838
- LaraHHGarza-TreviñoENIxtepan-TurrentLSinghDKSilver nanoparticles are broad-spectrum bactericidal and virucidal compoundsJ Nanobiotechnol2011930
- VeerapandianMYunKFunctionalization of biomolecules on nanoparticles: specialized for antibacterial applicationsAppl Microbiol Biotechnol20119051655166721523475
- ErnestVGajalakshmiSMukherjeeAChandrasekaranNEnhanced activity of lysozyme-AgNP conjugate with synergic antibacterial effect without damaging the catalytic site of lysozymeArtif Cells Nanomed Biotechnol201442533634323863117
- BrownANSmithKSamuelsTALuJObareSOScottMENanopar-ticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureusAppl Environ Microbiol20127882768277422286985
- LiuLHXuKJWangHYSelf assembled cationic peptide nano-particles as an efficient antimicrobial agentNat Nanotechnol2009445746319581900
- WangHXuKLiuLThe efficacy of self assembled cationic antimicrobial peptide nanoparticles against Cryptococcus neoformans for the treatment of meningitisBiomaterials2010312874288120044131
- HancockRESahlHGAntimicrobial and host-defense peptides as new anti-infective therapeutic strategiesNat Biotechnol2006241551155717160061
- PettyJTZhengJHudNVDicksonRMDNA-templated Ag nanocluster formationJ Am Chem Soc20041265207521215099104
- JonesMROsbergKDMacfarlaneRJLangilleMRMirkinCATemplated techniques for the synthesis and assembly of plasmonic nanostructuresChem Rev20111113736382721648955
- MorrisonDCJacobsDMBinding of polymyxin B to the lipid A portion of bacterial lipopolysaccharidesImmunochemistry197613813818187544
- SergeevBMKiryukhinMVRubtsovaMYPrusovANSynthesis of protein A conjugates with silver nanoparticlesColl J200365636638
- SoonhyangPHichamCJodyWNadeauJLAntimicrobial activity and cellular toxicity of nanoparticle–polymyxin B conjugatesNanotechnology20112218510121415471
- MunroCHSmithWEGarnerMClarksonJWhitePCCharacterization of the surface of a citrate-reduced colloid optimized for use as a substrate for surface-enhanced resonance Raman-scatteringLangmuir19951137123720
- Clinical and Laboratory Standards InstituteMethods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard8th edM07-A8Wayne, PACLSI2009
- GhoshINPatilSDSharmaTKSrivastavaSKPathaniaRNavaniNKSynergistic action of cinnamaldehyde with silver nanoparticles against spore-forming bacteria: a case for judicious use of silver nanoparticles for antibacterial applicationsInt J Nanomedicine201384721473124376352
- RudenSHilpertKBerditschMWadhwaniPUlrichASSynergistic interaction between silver nanoparticles and membrane permeabilizing antimicrobial peptidesAntimicrob Agents Chemo20095335383540
- BendaoudMVinogradovEBalashovaNVKadouriDEKachlanySCKaplanJBBroad-spectrum biofilm inhibition by Kingella kingae exopolysaccharideJ Bacteriol2011153879388621602333
- InbakandanDKumarCAbrahamLSKirubagaranRVenkatesanRKhanSASilver nanoparticles with anti microfouling effect: a study against marine biofilm forming bacteriaColloids Surf B Biointerfaces201311163664323907051
- O’TooleGAMicrotiter dish biofilm formation assayJ Vis Exp201147e2437
- O’TooleGAPrattLAWatnickPINewmanDKWeaverVBKolterRGenetic approaches to study of biofilmsMethods Enzymol1999319110910547784
- BarraudNBusonAJarolimekWRiceSAMannitol enhances antibiotic sensitivity of persister bacteria in P. aeruginosa biofilmsPLoS One2013812e8422024349568
- StoodleyHLCostertonJWStoodleyPBacterial biofilms: from the natural environment to infectious diseasesNat Rev Microbiol200429510815040259
- RadzigMANadtochenkoVAKoksharovaOAKiwiJLipasovaVAKhmelIAAntibacterial effects of silver nanoparticles on gram-negative bacteria: influence on the growth and biofilms formation, mechanisms of actionColloids Surf B Biointerfaces201310230030623006569
- EbyDMLuckariftHRJohnsonGRHybrid antimicrobial enzyme and silver nanoparticle coatings for medical instrumentsAppl Mater Interfaces2009115531560
- MockJJBarbicMSmithDRSchultzDASchultzSShape effects in plasmon resonance of individual colloidal silver nanoparticlesJ Chem Phys200211667556759
- DhawanASharmaVToxicity assessment of nanomaterials: methods and challengesAnal Bioanal Chem201039858960520652549
- SocratesGInfrared and Raman Characteristic Group Frequencies, Tables and ChartsChichester, UKJohn Wiley and Sons Ltd20013
- FormaggioFCrismaMRossiPThe first water-soluble 310-helical peptidesChem Eur J200064498450411192082
- TonioloCPoleseAFormaggioFKamphuisCMCircular dichroism spectrum of a peptide 310-helixJ Am Chem Soc19961827442745
- PathaniaRZlitniSBarkerCChemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targetingNat Chem Biol20091184985619783991
- DaugelaviciusRBakieneEBamfordDHStages of polymyxin B interaction with the Escherichia coli cell envelopeAntimicrob Agents Chemother200044112969297811036008
- BondarenkoOIvaskAKakinenAKurvetIKahruAParticle-cell contact enhances antibacterial activity of silver nanoparticlesPLoS One20138e6406023737965
- SauerKCamperAKEhrlichGDCostertonJWDaviesDGPseudomonas aeruginosa displays multiple phenotypes during development as a biofilmJ Bacteriol200218441140115411807075
- PalmerJFlintSBrooksJBacterial cell attachment, the beginning of a biofilmJ Ind Microbiol Biotechnol20073457758817619090
- MichaelBFrederikHFranziskaBWeilenmannHUEgliTAssessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometryAppl Environ Microbiol2007733283329017384309
- GiãoMSWilksSAAzevedoNFVieiraMJKeevilCWValidation of SYTO 9/propidium iodide uptake for rapid detection of viable but non-cultivable Legionella pneumophilaMicrob Ecol200958566219043657
- HéquetAHumblotVBerjeaudJMPradierCMOptimized grafting of antimicrobial peptides on stainless steel surface and biofilm resistance testsColloids Surf B Biointerfaces201184230130921310597
- VincentJLLaterrePFCohenJA pilot-controlled study of a polymyxin B-immobilized hemoperfusion cartridge in patients with severe sepsis secondary to intra-abdominal infectionShock200523540040515834304
- DaviesBCohenJEndotoxin removal devices for the treatment of sepsis and septic shockLancet Infect Dis201111657121183148