Abstract
Recent studies have indicated that statins induce osteogenic differentiation both in vitro and in vivo. The molecular mechanism of statin-stimulated osteogenesis is unknown. Activation of RhoA signaling increases cytoskeletal tension, which plays a crucial role in the osteogenic differentiation of mesenchymal stem cells. We thus hypothesized that RhoA signaling is involved in simvastatin-induced osteogenesis in bone marrow mesenchymal stem cells. We found that although treatment with simvastatin shifts localization of RhoA protein from the membrane to the cytosol, the treatment still activates RhoA dose-dependently because it reduces the association with RhoGDIα. Simvastatin also increased the expression of osteogenic proteins, density of actin filament, the number of focal adhesions, and cellular tension. Furthermore, disrupting actin cytoskeleton or decreasing cell rigidity by using chemical agents reduced simvastatin-induced osteogenic differentiation. In vivo study also confirms that density of actin filament is increased in simvastatin-induced ectopic bone formation. Our study is the first to demonstrate that maintaining intact actin cytoskeletons and enhancing cell rigidity are crucial in simvastatin-induced osteogenesis. The results suggested that simvastatin, which is an osteoinductive factor and acts by increasing actin filament organization and cell rigidity combined with osteoconductive biomaterials, may benefit stem-cell-based bone regeneration.
Introduction
Bone fractures are common in various accident types. Most bone fractures heal spontaneously; however, 5%–10% of bone fractions have delayed union or nonunion,Citation1 possibly necessitating bone grafts to facilitate healing. The ideal bone graft possesses osteoinductive and osteoconductive properties. Statins inhibit 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase and are used clinically to reduce circulatory cholesterol levels.Citation2,Citation3 In vitro studies have revealed that statins increase the expression of osteogenic marker genes in osteoblasts and bone-marrow-derived cells,Citation4–Citation9 and in vivo research has demonstrated that oral administration of statins enhance trabecular bone volume in ovariectomized ratsCitation10,Citation11 and improve bone healing by local delivery.Citation12,Citation13 Recent studies have indicated that statins exert bone anabolic effectsCitation4–Citation9 and can be used as osteoconductive agents in bone tissue engineering.Citation14–Citation16 However, the molecular mechanisms whereby statins enhance osteogenic differentiation in mesenchymal stem cells (MSCs) are not clear. This study sought to clarify the action mechanisms of statins on MSC osteogenic differentiation for enhancing the application of statins in stem-cell-based bone tissue engineering.
The mechanical properties of the environment play a critical role in the fate of stem cells by inducing cytoskeleton organization and increasing cell rigidity.Citation17–Citation22 For example, MSCs cultured on the hardest substrates revealed increased formation of stress fiber and caused osteogenic differentiation.Citation23,Citation24 In addition, disrupting cytoskeleton or decreasing cell rigidity reduces the expression of osteoblastic gene, including alkaline phosphatase (ALP) and osteopontin,Citation25,Citation26 and further promotes adipogenic differentiation by increasing peroxisome proliferator-activated receptor (PPAR) gamma.Citation18,Citation20,Citation27 The cytoskeletal organization and myosin-mediated cytoskeleton tension are regulated by RhoA and its effector, Rho-associated protein kinase (ROCK). Activation of RhoA causes human MSCs to undergo osteogenic differentiation by upregulating the expression of Runx2 gene; however, inactivation of RhoA causes cells to undergo adipogenic differentiation by increasing expression of the adipogenic marker gene PPAR gamma.Citation18,Citation20 Furthermore, inhibiting RhoA signaling promotes chondrogenic differentiation by increasing the expression of SOX-9 gene.Citation21,Citation22 Accordingly, the activation of RhoA and the resultant actin organization and cell rigidity may be necessary for osteogenic differentiation.
Whether RhoA signaling is related to the effects of statins on osteogenesis is unclear. One suggested association is mediated through changes in the posttranslational prenylation of Rho.Citation28 Statins reduce the synthesis of isoprenoid groups by inhibiting HMG-CoA reductase, causing decreased prenylation of small G proteins, including Rho.Citation29,Citation30 For example, mevastatin inhibits the differentiation of osteoclasts by reducing the geranylgeranylation of Rho and Rac.Citation31 Treatment with statin not only inhibits the prenylation of RhoA but also increases the cellular levels of activated RhoA.Citation32–Citation34 Statins are assumed to activate RhoA by reducing the binding of Rho guanine nucleotide dissociation inhibitor (RhoGDIα), a negative regulator of Rho GTPases.Citation32
In this study, we investigated how Rho/actin/cell rigidity signaling contributes to simvastatin-induced osteogenic differentiation in mouse bone marrow mesenchymal stem cells (mBMSCs). We first examined how simvastatin affects RhoA signaling, actin reorganization, cell rigidity, and osteogenesis. We then used the pharmacological agents, ROCKII protein inhibition, myosin II ATP hydrolysis inhibition, and actin polymerization disruption to evaluate how cytoskeletal mechanics alter simvastatin-induced osteogenesis in vitro. Finally, the in vivo effects of simvastatin were assessed using an ectopic bone formation model.
Materials and methods
Cell culture of mBMSCs
The murine bone marrow mesenchymal stem cell line D1 was purchased from ATCC (Manassas, VA, USA). The cells were maintained in a bone medium (Dulbecco’s Modified Eagles Medium containing 100 µg/mL of ascorbic acid, 100 µg/mL of nonessential amino acids, 100 µg/mL of penicillin/streptomycin, and 10% fetal bovine serum) in a 37°C incubator containing 5% CO2. During drug treatment, cells were cultured in a 2% FBS osteoinduction medium containing 10 mM glycerophosphate, 0.2 mM l-ascorbic acid-2-phosphate, and 100 nM dexamethasone in the bone medium.
Drug treatment
The simvastatin solution was prepared as described in our previous study.Citation13 The following pharmacological agents were used: 10 µM Y-27632 dihydrochloride (Tocris Bioscience, Bristol, UK), an inhibitor of p160 ROCKII activity; 10 µM blebbistatin (Tocris Bioscience), a selective inhibitor of the ATPase function of nonmuscle myosin II; and 0.5 µM cytochalasin D (Sigma-Aldrich Co., St Louis, MO, USA), which inhibits actin polymerization by capping the barbed end of F-actin polymers. Cells were pretreated with inhibitors (Y-27632 dihydrochloride, blebbistatin, and cytochalasin D) in the bone medium for 1 hour and cotreated with simvastatin and blockers in the 2% FBS osteoinduction medium. The extent of mineralization, ALP activity, expression of osteogenic gene, and osteogenic protein levels were analyzed at various time points.
ALP activity staining
The Sigma-Aldrich alkaline phosphatase kit (No 85; Sigma-Aldrich Co.) was used to detect ALP activity after treatment with simvastatin. The alkaline-dye solution was prepared according to manufacturer instructions. Cells were fixed with 10% formalin-saline at room temperature for 10 minutes. After being washed once with distilled water, the alkaline-dye mixture was added to each well of a 48-well plate and incubated for 5 minutes. The staining solution was removed, and the wells were washed with distilled water. The samples were then air-dried at room temperature and dissolved in ALP staining solution with 0.1N NaOH, and the absorbance were measured at 405 nm.
Alizarin Red S staining
The mineralization of D1 cells was assayed using Alizarin Red S staining. The cells were stained in Alizarin Red solution on Day 5 of culture in the osteoinduction medium. The cells were fixed with 10% formalin in phosphate buffered saline (PBS) for 5 minutes in 48-well plates, washed twice with distilled water, and stained with Alizarin Red solution for 5 minutes. The cells were then washed with distilled water, air-dried at room temperature, resuspended in 10% acetic acid, and quantified spectrophotometrically at 450 nm.
RNA extraction and real-time PCR
Real-time polymerase chain reaction (PCR) was used to analyze the expression of osteogenic genes. Total RNA of the D1 cells was extracted using ZR RNA MiniPrep (Zymo Research, Irvine, CA, USA) after 12, 24, and 48 hours of incubation in the osteoinduction medium. Total RNA (1 µg) was reverse-transcribed into cDNA by using oligo-dT primers and a High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Waltham, MA, USA). Real-PCR was performed using the Bio-Rad iQ5 real-time PCR detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and SYBR green (Applied Biosystems). The cDNA was amplified with BMP, Runx2, OC, and β-actin primers by using real-time PCR. The primers were as follows: BMP2 forward, 5′-AGCTGCAAGAGACACCCTTTG-3′; BMP2 reverse, 5′-AGCATGCCTTAGGGATTTTGGA-3′; Runx2 forward, 5′-CCCAGCCACCTTTACCTACA-3′; Runx2 reverse, 5′-TATGGAGTGCTGCTGGTCTG-3′; OC forward, 5′-GAGGGCAATAAGGTAGTGAACA-3′; OC reverse, 5′-AAGCCATACTGGTCTGATAGCTCG-3′; β-actin forward, 5′-CCAACCGTGAAAAGATGACC-3′; and β-actin reverse 5′-ACCAGAGGCATACAGGGACA-3′. The PCR conditions were as follows: 95°C for 30 seconds and 95°C for 4 minutes, followed by 40 cycles of 95°C for 10 seconds, 59°C for 15 seconds, and 72°C for 15 seconds. Melting curves were generated to confirm the specificity of the PCRs. The relative mRNA expression level was calculated from the threshold cycle (Ct) and normalized with β-actin. All real-time PCR experiments were performed in triplicate independent experiments.
Immunofluorescence staining
The D1 cells were seeded on glass coverslips. At the end of the experiment, cells were fixed for 15 minutes in 4% paraformaldehyde containing 0.5% Triton-X-100 and preincubated in 100 µL of blocking solution (2.5% bovine serum albumin in phosphate buffered saline) for 40 minutes. The cytoskeleton was detected using 0.5 µM Alexa Fluor 488-conjugated phalloidin (Molecular Probes, Invitrogen, Basel, Switzerland) and DAPI for 1 hour. Osteogenic proteins were labeled by incubation with goat anti-Runx2 (Santa Cruz Biotechnology Inc., Dallas, TX, USA), goat anti-BMP-2 (Santa Cruz Biotechnology Inc.), and rabbit antiosteocalcin (Santa Cruz Biotechnology Inc.) overnight. Cells were rinsed five times with PBST and stained with antigoat IgG conjugated with Alexa 638 (Molecular Probes) or antirabbit IgG conjugated with Alexa 638 (Molecular Probes) combined with 0.5 µM Alexa Fluor 488-conjugated phalloidin (Molecular Probes), and DAPI for 1 hour. Cells were rinsed five times in phosphate buffered saline with 0.01% Triton X-100 (PBST). Glass coverslips were fixed to the slides by using a mounting medium (35 µL) and sealed with nail polish. Images were observed using a confocal microscope (FluoView 1000; Olympus Corporation, Tokyo, Japan) at 60×.
RhoA pull-down assay
RhoA activity was measured as a function of RhoA-GTP binding to corresponding effector-binding domains fused with glutathione S-transferase (GST) in pull-down assays following the manufacturer protocol (EMD Millipore, Billerica, MA, USA). Briefly, cells were lysed in 300 µL of 50 mM Tris, pH 7.4, 10 mM MgCl2, 500 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxycholate, and protease inhibitors. Lysates (500–750 µg) were cleared at 1,000× g for 5 minutes, and the supernatants were rotated for 45 minutes with 30 µg of GST-RBD (GST fusion protein containing the Rho-binding domain of rho-tekin) bound to glutathione–Sepharose beads. Samples were washed with 50 mM Tris, pH 7.4, 10 mM MgCl2, 150 mM NaCl, 1% Triton X-100, and protease inhibitors. GST-RBD pull-downs (active-form RhoA) and lysates were detected using immunoblotting with a Rho-specific antibody (Cell Signaling Technology, Beverly, MA, USA).
Immunoprecipitations
For the immunoprecipitation experiment, D1 cells were lysed in buffer containing 200 mM Tris (pH 7.4), 1% Nonidet P-40, 0.2% SDS, protease inhibitor mix (Hoffman-La Roche Ltd., Basel, Switzerland), and phosphatase inhibitor mix (Hoffman-La Roche Ltd.). Equal amounts of total protein (500 µg) were subjected to immunoprecipitation using Catch and Release (EMD Millipore) with 2 µg of rabbit polyclonal RhoGDIα (Santa Cruz Biotechnology Inc.) according to manufacturer instructions. Immune complexes precipitated with RhoGDIα were subjected to SDS–polyacrylamide gel electrophoresis. After transferring proteins to polyvinylidene difluoride membranes, RhoA was detected using rabbit anti-RhoA (Santa Cruz Biotechnology Inc.). Immunoprecipitates were also immunoblotted, with RhoGDIα used as a loading control.
Immunoblotting
The following antibodies were used to assess protein levels: anti-RhoA (Cell Signaling Technology), antiphosphate MYPT (EMD Millipore), antiphosphomyosin light chain (MLC; Cell Signaling Technology), anti-MLC (Cell Signaling Technology), and anti-β-actin (Sigma-Aldrich Co.). The secondary antibodies used were horseradish peroxidase-conjugated goat antirabbit and antimouse IgG (Santa Cruz Biotechnology Inc.). Protein concentrations were assayed using the Bio-Rad Protein Assay Dye Reagent (Bio-Rad Laboratories, Hercules, CA, USA). Immunoprecipitates were separated on SDS gels. Samples of total protein (50 µg) were separated using 10% SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated with primary antibody. Immunocomplexes were detected using the appropriate horseradish peroxidase-conjugated secondary antibody and observed using a UVP AutoChemi Image and Analysis System (UVP, Upland, CA, USA) with enhanced chemiluminescence (Amersham-Pharmacia International, Piscataway, NJ, USA).
Atomic force microscopy analysis
D1 cells (1×10−5) were seeded on 6 cm dishes, incubated for 24 hours, and treated with RhoA stimulator or inhibitor for 30 minutes. Cells were detected using atomic force microscopy (Nano-wizard II; JPK Instruments, Berlin, Germany), and cell rigidity was detected using a cantilever (ArrowTL1; Oxford Instruments, Scotts Valley, CA, USA) with a 5 µm polystyrene sphere. The indentation area targeted cell nuclei. JPK data-processing software was used to analyze cell rigidity according to the Herz model.
Mouse model of ectopic bone formation
Type I collagen gel was prepared as described in a previous study.Citation35 D1 cells (1×106 per injection) were harvested and mixed with the collagen gel. Experimental groups included cells treated with 1 µM simvastatin (SIM), and 1 µM SIM mixed with Y27632 (10 µM), blebbistatin (Blebb) (10 µM), or cytochalasin D (CD) (0.5 µM), respectively, and untreated controls. The solution (150 µL), including type I collagen gel, D1 cells, and pharmaceuticals drugs, was subcutaneously injected into the dorsal side of 5-week-old male imprinting control region mice. All animal experiments were conducted following the Kaohsiung Medical University guidelines for animal experiments. Two weeks after injection, the animals were euthanized, and the masses at the injection sites were retrieved for detection of osteogenic gene and histological evaluation. All masses were frozen in liquid nitrogen after harvesting. Masses from each group of mice were ground into a fine powder, and RNA was extracted using a TRIzol reagent. For histological analysis, samples were embedded in paraffin and 8 µm sections were prepared. The sections were stained using Alizarin Red S and quantified using Image-Pro Plus 5.0 software (Media Cybernetics, Rockville, MD, USA).
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM) of three replicated, independent experiments. Significant differences between the groups were tested using one-way or two-way analysis of variance, and multiple comparisons were conducted using the Scheffe method. The thresholds for significance and high significance were P<0.05 and P<0.01, respectively.
Results
The osteogenic effects of mBMSCs with simvastatin treatment
We first examined the optimal dose of simvastatin stimulates osteogenesis in mesenchymal stem cells. The cell toxicity data showed that 5 µM SIM significantly enhanced cytotoxicity (). The osteogenic gene expression showed that simvastatin 1 uM significantly up-regulated Runx2, BMP-2 and OC gene expression within 12 hours and gradually increased until 48 hours (). The quantitative results from the ALP stain and Alizarin red staining showed that the simvastatin 0.5 and 1uM enhanced ALP activity at day 3 (), and mineralization at day 5 () in D1 cells. We used simvastatin 1uM to complete the following experiments.
Simvastatin treatment increased active RhoA levels in mBMSCs
Active RhoA was detected using pull-down assays with or without the treatment of simvastatin. Simvastatin increased the levels of active RhoA within 30 minutes of treatment, with peak levels reaching within 3 hours of treatment followed by a slow decrease for 24 hours (). The effect of simvastatin was dose-dependent (). We used a membrane protein extraction assay to determine whether simvastatin alters the membrane anchoring of RhoA. Simvastatin dose-dependently increased the amount of cytosolic RhoA and reduced membrane-bound RhoA (). To further investigate the mechanism of RhoA activation by simvastatin, we assessed the association of RhoA with RhoGDIα by using immunoprecipitation. The amount of RhoA bound to RhoGDIα was lower in simvastatin-treated cells than in untreated cells (). The treatment with simvastatin increased the level of phospho-MYPT, a ROCK substrate molecule, suggesting increased ROCK activity (). Simvastatin increased the phosphorylation of MLC ().
Figure 1 Simvastatin treatment increases active RhoA in D1 cells. Simvastatin constitutively activated RhoA, 0.5–24 hours after treatment.
Notes: Peak activity was reached at 3 hours (A), with RhoA activity increasing in a dose-dependent manner (B), as determined using Western blots of immunoprecipitates. Simvastatin dose-dependently increased RhoA protein levels in the cytosol (aqueous phase) and reduced RhoA content in the membrane (detergent phase) at 3 hours (C). The amount of RhoA that coimmunoprecipitated with RhoGDIα was proportional to the amount of RhoGDIα in the lysate, because simvastatin treatment reduced RhoGDIα-associated RhoA at 3 hours (D). Simvastatin enhanced the ROCK substrate of phospho-MYPT (E) and pMLC (F) at 3 hours. The results are the mean values of three independent measurements, error bars: SEM. *P<0.05; **P<0.01 vs control; #P<0.05, ##P<0.01 vs SIM (1 µm).
Abbreviations: SIM, simvastatin; ROCK, Rho-associated protein kinase; SEM, standard error of the mean; pMLC, phosphomyosin light chain; IP, immunoprecipitation.
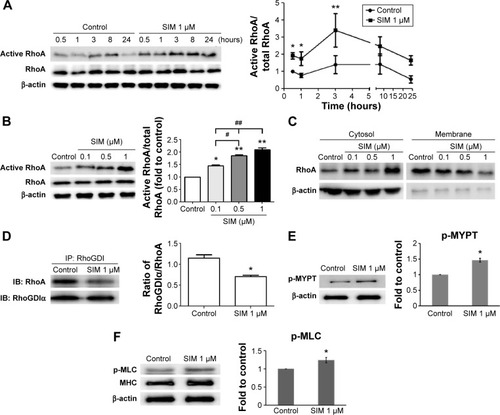
Simvastatin-induced osteogenic differentiation and effects of cytoskeletal alterations
Biochemical agents, including Y-27632, blebbistatin, and cytochalasin D, were used to determine whether RhoA/actin/cell rigidity signaling was involved in simvastatin-induced osteogenesis. Cells treated with Y-27632 or blebbistatin became stellate-shaped and lacked actin fibril organization. Cells treated with cytochalasin D exhibited disrupted actin fibril organization and were clustered into irregular aggregates, even in the cultures treated with simvastatin (). Confocal images revealed clear increases in focal adhesion after treatment with simvastatin and decreases after adding Y-27632, blebbistatin, and cytochalasin D (). These biochemical agents not only disrupted fibril organization but also reduced simvastatin-enhanced cell rigidity (). Next, we measured the mRNA and protein levels of osteogenic genes (Runx2, BMP-2, and OC) in D1 cells treated with simvastatin alone or in combination with Y-27632, blebbistatin, or cytochalasin D. We found that incubation with Y-27632 or blebbistatin or disrupting actin polymerization with cytochalasin D significantly reduced the simvastatin-induced gene expression of Runx2, BMP-2, and OC at 12 and 24 hours (). Confocal images showed that simvastatin-induced increases in Runx2 protein in nuclei were lower at 12 and 24 hours () and that simvastatin-induced increases in BMP-2 and OC protein levels were lower at 12 hours of culture in Y-27632, blebbistatin, or cytochalasin D (). These blockers also inhibited simvastatin-induced ALP activity on Day 3 and mineralization effects on Day 5 after treatment with simvastatin (). These results demonstrated that simvastatin-induced osteogenesis involves RhoA signaling, actin rearrangement, and increased cell rigidity.
Figure 2 Simvastatin increased actin filament formation, focal adhesions, and cellular tension.
Notes: Immunofluorescence staining showed actin cytoskeleton organization (A) in D1 cells with or without simvastatin, biochemical agents (Y-27632, Bleb, and CD), or simvastatin plus biochemical agents (SIM + Y-27632, SIM + Bleb, and SIM + CD) treatment and vinculin expression (a marker of focal adhesions) (B) with control, simvastatin, SIM + Y-27632, SIM + Bleb, and SIM + CD treatment at 12 hours. Cell rigidity was measured using AFM at 12 hours in control cells and those treated with simvastatin, SIM + Y-27632, SIM + Bleb, and SIM + CD, and was presented as Young’s modulus (green, phalloidin-labeled F-actin; blue, nucleus) (C). **P<0.01 vs control; ##P<0.01 vs SIM (1 µm).
Abbreviations: SIM, simvastatin; Bleb, blebbistatin; CD, cytochalasin D; AFM, atomic force microscopy.
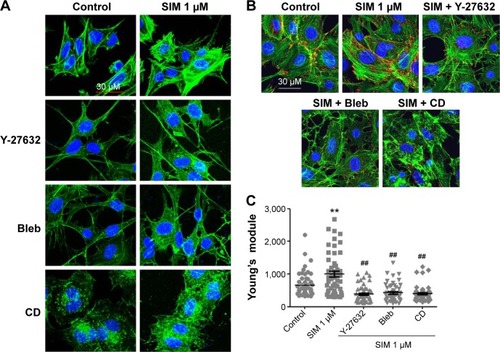
Figure 3 Simvastatin-induced osteogenic gene and protein expression was affected by cytoskeletal alteration agents.
Notes: Disrupting the actin filament organization or decreasing cell rigidity significantly reduced the simvastatin-induced upregulation of osteogenic genes, including Runx2, BMP-2, and OC, 12 (A) and 24 hours (B) after SIM treatment. Immunofluorescence staining revealed simvastatin-induced osteogenic transcription factor Runx2 in the nucleus at 12 (C) and 24 hours (D) and the osteogenic proteins BMP-2 (C) and OC (E) in the cytosol at 24 hours were significantly abrogated by the biochemical agents. Results are mean values of three independent measurements, error bars: SEM. *P<0.05; **P<0.01 vs control; #P<0.05, ##P<0.01 vs SIM (1 µM).
Abbreviations: SIM, simvastatin; Bleb, blebbistatin; CD, cytochalasin D; SEM, standard error of the mean; OC, osteocalcin.
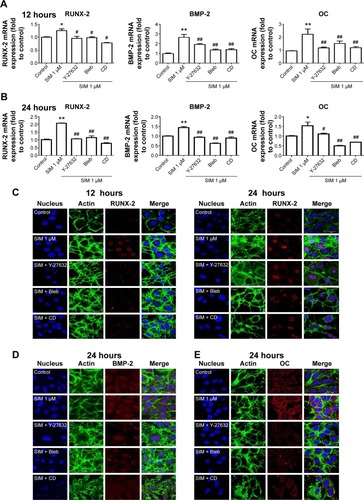
Figure 4 Simvastatin-induced ALP activity and mineralization was reduced by cytoskeletal alteration agents.
Notes: Disrupting actin filament organization or decreasing cell rigidity reduced simvastatin-induced ALP activity on Day 3 (A) and mineralization on Day 5 (B) after treatment. The results are the mean values of three independent measurements, error bars: SEM. **P<0.01 vs control; ##P<0.01 vs SIM (1 µM).
Abbreviations: SIM, simvastatin; Bleb, blebbistatin; CD, cytochalasin D; ALP, alkaline phosphatase; SEM, standard error of the mean.
Simvastatin-induced bone formation and effects of cytoskeletal alterations
We used an ectopic bone formation model in imprinting control region mice to confirm that Rho/actin/cell rigidity signaling is involved in simvastatin-induced osteogenesis. Confocal images revealed clear increases in density of actin fibril after treatment with simvastatin, and actin fibril was attenuated by Y-27632, blebbistatin, or cytochalasin D, 1 week after injection (). In addition, simvastatin-induced increases in expression of Runx2, BMP-2, and OC mRNA were also significantly reduced by the inhibitors, 1 week after injection (). Alizarin Red S images and quantification results demonstrated that simvastatin induced the mineralization of ectopic bone mass; this effect was decreased by treatment with Y-27632, blebbistatin, or cytochalasin D, 2 weeks after injection ().
Figure 5 Simvastatin-induced osteogenic differentiation in an animal model of ectopic bone formation was reduced by cytoskeletal alteration agents.
Notes: Immunofluorescence staining revealed that the F-actin, staining with phalloidin, was increased by simvastatin treatment and reduced by cotreatment with biochemical agents, scale bar represents 50 µm (A). Inhibiting actin filament organization or decreasing cell rigidity significantly inhibited simvastatin-induced osteogenic gene expression (B). Representative masses of ectopic bone formation were stained with Alizarin Red S, the 100× figures are represented as the boxed area in the 40× line, and the mineralization of masses was quantified using Image-Pro Plus software (C). The results are the mean values of three independent measurements, error bars: SEM. **P<0.01 vs control; #P<0.05; ##P<0.01 vs SIM (1 µM).
Abbreviations: SIM, simvastatin; Bleb, blebbistatin; CD, cytochalasin D; SEM, standard error of the mean; OC, osteocalcin.
Discussion
Although statins were developed as inhibitors of cholesterol synthesis, recent studies have indicated that these agents exert pleiotropic effects.Citation36 In this study, we demonstrated that simvastatin enhances osteogenic differentiation in cultured mBMSCs by activating the RhoA/ROCK pathway. We also revealed that this effect is not related to simvastatin-inhibited prenylation. Simvastatin also increases actin cytoskeleton organization, focal adhesion formation, and cellular tension. These simvastatin-induced osteogenic effects are significantly attenuated by blockers of the RhoA-signaling pathway in mBMSCs and in a mouse ectopic bone formation model. Our findings indicated that the treatment with simvastatin enhances osteogenic differentiation in mBMSCs.
Statins are known to block cholesterol synthesis by inhibiting the HMG-CoA reductase that converts HMG-CoA to mevalonate.Citation37 The mevalonate pathway provides the source of isoprenoid intermediates such as farnesylpyrophosphate and geranylgeranylpyrophosphate for the prenylation, cell membrane anchoring, and function of small G proteins.Citation38,Citation39 Therefore, statins inhibit the prenylation of small G proteins and block their related signaling pathways. However, recent studies have reported that statins increase active-form RhoA,Citation32–Citation34,Citation40 including increases in Rho-GTP in cardiac myofibroblastsCitation33 and Rho-GTP and Rac-GTP in THP-1 monocytes and endothelial cells.Citation34,Citation40 In human erythroleukemia cells, lovastatin was reported as reducing the association between Rho and RhoGDα, thereby increasing cytosolic Rho-GTP.Citation32 Notably, the present study found that treatment with simvastatin altered the distribution of RhoA from the membrane to the cytosol () but still activated RhoA (). These results suggested that simvastatin-induced activation of RhoA does not require its anchoring to the membrane. We further observed that the treatment with simvastatin reduces the association between Rho and RhoGDIα (). Accordingly, we suggest that the key mechanism whereby simvastatin activates RhoA is decreasing the Rho/RhoGα association.
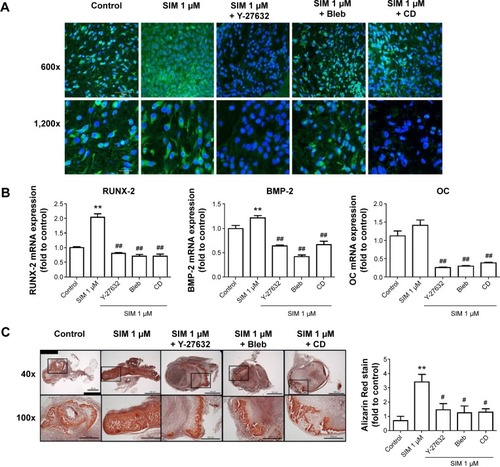
Small GTPase signaling occurs in various cellular events, including proliferation, differentiation, and cell survival.Citation41,Citation42 The constitutive expression of active RhoA is reported to upregulate expression of Runx2 mRNA and cause osteoblastic differentiation in human MSCs.Citation18,Citation20 The attenuation of RhoA activity upregulates expression of PPARγ mRNA and causes adipogenic differentiation.Citation18,Citation20 In this study, we investigated the role of RhoA in the osteogenic differentiation of BMSCs by transfecting constitutively active RhoA (RhoV14) and dominant negative RhoA (RhoV19) (Supplementary Materials and methods). We confirmed that expression of RhoV14 increased the expression of osteogenic marker genes, including Runx2, BMP-2, and OC, and enhanced mineralization (). These results indicated that RhoA signaling plays a crucial role in osteogenic differentiation in mBMSCs. Furthermore, our data demonstrated that the treatment with simvastatin increased the active form of RhoA, and cotreatments with RhoA blockers reduced simvastatin-induced osteogenic differentiation, indicating that simvastatin-induced osteogenic differentiation occurs, at least partly, through the activation of RhoA signaling.
Several studies have used the ROCK inhibitor Y-27632 to investigate the role of the RhoA signaling pathway in osteogenic differentiation, with conflicting results.Citation43,Citation44 Y-27632 has been reported as increasing mineralization in mouse calvarial osteoblastsCitation43,Citation44 and inducing ectopic bone formation in response to rhBMP-2.Citation45 However, ROCK activation has also been reported as enhancing osteogenic differentiation in MSCs under mechanical stimulationCitation18,Citation46–Citation49 or treatment with BMP-2.Citation50 These opposing effects of ROCK in osteogenic differentiation may be due to different roles played by ROCK during different stages of differentiation in MSCs and/or in MSCs from different species. In this study, we observed that Y-27632 reduced simvastatin-induced expression of osteogenic gene, ALP activity, and mineralization in mBMSCs (). Most importantly, our results are the first to demonstrate that Rho/ROCK signaling mediates simvastatin-enhanced osteogenic differentiation in mBMSCs.
Actin cytoskeletal organization and resultant contractility are regulated by RhoA/ROCK signaling, which is crucial in regulating stem cell differentiation.Citation18,Citation20,Citation51 In previous studies, it was found that stem cells that were cultured on a stiff matrix underwent osteogenic differentiation,Citation23,Citation24,Citation52 whereas those on a soft matrix underwent adipogenic differentiation.Citation53–Citation55 These results indicated that actin filament organization and actin-related cell rigidity directly affect the fate of stem cells. In addition, disrupting actin cytoskeleton or decreased cell rigidity because of cytochalism D or blebbistatin reduces osteogenic differentiation even under a rigid substrateCitation48,Citation56 but promotes adipogenic differentiationCitation18,Citation57 in human MSCs. In this study, we found that blocking actin filament organization or losing cell rigidity significantly reduced the simvastatin-enhanced expression of osteogenic marker gene and protein but significantly increased expression of adipogenic marker gene (). Furthermore, in our in vivo study, disrupting the actin filament or decreasing cell rigidity reduced simvastatin-enhanced actin filament density and bone formation. These results strongly indicated that maintaining actin filament intact and enhancing cell rigidity are required for the osteogenic differentiation of MSCs.
Conclusion
We found that simvastatin enhances the osteogenic differentiation of mBMSCs by activating the RhoA/actin/cell rigidity signaling pathway. Both disrupting actin cytoskeletal organization and losing cell rigidity decrease simvastatin-enhanced osteogenic differentiation in mBMSCs. Our study also confirmed that RhoA/actin/cell rigidity signaling plays a crucial role in simvastatin-induced bone formation in vivo. These results suggested that simvastatin is a potent osteoinductive factor that can increase actin filament organization and cell rigidity in MSCs and that combined use with osteoconductive materials may benefit stem-cell-based bone tissue engineering.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
The authors thank Professor Tang Ming-Jer from the Physiology Department of National Cheng Kung University, Tainan, Taiwan, for providing atomic force microscopy and technical support. We thank the Center for Research Resources and Development of Kaohsiung Medical University in protein identification using Olympus Confocal. We gratefully acknowledge support for this research provided by the Ministry of Economic Affairs, Ministry of Science and Technology, Kaohsiung Medical University Hospital and Kaohsiung Medical University Aim for the Top Universities Grant under grant numbers 102-EC-17-A-19-S1-176, MST 102-2314-B-037-021-MY2, KMUH-1I-01, KMUH-2R-35, KMUH-2R-36, and KMU-TP103B01, respectively.
Supplementary materials
Lactate dehydrogenase (LDH) assay
The LDH concentration was assayed utilizing a commercial kit (Cytotoxicity Detection Kit, Roche), which quantifies the LDH release from the cells into the culture medium. The D1 cells were seeded 5,000 cells per well in 96-well plates for 24 hours to promote adherence and then treated with simvastatin 0.1, 0.5, 1, and 5 µM for 24 hours. Absorbance was read at 490 nm by using a microplate reader.
Constitutively active and dominant negative RhoA transfection
RhoA-V14, RhoA-N19, and GFP recombinant plasmids (donated by Dr Tang Ming-Jer at National Cheng Kung University) were transfected into 90% confluent D1 cells by using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) according to the manufacturer protocol. In brief, the cells were cultured in a growth medium without antibiotics before transfection. Transfection was performed using Opti-MEM (250 µL) mixed with 4 µg of RhoA-V14, RhoA-N19, or GFP recombinant plasmids and Lipofectamine 2000 (6 µL). Following incubation at room temperature for 20 minutes, the mixture was added to the cells in six-well plates. After 4–6 hours, the medium was exchanged with the growth medium overnight and cells selected after using G418 for 1 week. After selection, cells were harvested to detect RhoA activity and antiphospho-MLC. The cells were cultured in the growth medium, and gene expression was detected on Days 1, 3, and 5. The cells were cultured in the osteoinduction medium and mineralization was measured on Day 5.
PPARγ primers
The cDNA was amplified using PPARγ primers with real-time PCR. The primers were PPARγ forward, 5′-TGGGTGAAACTCTGGGAGATTC-3′; and PPARγ reverse, 5′-GGAGCAGAAATGCTGGAGAAATC-3′.
Figure S1 Simvastatin enhanced osteogenesis in D1 cells.
Notes: The LDH cytotoxicity assay after simvastatin treatment at 24 hours (A). Simvastatin increased osteogenic gene expression, including in Runx-2 (B), BMP-2 (C), and osteocalcin (OC) (D) by real-time PCR. Simvastatin dose-dependently increased ALP stain on Day 3 (E) and Alizarin Red S stain on Day 5 (F). *P<0.05; **P<0.01; ##P<0.01 vs SIM (1 µM).
Abbreviations: SIM, simvastatin; ALP, alkaline phosphatase; LDH, lactate dehydrogenase; PCR, polymerase chain reaction.
Figure S2 RhoA transfection alters osteogenic differentiation in D1 cells.
Notes: Constitutively active forms of RhoA exhibited more active RhoA and phospho-MLC (A), and then increased osteogenic gene expression (B), and mineralization (C) to more than that of the dominant negative form of RhoA.
Abbreviations: pMLC, phosphomyosin light chain; OC, osteocalcin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Figure S3 RhoA-signaling blockers restored simvastatin-inhibited PPARγ mRNA levels in D1 cells.
Notes: RhoA-signaling blockers significantly restored simvastatin-inhibited PPARγ gene expression 12 hours after treatment. **P<0.01 vs control; ##P<0.01 vs SIM (1 µM).
Abbreviations: SIM, simvastatin; Bleb, blebbistatin; CD, cytochalasin D; PPARγ, peroxisome proliferator-activated receptor gamma.
Disclosure
The authors declare no conflicts of interest in this work.
References
- YelinEHFeltsWRA summary of the impact of musculoskeletal conditions in the United StatesArthritis Rheum19903357507552346529
- CorsiniABellostaSBaettaRFumagalliRPaolettiRBerniniFNew insights into the pharmacodynamic and pharmacokinetic properties of statinsPharmacol Ther199984341342810665838
- SchachterMChemical, pharmacokinetic and pharmacodynamic properties of statins: an updateFundam Clin Pharmacol200519111712515660968
- MaedaTMatsunumaAKawaneTHoriuchiNSimvastatin promotes osteoblast differentiation and mineralization in MC3T3-E1 cellsBiochem Biophys Res Commun2001280387487711162604
- SongCGuoZMaQSimvastatin induces osteoblastic differentiation and inhibits adipocytic differentiation in mouse bone marrow stromal cellsBiochem Biophys Res Commun2003308345846212914771
- HwangRLeeEJKimMHCalcyclin, a Ca2+ ion-binding protein, contributes to the anabolic effects of simvastatin on boneJ Biol Chem200427920212392124714973129
- BaekKHLeeWYOhKWThe effect of simvastatin on the proliferation and differentiation of human bone marrow stromal cellsJ Korean Med Sci200520343844415953866
- SolomonDHFinkelsteinJSWangPSAvornJStatin lipid-lowering drugs and bone mineral densityPharmacoepidemiol Drug Saf200514421922615386702
- MaritzFJConradieMMHulleyPAGopalRHoughSEffect of statins on bone mineral density and bone histomorphometry in rodentsArterioscler Thromb Vasc Biol200121101636164111597938
- HoMLChenYHLiaoHJSimvastatin increases osteoblasts and osteogenic proteins in ovariectomized ratsEur J Clin Invest200939429630319292885
- MundyGGarrettRHarrisSStimulation of bone formation in vitro and in rodents by statinsScience199928654461946194910583956
- SkoglundBForslundCAspenbergPSimvastatin improves fracture healing in miceJ Bone Miner Res200217112004200812412808
- TaiICFuYCWangCKChangJKHoMLLocal delivery of controlled-release simvastatin/PLGA/HAp microspheres enhances bone repairInt J Nanomedicine201383895390524143094
- SteinDLeeYSchmidMJLocal simvastatin effects on mandibular bone growth and inflammationJ Periodontol200576111861187016274305
- ThylinMRMcConnellJCSchmidMJEffects of simvastatin gels on murine calvarial boneJ Periodontol200273101141114812416771
- NyanMSatoDOdaMBone formation with the combination of simvastatin and calcium sulfate in critical-sized rat calvarial defectJ Pharmacol Sci2007104438438617721043
- DischerDEMooneyDJZandstraPWGrowth factors, matrices, and forces combine and control stem cellsScience200932459351673167719556500
- McBeathRPironeDMNelsonCMBhadrirajuKChenCSCell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitmentDev Cell20046448349515068789
- SunYChenCSFuJForcing stem cells to behave: a biophysical perspective of the cellular microenvironmentAnnu Rev Biophys20124151954222404680
- ArnsdorfEJTummalaPKwonRYJacobsCRMechanically induced osteogenic differentiation – the role of RhoA, ROCKII and cytoskeletal dynamicsJ Cell Sci2009122pt 454655319174467
- WoodsAWangGBeierFRhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesisJ Biol Chem200528012116261163415665004
- WoodsABeierFRhoA/ROCK signaling regulates chondrogenesis in a context-dependent mannerJ Biol Chem200628119131341314016565087
- TanSFangJYYangZNimniMEHanBThe synergetic effect of hydrogel stiffness and growth factor on osteogenic differentiationBiomaterials201435205294530624703716
- ZouaniOFKaliskyJIbarboureEDurrieuMCEffect of BMP-2 from matrices of different stiffnesses for the modulation of stem cell fateBiomaterials20133492157216623290467
- TakeuchiYSuzawaMKikuchiTNishidaEFujitaTMatsumotoTDifferentiation and transforming growth factor-beta receptor down-regulation by collagen-alpha2beta1 integrin interaction is mediated by focal adhesion kinase and its downstream signals in murine osteoblastic cellsJ Biol Chem19972724629309293169361011
- TomaCDAshkarSGrayMLSchafferJLGerstenfeldLCSignal transduction of mechanical stimuli is dependent on microfilament integrity: identification of osteopontin as a mechanically induced gene in osteoblastsJ Bone Miner Res19971210162616369333123
- FengTSzaboEDziakEOpasMCytoskeletal disassembly and cell rounding promotes adipogenesis from ES cellsStem Cell Rev201061748520148318
- AdamsonPMarshallCJHallATilbrookPAPost-translational modifications of p21rho proteinsJ Biol Chem19922672820033200381400319
- WongWWDimitroulakosJMindenMDPennLZHMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosisLeukemia200216450851911960327
- LaufsULiaoJKTargeting Rho in cardiovascular diseaseCirc Res200087752652811009552
- WooJTNakagawaHKrecicAMInhibitory effects of mevastatin and a geranylgeranyl transferase I inhibitor (GGTI-2166) on mononuclear osteoclast formation induced by receptor activator of NF kappa B ligand (RANKL) or tumor necrosis factor-alpha (TNF-alpha)Biochem Pharmacol2005691879515588717
- TurnerSJZhuangSZhangTBossGRPilzRBEffects of lovastatin on Rho isoform expression, activity, and association with guanine nucleotide dissociation inhibitorsBiochem Pharmacol200875240541317920041
- PorterKETurnerNAO’ReganDJBalmforthAJBallSGSimvastatin reduces human atrial myofibroblast proliferation independently of cholesterol lowering via inhibition of RhoACardiovasc Res200461474575514985071
- VecchioneCBrandesRPWithdrawal of 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors elicits oxidative stress and induces endothelial dysfunction in miceCirc Res200291217317912142351
- WangYHChiuWTWangYKDeregulation of AP-1 proteins in collagen gel-induced epithelial cell apoptosis mediated by low substratum rigidityJ Biol Chem2007282175276317085440
- LiaoJKLaufsUPleiotropic effects of statinsAnnu Rev Pharmacol Toxicol2005458911815822172
- GoldsteinJLBrownMSRegulation of the mevalonate pathwayNature199034362574254301967820
- XiaZTanMMWongWWDimitroulakosJMindenMDPennLZBlocking protein geranylgeranylation is essential for lovastatin-induced apoptosis of human acute myeloid leukemia cellsLeukemia20011591398140711516100
- ZhangFLCaseyPJProtein prenylation: molecular mechanisms and functional consequencesAnnu Rev Biochem1996652412698811180
- CordleAKoenigsknecht-TalbooJWilkinsonBLimpertALandrethGMechanisms of statin-mediated inhibition of small G-protein functionJ Biol Chem200528040342023420916085653
- BoureuxAVignalEFaureSFortPEvolution of the Rho family of ras-like GTPases in eukaryotesMol Biol Evol200724120321617035353
- BusteloXRSauzeauVBerenjenoIMGTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivoBioessays200729435637017373658
- KanazawaIYamaguchiTYanoSYamauchiMSugimotoTActivation of AMP kinase and inhibition of Rho kinase induce the mineralization of osteoblastic MC3T3-E1 cells through endothelial NOS and BMP-2 expressionAm J Physiol Endocrinol Metab20092961E139E14619001547
- HarmeyDStenbeckGNobesCDLaxAJGrigoriadisAERegulation of osteoblast differentiation by Pasteurella multocida toxin (PMT): a role for Rho GTPase in bone formationJ Bone Miner Res200419466167015005854
- YoshikawaHYoshiokaKNakaseTItohKStimulation of ectopic bone formation in response to BMP-2 by Rho kinase inhibitor: a pilot studyClin Orthop Relat Res2009467123087309519609629
- WangGWoodsASabariSPagnottaLStantonLABeierFRhoA/ROCK signaling suppresses hypertrophic chondrocyte differentiationJ Biol Chem200427913132051321414726536
- HamamuraKSwarnkarGTanjungNRhoA-mediated signaling in mechanotransduction of osteoblastsConnect Tissue Res201253539840622420753
- KhatiwalaCBKimPDPeytonSRPutnamAJECM compliance regulates osteogenesis by influencing MAPK signaling downstream of RhoA and ROCKJ Bone Miner Res200924588689819113908
- McMurrayRJGadegaardNTsimbouriPMNanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotencyNat Mater201110863764421765399
- WangYKYuXCohenDMBone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tensionStem Cells Dev20122171176118621967638
- Sarasa-RenedoATunc-CivelekVChiquetMRole of RhoA/ROCK-dependent actin contractility in the induction of tenascin-C by cyclic tensile strainExp Cell Res200631281361137016448650
- EnglerAJSenSSweeneyHLDischerDEMatrix elasticity directs stem cell lineage specificationCell2006126467768916923388
- TrappmannBGautrotJEConnellyJTExtracellular-matrix tethering regulates stem-cell fateNat Mater201211764264922635042
- FuJWangYKYangMTMechanical regulation of cell function with geometrically modulated elastomeric substratesNat Methods20107973373620676108
- ParkJSChuJSTsouADThe effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-betaBiomaterials201132163921393021397942
- ShihYRTsengKFLaiHYLinCHLeeOKMatrix stiffness regulation of integrin-mediated mechanotransduction during osteogenic differentiation of human mesenchymal stem cellsJ Bone Miner Res201126473073820939067
- ArnsdorfEJTummalaPJacobsCRNon-canonical Wnt signaling and N-cadherin related beta-catenin signaling play a role in mechanically induced osteogenic cell fatePLoS One200944e538819401766
