Abstract
Prevalence of allergy and allergic asthma are increasing worldwide. More than half of the US population has a positive skin prick test and approximately 10% are asthmatics. Many studies have been conducted to define immunological pathways underlying allergy and asthma development and to identify the main genetic determinants. In the effort to find missing pieces of the puzzle, new genomic approaches and more standardized ones, such as the candidate gene approach, have been used collectively. This article proposes an overview of the actual knowledge about immunological and genetic aspects of allergy and asthma. Special attention has been drawn to the challenges linked to genetic research in complex traits such as asthma and to the contribution of new genomic approaches.
Allergy
Allergy, also called hypersensitivity, is a reaction that occurs when the immune system responds to a harmless antigen.Citation1 According to results published by the National Health and Nutrition Examination Surveys in 2005, more than half of the US population (54.3%) tests positive to one or more allergens (a common antigen that gives rise to an immediate hypersensitivity response)Citation2 using skin prick tests between 1988 and 1994.Citation3 This survey also underlines the evidence of the growing prevalence of allergy in industrialized countries.
Allergies have been studied according to the different categories of allergens (indoor, outdoor, food, drug, etc) and according to the different allergic diseases (allergic rhinitis, atopic dermatitis, allergic asthma, etc). Prevalence of allergy to different types of antigen and prevalence of different allergic diseases ranges from 4% to 6% for food allergy,Citation4 to 10% for allergic asthma,Citation5 and up to 49% reported in one study for allergic rhinitis.Citation6
Immunology of allergy
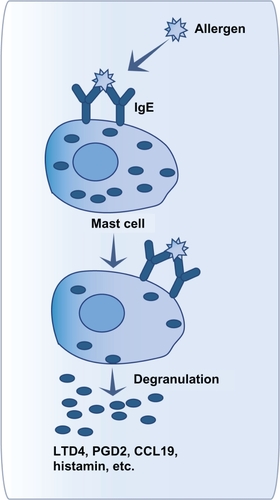
Hypersensitivity reactions have been classified by Coombs and Gell into four different types, characterized by different immunological mechanisms.Citation7 These types have been well described in a review by Averbeck et al.Citation1 A brief description of each type is as follows. Type I hypersensitivity refers to immediate hypersensitivity responses against foreign proteins that are common (pollen, grass, animal danders, etc). It can be observed in allergic rhinitis and allergic asthma (see section “immunology of asthma” for more detail).Citation1,Citation8 This type of hypersensitivity is characterized by immunoglobulin (Ig) E production during the sensitization phase, which will bind to FcɛRI receptors on mast cells and basophils. Upon renewed contact, the allergen will bind cellular IgE. Crosslinking of FcɛRI receptors will lead to degranulation of mast cells and basophils (see ). Mediators released that way will lead to immediate and, sometimes, delayed immune responses.Citation9,Citation10
Figure 1 Mast cell activation after allergen contact.
Type II hypersensitivity is a humoral response mediated by IgG or IgM that are produced against surface antigens on body cells. An example of type II hypersensitivity is drug-induced cytopenia caused by penicillin, cephalosporin, or transfusion reactions.Citation1,Citation11 Immune response occurs within minutes after antigen contact. In drug allergic subjects, IgG or IgM will be formed in response to drugs or their metabolites which accumulate in membrane structures. This binding will allow the killing of the target cells by three different ways.Citation1,Citation11 The first one is the activation of the classic complement pathway, which will lead to cytolysis. The second one is the antibody-dependant cell-mediated cytotoxicity, which will lead to lysis of target cells by natural killer cells. The last one is the recognition of Fc fragments of IgG and IgM by phagocytic cells, which will lead to opsonization of the target cells.
Type III hypersensitivity involves the formation of immune complexes that are not well cleared by innate immune cells as in malaria, rheumatoid arthritis, or farmer’s lung.Citation1,Citation12 This response can occur within four to six hours. The accumulation of immune complexes (antigens bound to antibodies) in vessels and tissues can be caused by antigen excess for subjects with immunosuppression or insufficient antibody production. It can also be caused by repeated antigen exposure which will lead to excess IgG antibody production. The presence of those persistent immune complexes will give rise to an inflammatory response due to leukocytes’ activation.Citation1,Citation12
Finally, type IV hypersensitivity is a delayed response principally mediated by T cells.Citation1,Citation13 The best known example of type IV hypersensitivity is contact allergy. The sensitization phase lasts 10–15 days and is asymptomatic. In contact allergy, a hapten, a low molecular weight molecule, will interact with skin cells or proteins. After this contact, Langerhans and dendritic cells will present antigen to naive T cells to stimulate their differentiation into CD4+ and CD8+ T cells and to induce production of memory T cells. A renewed contact between the skin and the antigen will stimulate sensitized memory T cells via antigen-presenting cells. Early arrival of CD4+ and CD8+ T cells and activation of keratinocytes, will all contribute to promote inflammatory reaction via cytokine secretion. Finally, this reaction will be controlled by regulatory T cells.Citation1,Citation13
This classification of allergic reactions has been widely accepted but some revised nomenclature was proposed, such as that of Johansson et alCitation14 who are part of the European Academy of Allergy and Clinical Immunology. This revised nomenclature, based on known immune mechanisms of allergic diseases and hypersensitivity, aimed to classify the hypersensitivity reactions according to the presence of allergy, IgE production, and atopy. This classification gives a more universal definition of hypersensitivity, regardless of the targeted tissue or organ.
Genetics of allergy
Twin studies have demonstrated the strong heritable component of allergic diseases and atopy, estimated at 33%–76%.Citation15,Citation16 Twin studies with monozygous mice revealed that, even with the same controlled environment, phenotypic variability can occur in allergic manifestations. These results demonstrated that environment explains about 30% of the phenotypic variability observed in allergy and that 70% is due to other factors, such as epigenetics.Citation17,Citation18
However, search of the genes involved in allergy have been mostly done when studying genetic factors of allergic diseases (). Indeed, as mentioned by Hong et al,Citation19 it is often difficult to evaluate the implication of genes on the development of allergy, using this method. However, Renkonen et alCitation20 have recently published a review of the genetics of allergic diseases and have found 39 genes associated with allergy.Citation20 In an effort to classify these genes according to their principal pathway, they concluded that 25 of them are involved in the same interaction pathway with another 70 proteins. Among all of these proteins, 20 are linked to the “host–virus interactions”. When looking to Gene Ontology categories, “response to stress” and “response to various stimuli” were also significantly enriched.Citation20 Another interesting finding of this study is the proportion of protein kinases in their results (23%). Indeed, those kinases may be involved in active allergen transport through intact epithelium.Citation20
Table 1 Prevalence and number of genes associated with allergy or most studied allergic diseasesCitation27–Citation29
To give a general idea of the genes identified by genetic studies on allergic diseases, here are some of the general pathways represented: the T helper type 2 (Th2) immune response and IgE switching cytokines and receptors (IL4RA, IL5, IL13, FCER1A, etc), the chemokines and chemokine receptors (CX3CR1, CCXCR1, CCR2, etc), the human leukocyte antigen (HLA; HLA-DPB1, HLA-DQB1, etc), and the lipoxygenase and cyclooxygenase pathways (CYSLTR1, LTC4S, etc).Citation6,Citation19–Citation26 shows the prevalence of allergy or some of the most studied allergic diseases and the number of genes already associated with each ones.
Asthma
One of the most studied allergic diseases is asthma. Asthmatic response is provoked by allergy in 75%–80% of all asthmatic cases.Citation29 According to the Global Initiative for Asthma,Citation30 asthma is defined as a chronic inflammatory disorder of the airways involving many cells and mediators. The principal associated symptoms are airway hyperresponsiveness and usually reversible airflow obstruction. These symptoms lead to recurrent episodes of wheezing, breathlessness, chest tightness, and coughing.
Clinical manifestations of asthma often appear during childhood.Citation31 However, some individuals show a late onset, sometimes after the age of 40 years.Citation32 According to articles published between 1987 and 1997, the prevalence of asthma varied between 0.5% and 6% depending on the regions of the world.Citation33–Citation37 Approximately 10 years later, it is estimated that 300 million people suffer from asthma in the world and that this number will reach 400 million people by 2025.Citation5 This represent a world prevalence of approximately 10% and a prevalence of 14.1% for Canada and of 10.9% for the United States.Citation5
Physiopathology and immunology of asthma
Asthma is a complex trait that is influenced by several genes as well as by the environment, thus it presents heterogeneous clinical manifestations always recognized as different subphenotypes. Researchers agree that asthma is not a single disease but rather an array of disorders that share common characteristics.Citation29,Citation38,Citation39 These characteristics are inflammation, intermittent bronchial obstruction, bronchial hyperreactivity, mucus hypersecretion, and hypertrophy and hyperplasia of smooth muscle.Citation8,Citation38
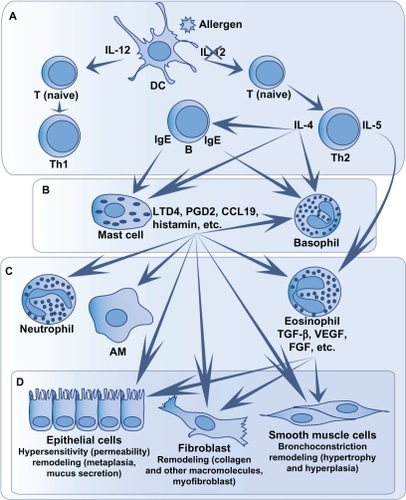
As mentioned above, in 75%–80% of casesCitation40,Citation41 these phenotypes are caused by an allergic response, which triggers a Th2 immune response.Citation29 It is a type I hypersensitivity reaction, that is an immediate exaggerated or harmful immune reaction.Citation8,Citation42 Interestingly, only 7% of allergic people develop asthma,Citation43 which can lead us to believe that they present a unique phenotype that distinguishes them from other allergic, but nonasthmatic, individuals. The main immune cells involved in asthma are CD4+ T cells, mast cells, and eosinophils.Citation44 The different steps of the inflammatory response in asthma, from contact with the allergen to remodeling, are illustrated in and explained later.
Figure 2 Immune response in asthma.
Contact with the allergen
When an allergen penetrates the airways, if not expulsed by the mucociliary barrier, it comes in contact with dendritic cells which will internalize and then digest the allergen. Some dendritic cells then migrate towards the lymph nodes to present the antigen to naive T cells ().Citation45,Citation46 This presentation happens through the major histocompatibility complex type II conjugated to the CD80 and CD86 costimulation molecules.Citation47
The introduction of the antigen to naive T cells triggers the differentiation of CD4+ T cells into Th1 or Th2. This differentiation is modulated by the cytokines present during the introduction of the allergen, eg, interleukin (IL)-12 ().Citation48 Evidence show, at least for mild to moderate asthma, that Th2 cells predominate.Citation49 Thus, after the differentiation of naive T cells in Th2 cells, the latter produce cytokines, including IL-4 which induces the production of IgE by B lymphocytes ().
Immediate response
When an individual has already been in contact with an allergen, its presence and the presence of IgE can activate mast cells and stimulate their degranulation just minutes after the contact with the allergen.Citation2,Citation8 The activation happens when the allergen binds with more than one IgE, which are linked to their high-affinity receptor (FcɛRI; ).Citation9,Citation10 Mast cells are key cells in immediate response. In fact, mast cell granules contain proinflammatory molecules such as histamine, tryptase and other proteases, tumor necrosis factor (TNF), and heparin.Citation2 New molecules are also produced and then released. These molecules are leukotrienes and prostaglandins, as well as cytokines, chemokines, and matrix metalloproteinases.Citation2,Citation50–Citation53 The releasing of these mediators triggers the development of immediate response symptoms such as coughing, bronchial spasms, smooth muscle contraction, oedema, mucus secretion, and infiltration of immune cells.Citation8,Citation54 Mast cells, through these mediators, contribute to the recruitment and activation of immune cells in the lungs, such as eosinophils, T cells, macrophages, basophils, neutrophils, structure cells (fibroblasts, smooth muscle and epithelial cells), and other mast cells.Citation52,Citation55,Citation56
Basophils, with the presence of FcɛRI receptors on their surface and the expression of Th2 cytokines, histamine and granules, can also play a role in immediate response. However, their role in asthma is less documented than that of mast cells.Citation57
Delayed response
Delayed, or late response, is not present in all asthmatics. It is mainly modulated by immune cells recruited by mast cells ().Citation8,Citation29 Among these cells, eosinophils are the main cells implicated in the development of this response.Citation58 Eosinophils produce Th2 cytokines,Citation59–Citation61 leukotrienes,Citation62–Citation64 and proteins that cause damage to airway cells, such as major basic protein, eosinophil cationic protein, eosinophil-derived neurotoxin, and eosinophil peroxidase.Citation65,Citation66
Chronic inflammation in airways
Chronic response in asthma is characterized by a persisting inflammation and the presence of structure alterations in the airways ().Citation67
Several immune cells play a role in this response. Mast cells participate in the chronic response through mediators that have an effect on bronchoconstriction and on airway remodeling causing contraction, hypertrophia and hyperplasia of smooth muscles as well as fibrogenesis.Citation50–Citation53,Citation68,Citation69 Dendritic cells that remain in the airways after coming in contact with the allergen repeatedly present the antigen to CD4+ T cells.Citation70 These cells, as well as the activated T cells, produce proinflammatory cytokines, maintain the chronicity of the inflammation and the eosinophilia.Citation8,Citation45,Citation46,Citation71 Eosinophils are indirectly involved in bronchial remodeling regarding collagen deposit, fibrogenesis, angiogenesis, and hyperplasia of smooth muscles in the peripheral airways via mediators secretion and other cells activation.Citation58,Citation72–Citation74
Structural cells also play an important role in maintaining the chronic response. The amount of mucous epithelial cells increases in asthmatics, thus participating in the bronchial obstruction, mainly in peripheral airways.Citation75,Citation76 Furthermore, epithelial cells, smooth muscle cells, and fibroblasts produce and/or store cytokines, chemokines, and other proinflammatory mediators.Citation77–Citation80 In asthmatics, fibroblasts also produce more collagen and other macromolecules for the extracellular matrix, and present an increased differentiation in myofibroblasts.Citation81,Citation82
Phenotypic variability in asthma
To complete this overview of the asthmatic allergic response, the phenotypic heterogeneity has to be taken into consideration. Indeed, apart from allergic asthma, which is the most common form, other types of asthma have been described.Citation8,Citation38,Citation83 One type is called nonallergic asthma, which develops independently from an allergic component.Citation32 More research still has to be conducted to better define biological pathways specific to this form of asthma.Citation84,Citation85 However, as with allergic asthma, the scientific community recognizes that IgE could be implicated in an inflammatory cascade, as well as CD4+ and CD8+ T cells, eosinophils, and mast cells.Citation86–Citation91
A second type of asthma is characterized by sensitivity to aspirin. It is estimated that approximately 10%–20% of adult asthmatics suffer from this particular type.Citation92 Aspirin sensitization is thought to be non-IgE mediated so could be considered as a nonallergic subphenotype of asthma.Citation92 A dysfunction of the eicosanoid metabolism is responsible for this type of asthma.Citation92,Citation93 A great number of individuals affected with this type of asthma will also resist a treatment with corticosteroids.Citation38
Asthmatics who do not respond to glucocorticoid treatments represent up to 10% of all patients affected with asthma.Citation94 This type of asthma can be induced by a diminution of the number of glucocorticoid receptors, by a misrecognition of the ligand by the receptors, by a decreased capacity of the receptors to link with DNA, or by an increase in the expression of certain proinflammatory transcription factors such as nuclear factor (NF)-κB.Citation94
Occupational asthma is exacerbated or induced by irritants or particles present in the workplace of the affected.Citation95 It is estimated that 9%–15% of all cases of asthma in adults are linked to the workplace and that up to 25% of new cases of adult asthma fall in the occupational asthma category.Citation96,Citation97
Finally, exercise-induced asthma affects 7%–15% of the general population and 3%–14% of athletes.Citation98 A study indicates that up to 39% of college athletes show one of the main symptoms of asthma, that is bronchoconstriction.Citation99 The two major hypotheses that could explain the induction of asthma after an effort are related to the augmentation of ventilation in the lungs.Citation100,Citation101 These two hypotheses are the osmotic hypothesis (dehydration by evaporation caused by the increase in ventilation) and the thermal hypothesis (airway cooling during exercise and rewarming after exercise).Citation101–Citation103 According to Anderson and Daviskas,Citation101 maybe a mix of these two hypotheses could better explain the phenotype rather than each one separately.
Asthma can also be classified according to inflammatory patterns. The three principal types are eosinophilic asthma, neutrophilic asthma, and paucigranulocytic asthma.Citation39 Paucigranulocytic asthma is defined as an asthma response without eosinophils or neutrophils.Citation29
Another way to classify asthma is according to the severity of the phenotype. This classification is used in the clinical treatment, but is also used in research in order to document the molecular and cellular biology variation related to the severity of the disease. Intermittent, mild, moderate, or severe asthma are part of this classification.Citation104 The definition criteria of these types of asthma include frequency of day and night symptoms, exacerbation, and maximum expiratory flow in one second ().Citation30
Table 2 The definition criteria used to classify asthma according to severity
Changes in principal cells involved in the inflammatory response have been observed according to the severity of asthma. For example, inflammation is located in the main airways in individuals with mild asthma, whereas it is also present in peripheral airways and alveoli in individuals with severe asthma or during exacerbation.Citation105 It is also interesting to note that a recruitment of Th1 and CD8+ T cells (cytotoxic T cells) has also been observed in the case of severe and chronic asthma and during exacerbation.Citation106–Citation108 Thus, other cell types involved in the Th1 response or in immune response regulation, such as alveolar macrophages (they inactivate dendritic cells and they can be activated or inhibited through different pathways),Citation109–Citation111 could play a role in the inflammatory response observed in asthma.Citation112,Citation113
Asthma genetics
Several studies demonstrated a strong familial structure in the prevalence of asthma.Citation114–Citation119 Studies conducted on twins on asthma and its subphenotypes (IgE, bronchial hyperreactivity, etc) supported the idea of a genetic component in the development of asthma.Citation116 Segregation analyses subsequently demonstrated that asthma does not follow a Mendelian transmission model. In fact, aggregation and segregation analyses have demonstrated that asthma is polygenic (several genes implicated), shows genetic heterogeneity (different combinations of genes appear in the families that are studied), and pleiotropy (some genes are implicated in the development of more than one phenotype). Moreover, environment plays also a role in the development of the phenotype.Citation116,Citation120 According to this, asthma is considered as a complex trait.
Many challenges appear in genetic research on asthma, such as (1) finding all the genes associated with asthma and with treatment responses (build a list of asthma genes), (2) defining the mechanisms that trigger the phenotypic heterogeneity observed in asthma (assess the contribution of each gene in the different asthma phenotypes), (3) understanding the biology of the mutated genes in the etiology of the disease (functional role of the associated genes), (4) understanding how the gene–gene and gene–environment interactions work (develop biological and mathematical analysis tools),Citation121 and (5) defining the impact of the epigenetic modulations on the development of asthma.Citation120
The ultimate goal beyond these challenges is to define principal pathways involved in asthma and also pathways specifically involved in asthma subphenotypes to find targets for new treatments.Citation122 According to the great phenotypic variability observed in asthma, a more precise description of pathways involved in asthma subphenotypes would increase our chance to develop more efficient treatments for people who do not respond to actual therapy. An ideal therapy should be specific enough to be taken orally without affecting the immune response in the whole body.Citation122 Actually, potential targets identified (for example, mediator antagonists and inhibitors of cytokines, p38 MAP kinase inhibitors, and anti-inflammatory cytokines) are maybe too specific to be very effective.Citation122 In the search for genes and pathways involved in asthma, association studies between genes or the entire genome and drug response phenotypes could also lead to pharmacogenetic therapies.Citation123 Indeed, characterizing the genetic profile of an asthmatic patient through genetic tests targeting specific genes of interest could help to select the appropriate treatment or to predict asthma developmentCitation123,Citation124 and allow improved prevention programs.
Genes associated with asthma
Several reviews have described the knowledge acquired on the genetics of asthma.Citation22,Citation24,Citation121,Citation125 classified the main genes associated with asthma according to their biological function into three principal categories: inflammation, remodeling, and other pathways. Most of the associated genes could be classified into the inflammation (32/61; 52%) or remodeling categories (12/61; 20%). Indeed, this classification not only underlines the great proportion of associated genes in the inflammatory response, but also underlines the need to better define the biological function for some of the other genes (other pathways; 17/61, 28%) to better understand their implication in asthma. The complete list of genes for all categories seen in is available in with ontology keywords.
Figure 3 Classification of the 61 main associated genes with asthma into biological functions.
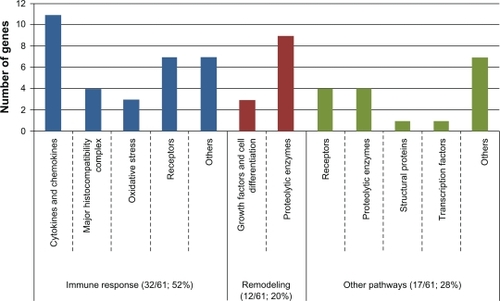
Table 3 Classification of the 61 main genes associated with asthma according to their respective ontology keywords
presents the same 61 genes classified according to the approach used to target them. As shown in this table, the main approach that has been used in the research of genetic determinants of asthma is the candidate gene approach. At the end of 2005, more than 100 genes had already been associated to asthma and another 54 associations have been performed in 2006 and 2007.Citation22,Citation24 Candidate gene association studies are based on the hypothesis of an association between the variants of a gene and asthma or one of its associated phenotypes. These phenotypes usually include respiratory capacity (wheezing, bronchial hyperreactivity, and pulmonary function measurements), immunological measures (total or specific blood IgE, allergy) or clinical criteria (atopic dermatitis, eczema, and rhinoconjunctivitis).Citation121 This approach implies a literature review on genes involved in the development of the trait, or closely related to biological pathways linked with physiopathology of the trait. The choice of candidate genes can be based on several types of information: (1) gene functions, (2) documented associations with the trait in other populations or with closely related phenotypes, (3) difference of expression of the gene for the trait or a closely related phenotype, (4) at least one of these criteria documented for an animal model, or (5) association of another gene in the same biological pathway with the trait or with closely related phenotypes.Citation22,Citation127 Consequently, this approach is biased according to the literature already published for the specific trait and the possibility of discovering new potentially interesting genes could be limited.Citation22,Citation127 This limitation may be even more important considering that the underlying biology of the development of most complex traits remains to be defined.Citation127
Table 4 List of the 61 main associated genes in genetic and genomic studies on asthma and classified according to their identification method
In order to go past this candidate gene selection bias, several approaches have been described. One of these is to select the candidate genes according to their known functions and to crosscheck the results with data obtained from genome-wide linkage, association, or expression studies.Citation127 These genomic approaches allow targeting of new genes without having to state a hypothesis regarding a specific gene or locus and, therefore, permit to increase the knowledge on the pathology of the trait.Citation22,Citation130
Before going further on the contributions of these techniques on the genetics of asthma, here is a brief description of each of them. Linkage analysis and genome-wide association studies (GWAS) are two techniques used to identify chromosomal loci associated in a studied trait. In both cases, analyses are based on two principles: equal transmission of alleles and increase in recombination for increased distances between two markers.Citation131 To identify those loci, the linkage analysis uses families to compare microsatellite markers transmission with phenotype transmission. Similarities between these two transmissions are translated into a statistical measure, the lod score. The lod score is greater when markers of a locus are more transmitted to sick children in studied families, and it is possible to hypothesize that genes near this locus are involved in the development of this disease.Citation132 For GWAS, familial or case–control cohorts can be used.Citation133 Contrary to linkage analysis, different genetic variant types can be used for GWAS as coding single nucleotide polymorphisms (SNPs), tagging SNPs, or copy number variants. presents a brief description of the methodology used for this technique. Linkage analysis has been very efficient to target genes responsible of monogenic trait development but,Citation132,Citation134 in complex traits, one disadvantage of linkage analysis is its impossibility to detect locus with modest effect on phenotype compared with GWAS.Citation22,Citation135 GWAS also has the advantage of targeting more specific chromosomal regions (≤500 bp) because of the great number of genetic variants (more than 500,000 variants) represented on a microarray (for examples, see: http://www.illumina.com/).Citation22 Moreover, new developments on genetics as HapMap international project, the development of new technologies as microarrays, and the possibility to bring together many samples owned by different researchers opened the way to GWAS.Citation121,Citation136
Figure 4 Illustration of the methodological steps for genome-wide association studies and genome-wide expression studies.
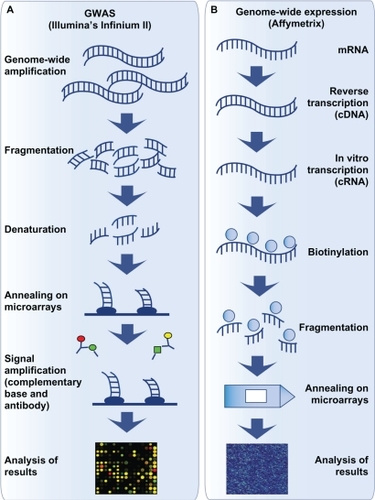
Genome-wide expression studies are another genomic approach that allow the identification of new genes and pathways involved in a target disease. presents the main technical steps to perform genome-wide expression studies using Affymetrix technology as example. These studies, as GWAS, used microarrays. The expression of more than 25,000 genes or transcripts can be measured in the same microarray (www.affymetrix.com). These are used to compare gene expression profiles of affected and nonaffected subjects, of treated and nontreated subjects, of measures for the same subjects at pretreatment and posttreatment, etc. Some advantages of genome-wide expression studies are accessibility of the microarray technology and analysis tools for researchers and clinicians, no sequencing step and possibility to study thousands of genes simultaneously.Citation137
About the contribution of these genome-wide techniques, the use of linkage studies has allowed targeting of genes involved in biological pathways that had not been studied in asthma before (). Several chromosomal regions have been associated with asthma using this method, but only a few have been replicated in several studies and populations (5q31–33, 6p21, 12q13–q24).Citation22,Citation138 GWAS and genome-wide expression studies are now also being used in order to target new genes or validate observations about already associated genes in asthma. Until now, more than 62 genome-wide expression studies have been performed on asthma or one of its subphenotypes (as found in PubMed using “asthma and microarray” and “asthma and gene chip” keywords and according to Rolph et alCitation139). Although most results obtained from expression microarrays are exploratory and must be validated through functional or association studies, a few genes have been targeted by these analyses ().Citation128 As for GWAS, only a few studies have been performed on asthma so far. The first was conducted during the summer of 2007 and allowed targeting of a chromosomal region that regulates the expression of the ORMDL3, GSDMB, and ZPBP2 genes.Citation140,Citation141 However, GWAS approach is becoming more popular and many studies are underway in the field of asthma research. For example, to increase statistical power of the studies, researchers on asthma or other lung diseases group their samples together. Indeed, two major consortia have been made and will soon publish GWAS results. They are called GABRIEL Project (http://www.gabriel-fp6.org/) and EVE Asthma Genetics Consortium (http://arrafunding.uchi-cago.edu/investigators/ober_c.shtml). GABRIEL includes independent asthma genetics studies of European ancestry and EVE includes independent ones from the United States. The scientific community, including these two consortia and others, actually work to plane a meta-global GWAS analysis. The great number of data researchers obtain from these studies will allow them to target new genes and pathways involved in asthma development or its subphenotypes. These may become new therapeutic candidates or may be part of a predictive genetic profile.Citation124
Replication discrepancies
However, no matter what kind of approach is used, researchers are confronted with the same question: why is there an absence of validation in several cases? These nonreplications can be linked to the statistical power of the samples used, or to other phenotypic, genetic, environmental, or epigenetic concerns.Citation121
To increase the chance to find polymorphisms associated with asthma and also replicate anterior results, statistical power of the sample is important. Genetic association studies are more powerful than genomic studies regarding number of subjects to number of genetic variants ratio. Indeed, with the great number of genetic variants or genes tested, the multiple testing problems are much more important in GWAS and genome-wide expression studies.Citation142,Citation143 It is known that for a GWAS, a sample of approximately 15,000 individuals would be necessary to obtain a statistical power of approximately 90% with a type I error of 5%.Citation144 This objective can be reached by grouping together all asthma genetic research studies. For expression studies, analyses showed that at least three to five samples per phenotypic group could be sufficient to find a difference of expression, depending on the objective of the study (ie, to look at greater or smaller differences of expression).Citation145
Another cause of replication discrepancies is the great phenotypic heterogeneity observed in asthma, as described in the section “Phenotypic variability in asthma”. This phenotypic heterogeneity can reflect genotypic heterogeneity, thus implying that the severity and onset of asthma could be influenced by the asthmatics’ genetic imprint.Citation146,Citation147 Thus, a precise characterization of the subjects remains essential. This precise phenotypic description of the sample helps to better select similar independent samples to replicate genetic association results and so helps to reduce the impact of differences in recruitment criteria and in population stratification between samples.Citation146 According to the fact that asthma is rather a collection of several diseases than a unique trait, the key of replication success may lie in association studies with asthma subphenotypes. The review article written by WenzelCitation39 presents several ways to categorize asthma subphenotypes that can be used for association and expression studies.
Gene–environment interactions are another source of variability since the environment can modulate the effect of a gene on the resulting phenotype.Citation148 Studies showed that gene–environment interactions had a greater effect on the phenotype than the environment and genes separately.Citation149 For example, some studies have demonstrated that a high exposure to endotoxin seems to switch the susceptibility effect of the CD14/-159C allele to a protective effect compared with the CD14/-159T allele.Citation150–Citation152 Another example of gene–environment interaction is the interaction between polymorphisms in the regulating regions of a gene and the cell differentiation cycle, leading to differences on the expression of a specific allele.Citation153
A part of the etiology of asthma could be explained by epigenetic changes. Epigenetics is the study of changes that are heritable and that modulate gene expression without directly altering gene sequence.Citation120 Those changes can be done through DNA methylation or posttranslational modification of histones: eg, acetylation, methylation, phosphorylation, and ubiquitylation.Citation154,Citation155 Studies have demonstrated that epigenetic changes are heritable for almost two subsequent generations (eg, methylation of a coat-color gene in mice).Citation156 Many studies in asthma underline the possible role of epigenetics in its development mostly for early onset asthma (for a review, see Miller and HoCitation120). For example, exposition to environmental tobacco smoke during prenatal development or during the first years of life could influence the development of asthma and may be transmitted across two generations.Citation157–Citation159
Gene–gene interactions are another source of variability. In complex traits, a single mutation often has a minor impact, but the combination of several mutations should increase the influences on a phenotype. In asthma, a synergy between a mutation in the IL4 (rs2243250), the IL13 (rs1800925), the IL4 R (rs1805010) and in the signal transducer and activator of transcription 6 (STAT6) (rs324011) genes was documented and increases by 10.8 times the risk of having a high level of IgE and by 16.8 times the risk of developing asthma.Citation160
The solution to these replication problems might partly lie in new genomic methods. In fact, new developments in the field of genetic and genomic research (for example, microarrays for GWAS and genome-wide expression studies), as well as knowledge acquired on the genome and its polymorphisms (as the HapMap project) allow the implementation of new analysis tools. Indeed, although there is still no consensus regarding the analysis methods that should be employed, new tools have been developed to overcome these problems. For example, many new software packages are developed or optimized regularly in Bioconductor, an open source software for the analysis and comprehension of genomic data, to better analyze and characterize data obtained from genome-wide studies (www.bioconductor.org). New genome-wide analysis tools are promising avenues to find new biological pathways, gene families or chromosomal loci involved in asthma and complex diseases, but candidate gene approach will remain a useful and powerful tool to refine our search for associated genes and to help better define the pathways involved in asthma development.
Acknowledgements
Catherine Laprise is the chair holder of the Canada Research Chair for genetic determinants in asthma (www.chairs.gc.ca) and is responsible for the inflammation and remodeling strategic group of the Respiratory Health Network (RHN) of the Fonds de la recherche en santé du Quebec (FRSQ).
Disclosure
The authors report no conflicts of interest in this work.
References
- AverbeckMGebhardtCEmmrichFTreudlerRSimonJCImmunologic principles of allergic diseaseJ Dtsch Dermatol Ges20075111015102817976144
- Platts-MillsTAEImmediate hypersensitivity (Type I)MaleDBrostoffJRothDBRoittIImmunology7th edPhiladelphia, PAElsevier2006423447
- ArbesSJJrGergenPJElliottLZeldinDCPrevalences of positive skin test responses to 10 common allergens in the US population: results from the third National Health and Nutrition Examination SurveyJ Allergy Clin Immunol2005116237738316083793
- LackGNew developments in food allergy: old questions remainJ Allergy Clin Immunol2004114112713015241355
- MasoliMFabianDHoltSBeasleyRGlobal burden of asthma Developed for the Global Initiative for Asthma. Medical Research Institute of New Zealand, Wellington, New Zealand and University of Southampton, Southampton, United Kingdom2004
- SettipaneRACharnockDREpidemiology of rhinitis: allergic and nonallergicClin Allergy Immunol200719233417153005
- GellPGHCoombsRRAThe classification of allergic reactions underlying diseaseCoombsRRAGellPGHClinical Aspects of ImmunologyLondon, UKBlackwell Science1963
- VerstraelenSBloemenKNelissenIWittersHSchoetersGVan Den HeuvelRCell types involved in allergic asthma and their use in in vitro models to assess respiratory sensitizationToxicol In Vitro20082261419143118603401
- GilfillanAMTkaczykCIntegrated signalling pathways for mast-cell activationNat Rev Immunol20066321823016470226
- MetzgerHThe receptor with high affinity for IgEImmunol Rev199212537481532373
- MaleDHypersensitivity (Type II)MaleDBrostoffJRothDBRoittIImmunology7th edPhiladelphia, PAElsevier2006449460
- HayFWestwoodOMRHypersensitivity (Type III)MaleDBrostoffJRothDBRoittIImmunology7th edPhiladelphiaElsevier2006461476
- BrittonWType IV hypersensitivityMaleDBrostoffJRothDBRoittIImmunology7th edPhiladelphia, PAElsevier2006477491
- JohanssonSGHourihaneJOBousquetJA revised nomenclature for allergy. An EAACI position statement from the EAACI nomenclature task forceAllergy200156981382411551246
- HoppRJBewtraAKWattGDNairNMTownleyRGGenetic analysis of allergic disease in twinsJ Allergy Clin Immunol19847322652706538209
- LichtensteinPSvartengrenMGenes, environments, and sex: factors of importance in atopic diseases in 7–9-year-old Swedish twinsAllergy19975211107910869404559
- Isidoro-GarciaMDavila-GonzalezIPascual de PedroMSanz-LozanoCLorente-ToledanoFInteractions between genes and the environment. Epigenetics in allergyAllergol Immunopathol (Madr)200735625425818047817
- GartnerKA third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals?Lab Anim199024171772406501
- HongXTsaiHJWangXGenetics of food allergyCurr Opin Pediatr200921677077619851108
- RenkonenJMattilaPParviainenVJoenvaaraSToppila-SalmiSRenkonenRA network analysis of the single nucleotide polymorphisms in acute allergic diseasesAllergy2010651404719796227
- BarnesKCAn update on the genetics of atopic dermatitis: scratching the surface in 2009J Allergy Clin Immunol201012511629e1e11 quiz 30–31.20109730
- OberCHoffjanSAsthma genetics 2006: the long and winding road to gene discoveryGenes Immun2006729510016395390
- WilliamsHRobertsonCStewartAWorldwide variations in the prevalence of symptoms of atopic eczema in the International Study of Asthma and Allergies in ChildhoodJ Allergy Clin Immunol19991031 Pt 11251389893196
- ZhangJParePDSandfordAJRecent advances in asthma geneticsRespir Res20089418197984
- PalikheNSKimSHParkHSWhat do we know about the genetics of aspirin intolerance?J Clin Pharm Ther200833546547218834360
- GueantJLGueant-RodriguezRMGastinIAPharmacogenetic determinants of immediate and delayed reactions of drug hypersensitivityCurr Pharm Des200814272770277718991696
- Castro-GinerFBustamanteMRamon GonzalezJA pooling-based genome-wide analysis identifies new potential candidate genes for atopy in the European Community Respiratory Health Survey (ECRHS)BMC Med Genet20091012819961619
- GomesERDemolyPEpidemiology of hypersensitivity drug reactionsCurr Opin Allergy Clin Immunol20055430931615985812
- HolgateSTPathogenesis of asthmaClin Exp Allergy200838687289718498538
- Global Initiative for AsthmaGlobal strategy for asthma management and prevention2008
- Global Initiative for AsthmaGlobal strategy for asthma management and prevention2006
- HumbertMDoes “intrinsic” asthma exist?Rev Mal Respir2000171 Pt 224525410902138
- KerkhofMDe GraafADrosteJHJPrevalentie van astmatische klachten in drie regio’s in NederlandTijdschr Soc Gezondheidsz199472181185
- Platts-MillsTACarterMCAsthma and indoor exposure to allergensN Engl J Med199733619138213849134881
- RijckenBKerkhofMde GraafAEuropees Luchtweg Onderzoek Nederland (ELON)GroningenRijksuniversiteit Groningen1996
- TurkeltaubPCGergenPJPrevalence of upper and lower respiratory conditions in the US population by social and environmental factors: data from the second National Health and Nutrition Examination Survey, 1976 to 1980 (NHANES II)Ann Allergy1991672 Pt 11471541867453
- WoolcockAJPeatJKSalomeCMPrevalence of bronchial hyperresponsiveness and asthma in a rural adult populationThorax19874253613683660290
- KileyJSmithRNoelPAsthma phenotypesCurr Opin Pulm Med2007131192317133120
- WenzelSEAsthma: defining of the persistent adult phenotypesLancet2006368953780481316935691
- CockcroftDWMurdockKYBerscheidBAGoreBPSensitivity and specificity of histamine PC20 determination in a random selection of young college studentsJ Allergy Clin Immunol1992891 Pt 123301730837
- PostmaDSKoppelmanGHMeyersDAThe genetics of atopy and airway hyperresponsivenessAm J Respir Crit Care Med20001623 Pt 2S118S12310988165
- MaleDBrostoffJRothDRoittIImmunology7th edLondonMosby2006
- BeasleyRPekkanenJPearceNHas the role of atopy in the development of asthma been over-emphasized?Pediatr Pulmonol Suppl200123149150
- KayABThe role of eosinophils in the pathogenesis of asthmaTrends Mol Med200511414815215823751
- ConstantSLBrogdonJLPiggottDAResident lung antigen-presenting cells have the capacity to promote Th2 T cell differentiation in situJ Clin Invest2002110101441144812438442
- JuliaVHesselEMMalherbeLGlaichenhausNO‘GarraACoffmanRLA restricted subset of dendritic cells captures airborne antigens and remains able to activate specific T cells long after antigen exposureImmunity200216227128311869687
- RieseRJChapmanHACathepsins and compartmentalization in antigen presentationCurr Opin Immunol200012110711310679409
- KuipersHHeirmanCHijdraDDendritic cells retrovirally overexpressing IL-12 induce strong Th1 responses to inhaled antigen in the lung but fail to revert established Th2 sensitizationJ Leukoc Biol20047651028103815316032
- AndersonGPThe immunobiology of early asthmaMed J Aust2002177SupplS47S4912225257
- DahlenBShuteJHowarthPImmunohistochemical localisation of the matrix metalloproteinases MMP-3 and MMP-9 within the airways in asthmaThorax199954759059610377203
- KaurDSaundersRBergerPAirway smooth muscle and mast cell-derived CC chemokine ligand 19 mediate airway smooth muscle migration in asthmaAm J Respir Crit Care Med2006174111179118816959919
- BraddingPWallsAFHolgateSTThe role of the mast cell in the pathophysiology of asthmaJ Allergy Clin Immunol200611761277128416750987
- WenzelSEBalzarSCundallMChuHWSubepithelial basement membrane immunoreactivity for matrix metalloproteinase 9: association with asthma severity, neutrophilic inflammation, and wound repairJ Allergy Clin Immunol200311161345135212789238
- HolsappleMPJonesDKawabataTTAssessing the potential to induce respiratory hypersensitivityToxicol Sci200691141316339788
- OgawaYCalhounWJThe role of leukotrienes in airway inflammationJ Allergy Clin Immunol20061184789798 quiz 799–800.17030228
- BraddingPHolgateSTImmunopathology and human mast cell cytokinesCrit Rev Oncol Hematol199931211913310451798
- Nouri-AriaKTIraniAMJacobsonMRBasophil recruitment and IL-4 production during human allergen-induced late asthmaJ Allergy Clin Immunol2001108220521111496235
- Nissim Ben EfraimAHLevi-SchafferFTissue remodeling and angiogenesis in asthma: the role of the eosinophilTher Adv Respir Dis20082316317119124368
- LampinenMCarlsonMHakanssonLDVengePCytokine-regulated accumulation of eosinophils in inflammatory diseaseAllergy200459879380515230810
- LeeJJDiminaDMaciasMPDefining a link with asthma in mice congenitally deficient in eosinophilsScience200430556911773177615375267
- Schmid-GrendelmeierPAltznauerFFischerBEosinophils express functional IL-13 in eosinophilic inflammatory diseasesJ Immunol200216921021102712097410
- AdelrothEMorrisMMHargreaveFEO’ByrnePMAirway responsiveness to leukotrienes C4 and D4 and to methacholine in patients with asthma and normal controlsN Engl J Med198631584804843526153
- LeeERobertsonTSmithJKilfeatherSLeukotriene receptor antagonists and synthesis inhibitors reverse survival in eosinophils of asthmatic individualsAm J Respir Crit Care Med200016161881188610852761
- MaromZShelhamerJHBachMKMortonDRKalinerMSlow-reacting substances, leukotrienes C4 and D4, increase the release of mucus from human airways in vitroAm Rev Respir Dis198212634494517125334
- FrigasEMotojimaSGleichGJThe eosinophilic injury to the mucosa of the airways in the pathogenesis of bronchial asthmaEur Respir J Suppl199113123s135s1953909
- JacobsenEATaranovaAGLeeNALeeJJEosinophils: singularly destructive effector cells or purveyors of immunoregulation?J Allergy Clin Immunol200711961313132017481717
- Van HoveCLMaesTJoosGFTournoyKGChronic inflammation in asthma: a contest of persistence vs resolutionAllergy20086391095110918616676
- BrightlingCEBraddingPSymonFAHolgateSTWardlawAJPavordIDMast-cell infiltration of airway smooth muscle in asthmaN Engl J Med2002346221699170512037149
- PlanteSSemlaliAJoubertPMast cells regulate procollagen I (alpha 1) production by bronchial fibroblasts derived from subjects with asthma through IL-4/IL-4 delta 2 ratioJ Allergy Clin Immunol200611761321132716750993
- LipscombMFMastenBJDendritic cells: immune regulators in health and diseasePhysiol Rev20028219713011773610
- Wills-KarpMLuyimbaziJXuXInterleukin-13: central mediator of allergic asthmaScience19982825397225822619856949
- ChoJYMillerMBaekKJInhibition of airway remodeling in IL-5-deficient miceJ Clin Invest2004113455156014966564
- HumblesAALloydCMMcMillanSJA critical role for eosinophils in allergic airways remodelingScience200430556911776177915375268
- MunitzABacheletILevi-SchafferFReversal of airway inflammation and remodeling in asthma by a bispecific antibody fragment linking CCR3 to CD300aJ Allergy Clin Immunol200611851082108917088133
- OrdonezCLKhashayarRWongHHMild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expressionAm J Respir Crit Care Med2001163251752311179133
- Perez-VilarJMucin granule intraluminal organizationAm J Respir Cell Mol Biol200736218319016960124
- BergeronCBouletLPStructural changes in airway diseases: characteristics, mechanisms, consequences, and pharmacologic modulationChest200612941068108716608960
- SmitJJLukacsNWA closer look at chemokines and their role in asthmatic responsesEur J Pharmacol20065331–327728816464446
- JohnsonSRKnoxAJSynthetic functions of airway smooth muscle in asthmaTrends Pharmacol Sci19971882882929277132
- PanettieriRAJrAirway smooth muscle: an immunomodulatory cellJ Allergy Clin Immunol2002110Suppl 6S269S27412464935
- BosseYParePDSeowCYAirway wall remodeling in asthma: from the epithelial layer to the adventitiaCurr Allergy Asthma Rep20088435736618606090
- DubeJChakirJLavioletteMIn vitro procollagen synthesis and proliferative phenotype of bronchial fibroblasts from normal and asthmatic subjectsLab Invest19987832973079520943
- CarlsenKHKowalskiMLAsthma, allergy, the athlete and the OlympicsAllergy200863438338618315726
- HumbertMMenzGYingSThe immunopathology of extrinsic (atopic) and intrinsic (non-atopic) asthma: more similarities than differencesImmunol Today1999201152853310529782
- CorriganCMechanisms of intrinsic asthmaCurr Opin Allergy Clin Immunol200441535615090920
- JayaratnamACorriganCJLeeTHThe continuing enigma of non-atopic asthmaClin Exp Allergy200535783583716008664
- HumbertMGrantJATaborda-BarataLHigh-affinity IgE receptor (FcepsilonRI)-bearing cells in bronchial biopsies from atopic and nonatopic asthmaAm J Respir Crit Care Med19961536 Pt 1193119378665058
- YingSHumbertMMengQLocal expression of epsilon germline gene transcripts and RNA for the epsilon heavy chain of IgE in the bronchial mucosa in atopic and nonatopic asthmaJ Allergy Clin Immunol2001107468669211295659
- YingSMengQZeibecoglouKEosinophil chemotactic chemokines (eotaxin, eotaxin-2, RANTES, monocyte chemoattractant protein-3 (MCP-3), and MCP-4), and C-C chemokine receptor 3 expression in bronchial biopsies from atopic and nonatopic (Intrinsic) asthmaticsJ Immunol1999163116321632910570327
- HumbertMDurhamSRYingSIL-4 and IL-5 mRNA and protein in bronchial biopsies from patients with atopic and nonatopic asthma: evidence against “intrinsic” asthma being a distinct immunopathologic entityAm J Respir Crit Care Med19961545149715048912771
- YingSHumbertMBarkansJExpression of IL-4 and IL-5 mRNA and protein product by CD4+ and CD8+ T cells, eosinophils, and mast cells in bronchial biopsies obtained from atopic and nonatopic (intrinsic) asthmaticsJ Immunol19971587353935449120316
- SzczeklikASanakMThe broken balance in aspirin hypersensitivityEur J Pharmacol20065331–314515516457808
- KimSHParkHSPathogenesis of nonsteroidal antiinflammatory drug-induced asthmaCurr Opin Allergy Clin Immunol200661172216505607
- ItoKChungKFAdcockIMUpdate on glucocorticoid action and resistanceJ Allergy Clin Immunol2006117352254316522450
- DykewiczMSOccupational asthma: current concepts in pathogenesis, diagnosis, and managementJ Allergy Clin Immunol20091233519528 quiz 529–53019281900
- BalmesJBecklakeMBlancPAmerican Thoracic Society Statement: occupational contribution to the burden of airway diseaseAm J Respir Crit Care Med2003167578779712598220
- TarloSMBalmesJBalkissoonRDiagnosis and management of work-related asthma: American College Of Chest Physicians Consensus StatementChest2008134Suppl 31S41S18779187
- LecomteJAsthma and exerciseRev Med Brux2002234A206A21012422436
- ParsonsJPKaedingCPhillipsGJarjouraDWadleyGMastronardeJGPrevalence of exercise-induced bronchospasm in a cohort of varsity college athletesMed Sci Sports Exerc20073991487149217805078
- WeilerJMBoniniSCoifmanRAmerican Academy of Allergy, Asthma and Immunology Work Group report: exercise-induced asthmaJ Allergy Clin Immunol200711961349135817433829
- AndersonSDDaviskasEThe mechanism of exercise-induced asthma is ..J Allergy Clin Immunol2000106345345910984363
- AndersonSDIs there a unifying hypothesis for exercise-induced asthma?J Allergy Clin Immunol1984735 Pt 26606656715730
- McFaddenERJrHypothesis: exercise-induced asthma as a vascular phenomenonLancet199033586948808831969985
- BuistASDefinitionsBarnesPJDrazenJMRennardSIThomsonNCAsthma and COPD2nd edSan Diego, CAAcademic Press, Elsevier200937
- KraftMMartinRJWilsonSDjukanovicRHolgateSTLymphocyte and eosinophil influx into alveolar tissue in nocturnal asthmaAm J Respir Crit Care Med199915912282349872843
- HamzaouiAChaouchNGrairiHAmmarJHamzaouiKInflammatory process of CD8+ CD28− T cells in induced sputum from asthmatic patientsMediators Inflamm20052005316016616106102
- O’SullivanSMAsthma death, CD8+ T cells, and virusesProc Am Thorac Soc20052216216516113486
- TruyenECoteurLDilissenEEvaluation of airway inflammation by quantitative Th1/Th2 cytokine mRNA measurement in sputum of asthma patientsThorax200661320220816449261
- GordonSAlternative activation of macrophagesNat Rev Immunol200331233512511873
- HoltPGOliverJBilykNDownregulation of the antigen presenting cell function(s) of pulmonary dendritic cells in vivo by resident alveolar macrophagesJ Exp Med199317723974078426110
- MacLeanJAXiaWPintoCEZhaoLLiuHWKradinRLSequestration of inhaled particulate antigens by lung phagocytes. A mechanism for the effective inhibition of pulmonary cell-mediated immunityAm J Pathol199614826576668579128
- BissonnetteEAlveolar macrophages in the pathogenesis of asthmaRecent Res Devol Allergy Clin Immunol20001129141
- Peters-GoldenMThe alveolar macrophage: the forgotten cell in asthmaAm J Respir Cell Mol Biol20043113715208096
- CookeRAvan der VeerAJHuman sensitizationJ Immunol19161201305
- DrinkwaterHMendelian hereditary in asthmaBr Med J1909188
- LosHPostmusPEBoomsmaDIAsthma genetics and intermediate phenotypes: a review from twin studiesTwin Res200142819311665340
- RackemannFMStudies in asthma. II. An analysis of two hundred and thirteen cases in which the patients were relieved for more than two yearsArch Intern Med192841346369
- SchwartzMHeredity in bronchial asthma; a clinical and genetic study of 191 asthma probands and 50 probands with Baker’s asthmaActa Allergol Suppl (Copenh)19522128812985005
- SpainWCCookeRAStudies in specific hypersensitiveness. XI. The familial occurrence of hayfever and bronchial asthmaJ Immunol19249521569
- MillerRLHoSMEnvironmental epigenetics and asthma: current concepts and call for studiesAm J Respir Crit Care Med2008177656757318187692
- VercelliDDiscovering susceptibility genes for asthma and allergyNat Rev Immunol20088316918218301422
- BarnesPJNew drugs for asthmaNat Rev Drug Discov200431083184415459674
- KazaniSWechslerMEIsraelEThe role of pharmacogenomics in improving the management of asthmaJ Allergy Clin Immunol1252295302 quiz 303–304.20159237
- KoppelmanGHte MeermanGJPostmaDSGenetic testing for asthmaEur Respir J200832377578218757702
- MoffattMFGenes in asthma: new genes and new waysCurr Opin Allergy Clin Immunol20088541141718769193
- MadoreAMPerronSTurmelVLavioletteMBissonnetteEYLapriseCAlveolar macrophages in allergic asthma: an expression signature characterized by heat shock protein pathwaysHum Immunol201071214415019913588
- ZhuMZhaoSCandidate gene identification approach: progress and challengesInt J Biol Sci20073742042717998950
- IzuharaKSaitoHMicroarray-based identification of novel biomarkers in asthmaAllergol Int200655436136717130677
- LapriseCSladekRPontonABernierMCHudsonTJLavioletteMFunctional classes of bronchial mucosa genes that are differentially expressed in asthmaBMC Genomics2004512115038835
- Willis-OwenSACooksonWOMoffattMFGenome-wide association studies in the genetics of asthmaCurr Allergy Asthma Rep2009913919063818
- GriffithsAJFGelbartWMMillerJHLewontinRCLes applications de la technologie de l’ADN recombinantDe Boeck UniversitéAnalyse génétique moderneParis2001341412
- BennettPDemystified ... microsatellitesMol Pathol200053417718311040939
- LairdNMLangeCFamily-based methods for linkage and association analysisAdv Genet20086021925218358323
- RiceJPSacconeNLCorbettJModel-based methods for linkage analysisAdv Genet20086015517318358320
- RischNMerikangasKThe future of genetic studies of complex human diseasesScience19962735281151615178801636
- International HapMap ConsortiumA haplotype map of the human genomeNature200543770631299132016255080
- MocellinSRossiCRPrinciples of gene microarray data analysisAdv Exp Med Biol2007593193017265713
- DenhamSKoppelmanGHBlakeyJMeta-analysis of genome-wide linkage studies of asthma and related traitsRespir Res200893818442398
- RolphMSSisavanhMLiuSMMackayCRClues to asthma pathogenesis from microarray expression studiesPharmacol Ther20061091–228429416203040
- MoffattMFKabeschMLiangLGenetic variants regulating ORMDL3 expression contribute to the risk of childhood asthmaNature2007448715247047317611496
- VerlaanDJBerlivetSHunninghakeGMAllele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune diseaseAm J Hum Genet200985337739319732864
- RiceTKSchorkNJRaoDCMethods for handling multiple testingAdv Genet20086029330818358325
- ZieglerAKonigIRThompsonJRBiostatistical aspects of genome-wide association studiesBiom J200850182818217698
- IoannidisJPTrikalinosTAKhouryMJImplications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseasesAm J Epidemiol2006164760961416893921
- ReimersMStatistical analysis of microarray dataAddict Biol2005101233515849016
- ChanockSJManolioTBoehnkeMReplicating genotype-phenotype associationsNature2007447714565566017554299
- GuerraSMartinezFDAsthma genetics: from linear to multifactorial approachesAnnu Rev Med20085932734117845134
- MartinezFDGene-environment interactions in asthma: with apologies to William of OckhamProc Am Thorac Soc200741263117202288
- ValdarWSolbergLCGauguierDGenetic and environmental effects on complex traits in miceGenetics2006174295998416888333
- EderWKlimeckiWYuLOpposite effects of CD 14/-260 on serum IgE levels in children raised in different environmentsJ Allergy Clin Immunol2005116360160716159630
- SimpsonAJohnSLJuryFEndotoxin exposure, CD14, and allergic disease: an interaction between genes and the environmentAm J Respir Crit Care Med2006174438639216614348
- Zambelli-WeinerAEhrlichEStocktonMLEvaluation of the CD14/-260 polymorphism and house dust endotoxin exposure in the Barbados Asthma Genetics StudyJ Allergy Clin Immunol200511561203120915940135
- CameronLWebsterRBStrempelJMTh2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammationJ Immunol2006177128633864217142763
- KrebsJEMoving marks: dynamic histone modifications in yeastMol Biosyst20073959059717700858
- NightingaleKPO’NeillLPTurnerBMHistone modifications: signalling receptors and potential elements of a heritable epigenetic codeCurr Opin Genet Dev200616212513616503131
- WaterlandRAJirtleRLTransposable elements: targets for early nutritional effects on epigenetic gene regulationMol Cell Biol200323155293530012861015
- AlatiRAl MamunAO’CallaghanMNajmanJMWilliamsGMIn utero and postnatal maternal smoking and asthma in adolescenceEpidemiology200617213814416477253
- LiYFLangholzBSalamMTGillilandFDMaternal and grandmaternal smoking patterns are associated with early childhood asthmaChest200512741232124115821200
- MagnussonLLOlesenABWennborgHOlsenJWheezing, asthma, hayfever, and atopic eczema in childhood following exposure to tobacco smoke in fetal lifeClin Exp Allergy200535121550155616393320
- KabeschMSchedelMCarrDIL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthmaJ Allergy Clin Immunol2006117226927416461126