?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
To gain a deeper understanding of the physicochemical phenomenon of self-assembled nanoparticles of different generations and ratios of poly (amidoamine) dendrimer (PAMAM) dendrimer and a short-stranded DNA (antisense oligonucleotide), multiple methods were used to characterize these nanoparticles including photon correlation spectroscopy (PCS); zeta potential measurement; and atomic force microscopy (AFM). PCS and AFM results revealed that, in contrast to larger molecules of DNA, smaller molecules produce more heterodisperse and large nanoparticles when they are condensed with a cationic dendrimer. AFM images also showed that such nanoparticles were spherical. The stability of the antisense content of the nanoparticles was investigated over different charge ratios using polyacrylamide gel electrophoresis. It was clear from such analyses that much more than charge neutrality point was required to obtain stable nanoparticles. For cell uptake, self-assembled nanoparticles were prepared with PAMAM G5 and 5’-FITC labeled antisense and the uptake experiment was carried out in T47D cell culture. This investigation also shows that the cytotoxicity of the nanoparticles was dependent upon the generation and charge ratio of the PAMAM dendrimer, and the antisense concentration had no significant effect on the cytotoxicity.
Introduction
Gene therapy is a promising approach for the treatment of both acquired (eg, AIDS and cancers) and inherited (cystic fibrosis, hemophilia) diseases which are lacking effective drug treatments.Citation1–Citation4 During the last ten years many targets of gene therapy have been identified although there is still no satisfactory working vector to deliver genetic materials safely and efficiently in vivo.Citation5 Gene delivery vectors can be roughly divided into two categories: viral and non-viral. Viral vectors are efficient in delivering a gene to the target cells although there are problems related to the safety and large-scale production. Moreover, there are problems with viral vectors where the synthetic oligo- or polynucleotides (such as siRNAs or antisense oligodeoxynucleotides [ODNs]) cannot be easily inserted into the viruses and delivered satisfactorily. Non-viral (ie chemical) vectors are attractive because of their lower immunogenicity, greater safety and ease of preparation (for reviews, seeCitation6,Citation7).In addition, chemical vectors can be formulated via a simple routine pharmaceutical process, and are increasingly used in experiments in vitro, in vivo and in clinical trials.Citation8
Among chemical vectors, dendrimers are uniquely efficient in the delivery of genetic materials into cells, especially in vitro.Citation9,Citation10 The size, shape and surface functionality of the dendrimer can be precisely controlled by designing an appropriate synthesis and modification process. The final products of a well-designed synthesis process have monodisperse structures. This is important especially when the hetero- or monodispersity of the used materials could greatly impact any responses in their medical or non-medical applications. Monodisperse raw materials often show very predictable consequences in their applications.
Since its introduction by Tomalia and colleagues in 1984,Citation11 the poly(amido amine) dendrimer (PAMAM) has become widely used in many fields.Citation12–Citation15 One of these being as a gene delivery vehicle and as such it has become one of the most successful vectors in the field of in vitro gene delivery.Citation16–Citation19 PAMAM dendrimers are a group of symmetrical hyper-branched macromolecules which, at generations higher than five, have a three-dimensional spheroid or elliptical shape with a diameter of approximately 6–12 nM, depending on the core structure, surface groups and generation (for more details of nomenclatures about PAMAM dendrimers, see).Citation12 Full generations of PAMAM have primary amino groups at their surface which are protonated at a physiological pH; therefore they can interact with the phosphate groups of nucleic acid strings, and condense them into more stable and protected particles from tens to many hundreds of nanometers in size (self-assembled nanoparticles of PAMAM/ANS). However, one problem with using PAMAMs especially in vitro is that it is toxic at the higher generations which are used for the efficient delivery of nucleic acids.Citation20,Citation21 For example, Polyfect® (which is a generation 6 PAMAM) and Superfect® (fractured generation 6 PAMAM) have been proved to have noticeable cyto- and genotoxicities in vitro.Citation22 At the same time, lower generations of PAMAM such as generation 5, while having the ability to condense both DNA or RNA, have a lower toxicity.Citation23,Citation24 However, very little is known about these lower generations. In order to elucidate more about their interaction with nucleic acids we selected, (based on previous work by a member of our group), an antisense oligonucleotide (ANS) designed against epidermal growth factor receptor (EGFR).Citation24 Some of its characteristics were then evaluated prior to biological application. The relationship between EGFR and the poor prognosis of some types of cancers, especially breast, ovarian, head and neck cancer, has been established and the overexpression of this receptor is known to be correlated to the division, growth and migration of tumor cells.Citation25 On the other hand, recent clinical trials for example using Mipomersen® and previously commercialized Vitravene®, an antisense drug, show that antisense therapy still has value in the field of genetic research and the therapeutic downregulation of a gene function.Citation26,Citation27 Antisense oligodeoxynucleotides (ANS-ODNs) are sequences of short single-stranded ODNs, often 19–25 bases in length. They can hybridize to a specific mRNA in the cytoplasm or nucleus via the Watson-Crick base pairing mechanism, which leads to the blocking of mRNA translation, then sensitization of target mRNA to specific RNase-H, which finally leads to degradation of the mRNA.Citation28 ANS-ODNs are hydrophilic macromolecules that need to be properly delivered like any other genetic material to the defined cells and to the cytoplasm. The physicochemical properties of these smaller macromolecules seem to be different from those of their heavier family members; for example DNAzyme, ribozyme or other kinds of gene therapeutics (eg, plasmids), when they interact with chemical vectors. Moreover, the physicochemical characterization of non-viral vectors is an important step before any in vivo or in vitro application.
Thus the main objectives of this study were to investigate the physicochemical properties (size, morphology and zeta potential) which are important for the biological activity of self-assembled PAMAM/ANS-ODN nanoparticles, at different generations of PAMAM, to evaluate the cytotoxicity of the prepared nanoparticles at different charge ratios, and to determine the impact of this vector on the cell uptake of self-assembled nanoparticles in T47D cell culture. The difference between the interaction of big double-stranded DNA molecules and small single-stranded DNA (EGFR ANS) with PAMAM dendrimer is also discussed.
Method
Materials
Polyamidoamine dendrimer generations two, three, four and five (PAMAM G2, G3, G4, G5), which have ethylene-diamine as an initiator core, 16, 32, 64 and 128 primary amino groups at their surface and a molecular weight of 3256, 6909, 14215 and 28826 Da respectively, were synthesized according to the previously reported method.Citation29 Briefly, this synthesis was done via iterative reactions of Michael addition of methylacrylate to the ethylenediamine for half generations, followed by exhaustive amidation of half generations by high excess methanolic ethylenediamine at the next step for full generations. The product at each step of the synthesis was purified by three methods: azeotropic distillation, high vacuum overnight, and ultrafiltration with defined molecular weight cut-offs, followed by the freeze drying of the results. They were later characterized by,Citation1 proton nuclear magnetic resonance (H-NMR) and Fourier-transform infrared spectrometry (FTIR). The sequence of phosphorothioated EGFR ANS 5′(TTT CTT TTC CTC CAG AGC CCG)3′ was according to that reported by Petch and colleagues.Citation28 5′-FITC labeled phosphorothioated EGFR ANS FITC-5′(TTT CTT TTC CTC CAG AGC CCG)3’ was used for cell uptake investigations. All sequences were purchased from TIB Molbiol GmbH, Berlin, Germany or Bioneer Daejeon, Korea and used as received. All solvents and reagents were analytical or high pressure liquid chromatography (HPLC) grade and purchased from Merck KGaA, Darmstadt, Germany.
Preparation of self-assembled nanoparticles for particle size and zeta potential analysis
Self-assembled nanoparticles of PAMAM with ANS were prepared in 5% (w/v) dextrose, pH 7, at different molar ratios of PAMAM primary amino groups to ANS phosphate groups (N/P ratio). Separate stock solutions of 5 mg/mL of PAMAM for different generations in 5% dextrose were prepared. Stock solution of 100 μM of EGFR ANS in diethylpyrocarbonate (DEPC) treated water was constructed. For preparation of N/P ratio of ten, 5 μL of PAMAM solution containing 1 μM primary amine was diluted to 50 μL by 5% dextrose and added drop-wise to 10 μM of ANS solution diluted with 5% dextrose. For N/P ratios of 2.5, 5 and 20 the volume of ANS was fixed at the same volume of N/P 10, while the proper relative volume was taken from PAMAM solution. In the case of N/P ratio of 0.5, the stock solution was diluted by a factor of 10 to make it possible to take the exact volume. Then the nanoparticles were mixed by pipetting up and down 15 times and then incubated for 10–15 minutes at room temperature. Self-assembled nanoparticles of polyethyleneimine (PEI)/ANS were prepared according to the method described by Boussif and colleagues.Citation30
For particle size analysis, 100 μl of nanoparticles was diluted 1:10 in 5% dextrose before the particle size and/or zeta potential of nanoparticles was evaluated using Malvern Nanosizer, ZN series, Malvern Instruments Ltd, Malvern, UK after substantially vortexing of the mixture before measurement.
Atomic force microscopy (AFM) analysis
For AFM analysis, different N/P ratios of PAMAM G5 were selected and diluted 1:100 with double-distilled H2O. After 10 minutes incubation at room temperature, 5 μL of each formulation was then added on a freshly cleaved mica sheet. After 5 minutes incubation at room temperature, the sheet was washed twice with 100 μL of double-distilled H2O. The prepared samples were first dried from the edge of the mica sheet by using a paper tissue, then by exposure to a gentle air flow for 10 minutes. Thereafter, the samples were subjected immediately to AFM study using a DME DualScope/Rasterscope™ SPM (Danish micro engineering Herlev, Denmark). The AFM studies were performed at AC mode, spring constant of 2.8 N/m with resonance frequency of 100 KHz, speed of 30–40 μm/s, and force constant of 42 N. Processing of topographic or phase images were carried out using DME SPM software version 2.1.1.2.
Polyacrylamide gel electrophoresis (PAGE) analysis
PAMAM G5/ANS nanoparticles at various N/P ratios were prepared as described earlier, and then diluted in double-distilled H2O. Nanoparticles (16 μL) containing 1.6 μg of ANS were mixed with 4 μL of urea loading dye (480 mg urea, 40 μL of 0.5 M ethylenediaminetetraacetic acid (EDTA), 2 μL of 1 M Tris pH 7.5 and 140 mg bromphenol blue per 1 mL of dye solution). After mixing, the suspension was loaded in each well of 16% polyacrylamide gel which was electrophoresed for 90 minutes at 250 volts. The gel was then soaked in 0.5% ethidium bromide solution for 20 minutes then visualized under UV illumination using Bio Doc it (UVPco, UK).
Cell culture
T47D cells (National Cell Bank of Iran, Pasteur Institute, Human breast cancer passage number between 15 and 40) were cultured in RPMI 1640 (Gibco,® Invitrogen Ltd, Paisley UK) supplemented with 10% fetal bovine serum (FBS) (Gibco), streptomycin (100 ng/mL), and penicillin (100 units/mL) in 37°C, 5% humidified CO2 incubator. The cells were passaged 2–3 times per week.
Cell uptake experiments
Cellular uptake of EGFR AS/PAMAM G5 nanoparticles was investigated using 5’end FITC-labeled EGFR ANS. Two days before the experiment, the cells were seeded in 6-well plates 4 × 105 cells/well in 3 mL of the medium. Then on the day of transfection, the medium was removed, the cells were washed with phosphate buffered saline (PBS), and 400 μL of nanoparticles diluted 1:5 in FBS-free RPMI, at different N/P ratios for PAMAM G5 were added to the cells. The final concentration of the ANS was 1 μM. The cells were incubated for 4 hours at 37°C. After incubation, the medium of each well was removed and washed with PBS and analyzed by inverted fluorescent microscope (Olympus IX50, Olympus Inc.) using 100X and 400X magnifications.
Cytotoxicity assays
The cytotoxicity of different generations of PAMAM nanoparticles was studied using MTT assay (3-[4,5-dimethylthiazol-2- yl]-2,5-diphenyltetrazolium bromide [Sigma Aldrich Munich, Germany]). PAMAM G5 at different N/P ratios and two different ANS concentrations (0.5 and 1 μM), and PAMAM G2-G4 at fixed N/P ratio of 10 were prepared according to the described method. After mixing and incubation of the proper dilution of PAMAM and ANS solution, the nanoparticles were diluted 1:10 in FBS-free RPMI medium. T47D cells (1 × 104 cells/well) were seeded in 96-well plates 48 hours before transfection. On the day of transfection, the medium was removed from the wells and 180 μL of freshly prepared nanoparticles were incubated with the cells. Proper control of FBS-free RPMI 1640, ANS alone, and PAMAM G2, G3, G4 and G5 alone at exact concentrations that were used for preparing different N/P ratios were used but without the addition of ANS. The incubation time was 4 hours, after which the medium was replaced with growth medium after washing of the cells by PBS. After 48 hours incubation, 25 μL of MTT dye (4 mg/mL) was added to each well and the cells were incubated for a further 3 hours at 37°C. Then the medium was removed and 100 μl dimethyl sulphoxide (DMSO) was added to each well. The plates were shaken for 30 seconds and the absorbance of Formazan dye was recorded at 540 nM using a microplate reader (Sunrise,™ Tecan, Grödig, Austria). The viability of the cells was calculated by employing of the following equation:
where Viability is the cell viability as a percentage of control, ODtreatment, ODcontrol, and ODblank are the measured optical density of the treated, RPMI and blank wells at a wavelength of 540 nm, respectively.
Statistical analysis
For comparison of two means, Student t-test was applied, and for more than two means, one way analysis of variance (ANOVA) was selected with Bonferroni post hoc. P value < 0.05 was considered as significantly different in all cases, unless otherwise mentioned.
Results
Size and zeta potential analysis of the nanoparticles
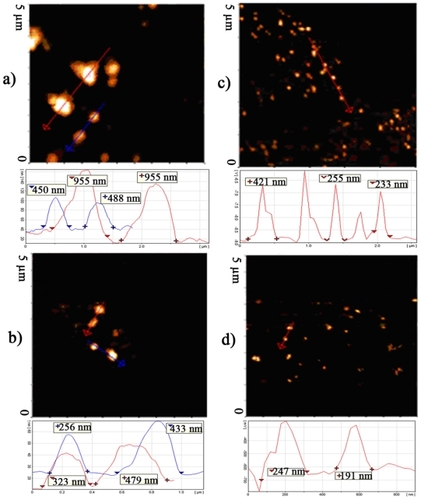
Self-assembled nanoparticles of the G2 through G4 of PAMAM with ANS at N/P ratio of 10 had a peak between the ranges of 40–500 nm. For G5 at different N/P ratios a peak in the range of 40–1100 nm was repeatable ( and ). The higher the N/P ratio, the narrower the observed particle distribution (). For example, for generation 5 the particles larger than 800 nm, present at the N/P ratio of 0.5 were not observed at higher N/P ratios ( and ). In some cases, especially for the lower generations, the graph was multimodal instead of a single peak. Various repeats of the measurements revealed that by increasing the charge ratios, the probability of observing multiple peaks decreased.
Figure 1 Size distribution by intensity of PAMAM/ANS nanoparticles (a and b) and Zeta potential graphs of different N/P ratios of PAMAM G3 (
 ), and G5 (▪) in 5% dextrose (c). Fixed concentrations of 500 nM of ANS in 5% dextrose were mixed with different generations of PAMAM at N/P ratio of 10 a). Part a) shows all size distributions at the same intensity scale in one graph. A peak of larger aggregates of G2/ANS nanoparticles can be observed in part a). Nanoparticles of various N/P ratios of PAMAM G5 which were formulated in 5% dextrose are shown in part b). In part c), the represented data are means of two (without error bar) or three (mean ± SD) measurements.
), and G5 (▪) in 5% dextrose (c). Fixed concentrations of 500 nM of ANS in 5% dextrose were mixed with different generations of PAMAM at N/P ratio of 10 a). Part a) shows all size distributions at the same intensity scale in one graph. A peak of larger aggregates of G2/ANS nanoparticles can be observed in part a). Nanoparticles of various N/P ratios of PAMAM G5 which were formulated in 5% dextrose are shown in part b). In part c), the represented data are means of two (without error bar) or three (mean ± SD) measurements.Abbreviations: PAMAM, poly(amidoamine) dendrimer; SD, standard deviation; ANS, antisense oligonucleotide.
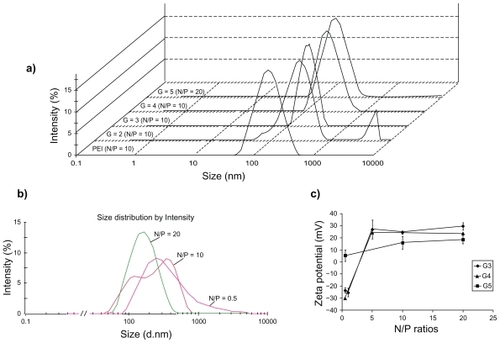
Table 1 Size distribution of nanoparticles determined by AFM and PCS methods. For AFM, n particles in different images were measured by the software of the device and then expressed as mean ± SD
The zeta potential of nanoparticles showed an ascending pattern from lower to higher N/P ratios which finally reached a plateau at higher charge ratios (). For G5 at N/P ratio of 0.5, the zeta potential was near to positive (5.2 ± 4.8 mV) and at N/P ratios of 10 and 20 the average surface charge of particles was 15.7 ± 5.1 mV and 18.4 ± 3.4 mV, respectively (, n = 3).
Despite the results observed for PAMAM, nanoparticles of PEI/ANS showed a strongly positive charge of 42.3 ± 5.9 (mean ± standard deviation [SD]) at N/P of 6.
AFM experiment
are representative of the AFM phase or topographic images of nanoparticles and surface profile of PAMAM G5/ANS at charge ratios of 0.5, 5, 10 and 20, respectively. Particles at different charge ratios were measured in AFM pictures using the DME software, summarized in . Similar to the PCS results, the nanoparticles at a charge ratio of 0.5 were bigger than at higher N/P ratios ( and ). shows that the smallest size distribution belongs to the N/P ratio of 20, in which nearly all the particles had a size distribution of less than 300 nm. Particles at different ratios had both spherical and rod-like shapes but AFM evaluation suggested that at higher charge ratios the spherical shape became the predominant morphology.
PAGE analysis
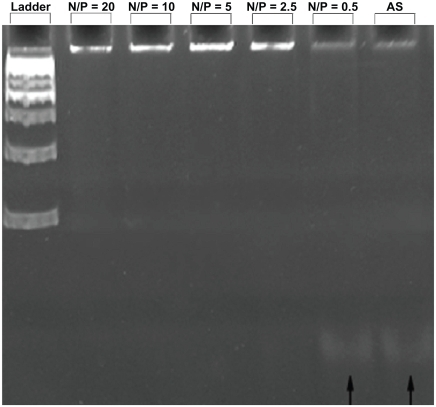
PAGE experiments showed that N/P ratios higher than 2.5 were necessary in order to stabilize and retard the ANS content of PAMAM G5/ANS self-assembled nanoparticles. shows that the lower N/P ratio of 0.5 could also move through the gel at almost the same speed as the naked ANS. But at N/P ratios of 2.5 or higher, almost all of the ANS cargo remained in the wells.
Cellular uptake of nanoparticles by human breast cancer T47D cells
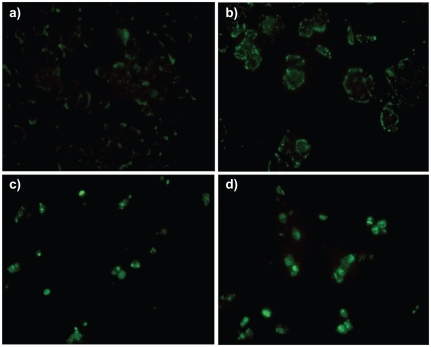
shows the cellular uptake of self-assembled nanoparticles of PAMAM G5 with FITC-tagged-ANS at different N/P ratios. After 4 hours incubation, the majority of naked FITC-ANS were attached to the cell surface which can be seen as pale green spots in and only a negligible amount of them entered the cytoplasm. At an N/P ratio of 0.5, PAMAM/ANS nanoparticles were not effectively taken up and many were bound to the cell surface (). But for N/P ratios of 10 and 20, the cytoplasm and nucleus of the T47D cells were the main location of the FITC-ANS/PAMAM G5 complexes ().
Toxicity of nanoparticles
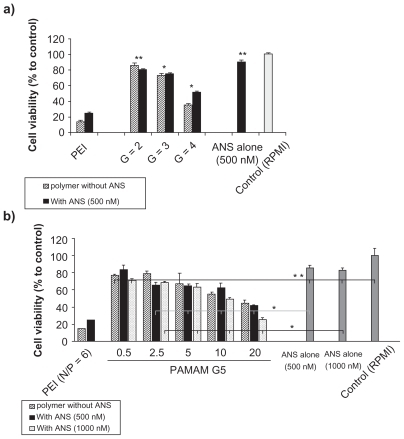
The toxicity of the nanoparticles was dependent on PAMAM generations and N/P ratio. The cell viability of PAMAM G2 and G3 nanoparticles was over 80%, whereas after exposure to G4 and G5 self-assembled nanoparticles the cell viability was approximately 60%, depending on the applied N/P ratio. The toxicity of G2 with or without ANS was not more than that of ANS alone (). At equal amine concentrations, significantly higher toxicity was observed as the generation increased (). Concentrations of 500 and 1000 nM of naked ANS had no significant toxicity on T47D cells, whereas the naked carrier caused toxicity proportional to the concentration. There was no significant difference between the toxicity of these two concentrations of ANS, whether it was naked or condensed by PAMAM G5. The result was the same for different N/P ratios in the case of PAMAM G5, except for N/P of 20 (One-way ANOVA, Bonferroni post-hoc, P value < 0.05). At this ratio, higher ANS concentrations had greater toxicity than the lower concentrations (). Moreover, for each charge ratio of G5, there was no difference between the toxicity of self-assembled nanoparticles and polymer alone at the same concentration, again except in the case of the charge ratio of 20 (t-test, P value ≥ 0.05, for N/P of 20 P value < 0.001). It should be noted that all nanoparticles formed at more than charge neutrality had greater toxic effects at both concentrations of ANS than the naked ANS (). PEI/ANS nanoparticles were significantly more toxic at N/P ratio 6 (which has been reported as the efficient N/P ratio in transfection) than PAMAM G5 even at higher N/P ratios.
Figure 5 MTT assay graphs of PAMAM/ANS nanoparticles. In Figure 5a, multiple generations of PAMAM at fixed a N/P ratio of 10 (black filled columns) are compared with their corresponding PAMAM concentration without ANS (hatched columns), polyethyleneimine (PEI) at N/P ratio of 6, and negative control of FBS-free RPMI.
Abbreviations: PAMAM, poly(amidoamine) dendrimer. ANS, antisense oligonucleotide; N/P, PAMAM, primary amino groups to ANS phosphate groups; FBS, fetal bovine albumin; PEI, polyethyleneimine.
Figure 5b shows PAMAM G5 at different charge ratios of 0.5, 2.5, 5, 10, and 20. Their cytotoxicity was evaluated at two different concentrations of 500 nM (black filled columns) and 1000 nM (dotted columns) and also in comparison with their corresponding dendrimer concentrations without ANS (hatched columns). The PEI (N/P of 6) and naked ANS (500 nM) were positive and FBS-free RPMI was negative controls. All represented data are means of eight different measurements relative to the negative control (RPMI) ± standard error of mean. *was significantly different when each treatment was compared with the ANS alone concentration treatment (t-test, P value < 0.05). **No statistical difference (One-way ANOVA or Student t-test, P value ≥ 0.05).
Abbreviations: PAMAM, poly(amidoamine) dendrimer; ANS, antisense oligonucleotide; N/P, PAMAM, primary amino groups to ANS phosphate groups; FBS, fetal bovine albumin; PEI, polyethyleneimine; ANOVA, analysis of variance.
Discussion
In non-viral gene delivery, the binding and protection of nucleic acids is a critical step in the delivery of gene therapeutics to specific targets.Citation31,Citation32 Except for some special routes of delivery, of these unstable hydrophilic macromolecules, (eg, conjugation of macromolecules to the cell penetrating peptidesCitation26 or old methods of sonoporation, electroporation, and applying hydrodynamic pressure), condensation with a polycation is an important and commonly used route for their delivery to the cytoplasm or nucleus of the target cells.Citation33,Citation34
The polycationic dendrimer of PAMAM in all generations can interact with DNA molecules and form nanoparticles which have some partially N/P ratio dependent features.Citation35–Citation37
The PCS results showed that from the lower N/P ratios to the higher N/P ratios, the deviation from the mean size of the nanoparticles decreased, and the same tendency can be observed for the AFM results (). An insufficient amount of counter ionization of the dendrimer, at lower N/P ratios can lead to inefficient compaction of the oligonucleotides. This effect, accompanied by a lower surface charge density of the resulting nanoparticles of the dendrimer and ANS, can cause the nanoparticles to easily produce large aggregates in the solution. These can be detected by PCS graphs ( graph of N/P = 0.5) and AFM pictures (). Several research groups have reported such behavior for heavy and large DNA molecules. Fant and colleaguesCitation35 studied the condensation of plasmid luciferase (4331 bp) by PAMAM G5 as a synthetic histone model. They also reported large aggregates for ANS at lower N/P ratios, using the PCS method. In our study, the PCS and AFM images showed that the aggregation of particles at lower charge ratio of dendrimer to small DNA occurred quite often, and sometimes such large aggregates were observed at N/P ratios of 10 and 20 (see , and large aggregates at higher N/P in , N/P = 10 of G2). It is obvious that the ionic strength and pH of the solution play a prominent role in the compaction of polynucleotides with polycations.Citation31,Citation32 Higher concentrations of cations in the solution can greatly affect the compaction efficiency of polycationic polymer, and lower pH can induce more protonation of primary amines of polycations, such as PAMAM. In comparison with the other solutions, eg, 150 mM NaCl,Citation35 or 10–20 mM HEPES,Citation17 and distilled water,Citation19 the dextrose/water solution that was used in this study had a moderate ionic strength and pH 6.5 at which nearly all the primary amines of the dendrimer are protonated. Therefore, it was expected that the resulting self-assembled nanoparticles would have a smaller size in comparison to those in the previously reported experiments of PAMAM with DNA.Citation19 But the PCS graphs and AFM images were not consistent with this hypothesis.
In their study on salmon testes DNA condensation by PAMAM G2 and G3 using small angle x-ray scattering, Su and colleaguesCitation36 observed that the main structure for G2/DNA aggregations was hexagonal, while for the G3/DNA complexes a square lattice was observed. Large DNA molecules cannot bend around these lower generations of PAMAM because of the high energy required for this to occur. However, for higher generations (G4 and bigger), DNA molecules of 1–50 kbp always have a final compaction morphology of toroids and perfectly hexagonal lattice structures, which constitute the majority (nearly 80%) of particle morphologies.
In another study, Carnerup and colleaguesCitation37 reported that the size of these nanoparticles was around 100 nm when they condense the luciferase plasmid DNA with PAMAM G1, G2 and G4 at more than neutrality charge ratios. However, for higher generations (in their study G4), this size is a result of the DNA double helix minimum energy and total energy cost effectiveness, which causes the DNA helix to bend around the dendrimer molecules when it is compacted by them.Citation32,Citation37,Citation38 In the case of small DNA molecules, eg, ANS, it seems that it is difficult to achieve a uniform hexagonal lattice formation and the morphology of toroids even for PAMAM G5 in view of the thermodynamic and kinetic theory of toroid formation. Citation32 As a result, there was a considerable heterogeneity in the particle size estimated by PCS or visualized by AFM methods (– and ), which are not in accord with the previously reported monodisperse data in the case of PAMAM,Citation39 (and our experiments on compaction of plasmid EGFP by PAMAM G5). However, detailed studies using x-ray scattering are needed to confirm the internal structure of the self-assembled nanoparticles of ANS/PAMAM.
Gel electrophoresis can show the efficiency of the compaction and protection of ANS by polycations. Yoo and colleaguesCitation40 evaluated the gel retardation of an 18-mer ANS ODN and PAMAM G5 at N/P ratios of 35 and 60. They reported that at these N/P ratios the ANS/PAMAM G5 nanoparticles are stable in the agarose gel. But our results show that there is no need for a very high N/P ratio for retardation of ANS by PAMAM G5. A 2.5- fold excess of dendrimer amine versus phosphate group was sufficient to stabilize these ionic complexes toward electrophoretic mobility.
Lower charge ratios of PAMAM G5/ANS cannot affect the cell surface, and the particles, although attached, were not able to effectively enter the cells. It is obvious from that an excess of cationic dendrimer is necessary to cause the uptake of nanoparticles. Surface charge and particle size are important factors determining how the nanoparticles interact with the biological environment and membranes.Citation41 Interaction of PAMAM G5 with ANS forms larger particles, and because of their size, their interaction with the cell surface, and consequently the cell uptake behavior, could be different from that of big DNA/PAMAM self-assembled nanoparticles. Recently, many articles have proved the formation of holes at the lipid bilayer model membrane and cell surface membrane following exposure to even non-cytotoxic concentrations of cationic nanoparticles and specially PAMAM dendrimers.Citation42–Citation45 These transient pores, which form quickly (within micro- to milliseconds),Citation42 exist long enough that even large nanoparticles (such as PAMAM/ANS nanoparticles) can easily enter the cells. Obviously, higher charge ratios have more impact on the cell membrane, because the probability of the interaction of the dendrimers with the membrane will increase. It is possible that the lower charge of cationic particles at the N/P ratio of 0.5 was not capable of inducing transient nanopores in the lipid bilayer of cell membrane, which is one of the mechanisms responsible for cell uptake of cationic nanoparticles.Citation46,Citation47 Another explanation for this observation could be the effect of a higher charge ratio causing higher affinity to negatively charged cell surfaces. This affinity can induce more endocytosis of the particles. The mechanism of uptake needs to be clarified, but it is possible that the hole formation may be the main mechanism, since nanoparticles attach to the cell membrane at the lower charge ratio, but cannot enter the cells (). For large particles of PAMAM/ANS, other mechanisms of uptake, such as macropinocytosis, should be considered in further investigations as well.
The toxicity of cationic dendrimers, polymers and their nanoparticles are well-known.Citation48 In this paper we have shown that the toxicity of self-assembled nanoparticles depends on both the PAMAM concentration and generation. At the same amine molarities, higher generations have higher cytotoxicity compared with the lower generations (). Yoo and colleaguesCitation40 reported the toxicity of naked PAMAM and PAMAM/ANS nanoparticles in the absence of serum. The observed cytotoxicity of naked PAMAM G5 and PAMAM G5/ANS (500 nM ANS concentration) was the same as in our experiment (around 60% cell viability), but at 1 μM concentration it was different. Naked ANS concentration at selected amounts had no effect on the cell viability. The effect of ANS on the target gene is concentration dependent, but only in the case that it can easily enter the cells and reach its target. Lower N/P ratios cannot effectively cause cell uptake (), so there is no difference between 500 nM and 1 μM ANS at this ratio. At higher N/P ratios (10 and 20), at which the ANS cargo can enter the cells effectively (), the difference between the cytotoxicity of two ANS concentrations is obvious (). Further investigations of the other ANS concentrations are necessary to conclusively prove this effect. In addition, further studies on these nanoparticles to identify the final effect of ANS and dendrimers are needed.Citation49
Conclusion
In this study we have shown that cationic PAMAM G5 dendrimers form larger nanoparticles with ANS than with plasmid DNA and can deliver ANS oligonucleotides inside the cells, with moderate toxicity on the cell culture. The destiny of these gene nanoparticles when they enter to the cells should be investigated further. Moreover, there are no comprehensive studies of the effect of PAMAM/ANS nanoparticles on the genomic content of the cells, and this also requires further investigation.
Acknowledgment
This research has been supported by Tehran University of Medical Sciences and Health Services grant No. 4404 to Ismaeil Haririan. We also thank Vivian Michael Paganuzzi, MA, for help in improving language of the article and Marika Ruponen (University of Kuopio), for her helpful comments about the manuscript.
Disclosure
The authors report no conflicts of interest relevant to this research.
References
- GuinnBAMulherkarRInternational progress in cancer gene therapyCancer Gene Ther2008151276577518846114
- StrayerDSAkkinaRBunnellBACurrent status of gene therapy strategies to treat HIV/AIDSMol Ther200511682384215922953
- AtkinsonTJCystic fibrosis, vector-mediated gene therapy, and relevance of toll-like receptors: A review of problems, progress, and possibilitiesCurr Gene Ther20088320120718537594
- YoujinSJunYThe treatment of hemophilia A: From protein replacement to AAV-mediated gene therapyBiotechnol Lett200931332132818979215
- RollandAGene medicines: The end of the beginning?Adv Drug Deliv Rev200557566967315757753
- KataokaKHarashimaHGene delivery systems: viral vs non-viral vectorsAdv Drug Deliv Rev2001523151
- ItakaKKataokaKRecent development of nonviral gene delivery systems with virus-like structures and mechanismsEur J Pharm Biopharm200971347548318955136
- The Journal of Gene Medicine Clinical Trial site Available from: http://www.wiley.co.uk/genmed/clinical/Accessed February 2010
- DufesCUchegbuIFSchtzleinAGDendrimers in gene deliveryAdv Drug Deliv Rev200557152177220216310284
- LeeCCMacKayJAchetJMJSzokaFCDesigning dendrimers for biological applicationsNat Biotechnol200523121517152616333296
- TomaliaDABakerHDewaldJNew class of polymers:Starburst dendritic macromoleculesPolym J1984171117132
- SvensonSTomaliaDADendrimers in biomedical applications: reflections on the fieldAdv Drug Deliv Rev200557152106212916305813
- ChengYWangJRaoTHeXXuTPharmaceutical applications of dendrimers: Promising nanocarriers for drug deliveryFront Biosci20081341447147117981642
- TomaliaDAReynaLASvensonSDendrimers as multi-purpose nanodevices for oncology drug delivery and diagnostic imagingBiochem Soc Trans2007351616717233602
- EsfandRTomaliaDAPoly(amidoamine) (PAMAM) dendrimers: From biomimicry to drug delivery and biomedical applicationsDrug Discov Today20016842743611301287
- HaenslerJSzokaFCPolyamidoamine cascade polymers mediate efficient transfection of cells in cultureBioconjugate Chem199345372379
- TangMXRedemannCTSzokaFCIn vitro gene delivery by degraded polyamidoamine dendrimersBioconjugate Chem199676703714
- EichmanJDBielinskaAUKukowska-LatalloJFBakerJThe use of PAMAM dendrimers in the efficient transfer of genetic material into cellsPharm Sci Technol Today20003723224510884679
- BielinskaAKukowska-LatalloJFJohnsonJTomaliaDABakerJRegulation of in vitro gene expression using antisense oligonucleotides or antisense expression plasmids transfected using starburst PAMAM dendrimersNucleic Acids Res19962411217621828668551
- DuncanRIzzoLDendrimer biocompatibility and toxicityAdv Drug Deliv Rev200557152215223716297497
- MalikNWiwattanapatapeeRKlopschRDendrimers: Relationship between structure and biocompatibility in vitro, and preliminary studies on the biodistribution of 125I-labelled polyamidoamine dendrimers in vivoJ Control Release2000651213314810699264
- HollinsAJOmidiYBenterIFAkhtarSToxicogenomics of drug delivery systems: Exploiting delivery system-induced changes in target gene expression to enhance siRNA activityJ Drug Target2007151838817365277
- ZinselmeyerBHMackaySPSchatzleinAGUchegbuIFThe lower-generation polypropylenimine dendrimers are effective gene-transfer agentsPharm Res200219796096712180548
- HollinsAJBenboubetraMOmidiYEvaluation of generation 2 and 3 poly(propylenimine) dendrimers for the potential cellular delivery of antisense oligonucleotides targeting the epidermal growth factor receptorPharm Res200421345846615070097
- ArteagaCLEpidermal growth factor receptor dependence in human tumors: More than just expression?Oncologist20027Suppl 4313912202786
- JulianoRLIntracellular delivery of oligonucleotide conjugates and dendrimer complexesAnn NY Acad Sci20061082182617145920
- ISIS Co. official web page Available from: http://www.isispharm.com/Pipeline/index.htmAccessed February 2010
- PetchAKSohailMHughesMDMessenger RNA expression profiling of genes involved in epidermal growth factor receptor signalling in human cancer cells treated with scanning array-designed antisense oligonucleotidesBiochem Pharmacol200366581983012948863
- TomaliaDABakerHDewaldJDendritic macromolecules: synthesis of starburst dendrimersMacromolecules198619924662468
- BoussifOLezoualc’hFZantaMAA versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimineProc Natl Acad Sci U S A19959216729773017638184
- BloomfieldVADNA condensation by multivalent cationsBiopolymers19974432692829591479
- HudNVVilfanIDToroidal DNA condensates: Unraveling the fine structure and the role of nucleation in determining sizeAnnu Rev Biophys Biomolec Struct2005341295318
- TangMXSzokaFCThe influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexesGene Ther1997488238329338011
- KabanovVASergeyevVGPyshkinaOAInterpolyelectrolyte complexes formed by DNA and astramol poly(propylene imine) dendrimersMacromolecules2000332695879593
- FantKEsbjornerEKLincolnPNordenBDNA condensation by PAMAM dendrimers: Self-assembly characteristics and effect on transcriptionBiochemistry20084761732174018189415
- SuCJChenHLWeiMCPengSFSungHWIvanovVAColumnar mesophases of the complexes of DNA with low-generation poly(amido amine) dendrimersBiomacromolecules20091077378319296683
- CarnerupAMAinalemMLAlfredssonVNylanderTWatching DNA condensation induced by poly(amido amine) dendrimer with time-resolved cryo-TEMLangmuir20092521124661247019856988
- KunzeKKNetzRRSalt-induced DNA-histone complexationPhys Rev Lett2009208543894392
- NavarroGTros de ILarduyaCActivated and non-activated PAMAM dendrimers for gene delivery in vitro and in vivoNanomed Nanotechnol Biol Med200953287297
- YooHSazaniPJulianoRLPAMAM dendrimers as delivery agents for antisense oligonucleotidesPharm Res199916121799180410644065
- PeetlaCLabhasetwarVEffect of molecular structure of cationic surfactant on biophysical interaction of surfactant-modified nanoparticles with a model membrane and cellular uptakeLangmuir2009252369237719161268
- ChenJHesslerJAPutchakayalaKCationic nanoparticles induce nanoscale disruption in living cell plasma membranesJ Phys Chem B200911332111791118519606833
- GinzburgVVBalijepalliSModeling the thermodynamics of the interaction of nanoparticles with cell membranesNano Lett20077123716372217983249
- LeeHLarsonRGMolecular dynamics simulations of PAMAM den-drimer- induced pore formation in DPPC bilayers with a coarse-grained modelJ Phys Chem B200611037182041821116970437
- LeeHLarsonRGCoarse-grained molecular dynamics studies of the concentration and size dependence of fifth- and seventh-generation PAMAM dendrimers on pore formation in DMPC bilayerJ Phys Chem B2008112267778778418543869
- ParimiSBarnesTJPrestidgeCAPAMAM dendrimer interactions with supported lipid bilayers: a kinetic and mechanistic investigationLangmuir20082423135321353918980350
- LeroueilPRBerrySADuthieKWide varieties of cationic nanoparticles induce defects in supported lipid bilayersNano Lett20088242042418217783
- LvHZhangSWangBCuiSYanJToxicity of cationic lipids and cationic polymers in gene deliveryJ Control Release2006114110010916831482
- NomaniAFouladdelSHaririanIThe impact of poly (amido amine) dendrimer-oligonucleotide nanoparticles on T47D cells in cultureProceeding of 36th Annual Meeting of Controlled Release Society2009 Jul 18–22Copenhagen, DenmarkCRS society publication200981