Abstract
Introduction
Hepatocellular carcinoma (HCC) was the sixth most prevalent cancer worldwide. Long non-coding RNA TGFB2-OT1 has been proven to mediate inflammation and autophagy in vascular endothelial cells. However, its function in HCC is still unknown.
Methods
We analyzed the relationship between TGFB2-OT1 expression and the clinicopathological features of 202 HCC patients. RT-qPCR was used to analyze the TGFB2-OT1 expression in HCC cell lines and tissues. In vitro and in vivo assays were conducted to verify the effect of TGFB2-OT1 on the phenotype of HCC. RNA pull-down assays were applied to reveal the proteins binding to the TGFB2-OT1. Western-blot assays were conducted to analyze the protein expression in HCC cell lines.
Results
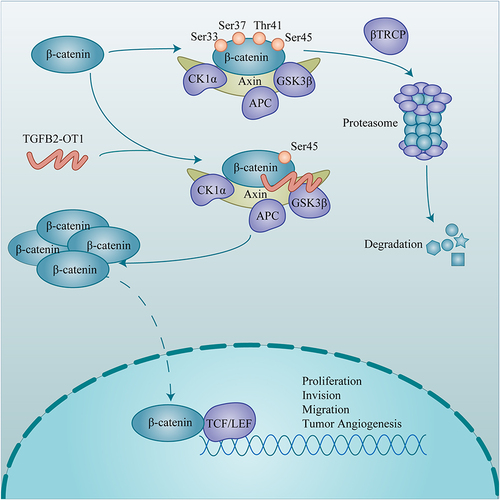
TGFB2-OT1 was found to be highly expressed in HCC samples and hepatoma cells. TGFB2-OT1 expression was significantly associated with age (P = 0.001), cirrhosis (P = 0.003), tumor size (P < 0.001), tumor encapsulation (P = 0.029), tumor protruding from the liver surface (P = 0.040), and alpha fetoprotein (AFP, P < 0.001) levels. TGFB2-OT1 promoted proliferation, migration, invasion, and angiogenesis in HCC cells, both in vitro and in vivo. TGFB2-OT1 binds to β-catenin and competitively impaired the binding of β-catenin to GSK3β, thus suppressing the phosphorylation of β-catenin at Ser33, Ser37, and Thr41.
Conclusion
TGFB2-OT1 is overexpressed in HCC and predicts the poor prognosis of HCC patients. TGFB2-OT1 impedes the phosphorylation of β-catenin and acts as an alternative activator of the Wnt/β-catenin pathway to promote the progression and angiogenesis of HCC.
Introduction
According to the World Health Organization’s global cancer statistics survey, primary liver cancer was the sixth most prevalent cancer and the third leading cause of tumor-related deaths worldwide in 2020.Citation1 China has a high prevalence of hepatocellular carcinoma (HCC) due to the high incidence of hepatitis B virus (HBV) infection.Citation2 A major cause of the high mortality of HCC is the lack of early diagnosis, which can be attributed to the non-specific early symptoms and rapid progression of HCC.Citation3 Despite the availability of multiple systemic therapeutic options,Citation4 the outcomes of HCC treatment remain unsatisfactory. Therefore, identifying specific molecules that influence the progression of HCC remains key for developing improved and targeted treatments for HCC.Citation5
Long non-coding RNAs (lncRNAs) are defined as non-protein-coding transcripts longer than 200 nucleotides in lengthCitation6 and have been confirmed to participate in important cellular functions in various cancers.Citation7–10 LncRNA TGFB2-OT1 is derived from the 3′ untranslated region (UTR) of the gene TGFB2. Previous studies have shown that TGFB2-OT1 acts as a competing endogenous RNA and binds to microRNAs, thereby promoting inflammation and autophagy in vascular endothelial cells (VECs) by enhancing the production of inflammatory factors.Citation11–13 However, TGFB2-OT1 has not been well-studied in the context of cancer, and its biological function in HCC is not clear.
In this study, we first examined the function of lncRNA TGFB2-OT1 in HCC along with its specific mechanisms in this type of cancer. We found that lncRNA TGFB2-OT1 competitively binds to β-catenin and dephosphorylates it, thus promoting the progression and angiogenesis of HCC.
Materials and Methods
Clinical HCC Samples and Information
Tumor tissues and adjacent normal liver tissues were obtained from 202 HCC patients (180 male and 22 female) who underwent liver resection at the Zhongnan Hospital of Wuhan University from 2015 to 2020. All patients were followed up for 100 days. Patients were followed up by telephone every month to record their survival and tumor recurrence. All patients voluntarily signed informed consent forms before the operation. None of the patients had received any pre-operative treatment like radiofrequency ablation, anhydrous alcohol injection, transcatheter arterial chemoembolization, or immunotherapy. The histopathological examination of samples was performed by two experienced pathologists. The relevant clinical data of these 202 patients were also recorded for statistical analysis, Patients were divided into two groups respectively based on RT-qPCR analysis of 202 HCC tissues. The patient whose TGFB2-OT1 relative expression was higher than the median, would be divided into high TGFB2-OT1 expression group, conversely, the low expression group.
RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR (RT-qPCR)
RNA was extracted from tissue samples and cells using TRIzol Reagent (Invitrogen, USA). Cytoplasmic and nuclear RNA purification were followed by the instruction of Cytoplasmic and Nuclear RNA Purification Kit (NORGEN, Thorold, ON, Canada). RNA concentration and purity were measured using NanoDrop ND2000 (Thermo Scientific, Massachusetts, USA). RNA reverse transcription was performed using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The SYBR Green PCR kit (Vazyme, Nanjing, China) was used for RT-qPCR on a CFX96TM Real-time System (Bio-Rad, California, USA). The 2−ΔΔCt (Ct=threshold cycle) method was used to calculate relative RNA expression levels normalized by 18S RNA. All experiments were performed in triplicate. The primer sequences are listed in Supplementary Table S1.
Cell Culture
Human hepatoma cell lines (Huh-7, HCCLM3, Hep3B, HepG2, and HepG2.215), an immortalized normal liver cell line (L02), and human umbilical vein endothelial cells (HUVECs) were purchased from the Shanghai Biological Cell Bank of Chinese Academy of Medical Sciences (Shanghai, China). All human cell lines were authenticated using short tandem repeat (or single nucleotide polymorphism) profiling and mycoplasma infection was ruled out. All hepatogenic cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, CA, USA) containing 10% fetal bovine serum (FBS) (VivaCell Bioscience, Shanghai, China) and 1% penicillin/streptomycin (Gibco, CA, USA) at 37°C in 5% CO2. HUVECs were cultured in DMEM containing 10% FBS and 1% endothelial cell growth supplement (BD Biosciences, USA) at 37°C in 5% CO2.
Fluorescence in situ Hybridization (FISH)
A TGBF2-OT1 probe was designed and produced by AXL-bio (Guangzhou, China) and Deconstruction Biotechnology (Hubei, China). 4% paraformaldehyde treated Huh-7 and HCCLM3 cells were permeated with 0.5% Triton X-100 for 20 minutes (min) and washed with 2× SSC for 10 mins, the cells were incubated with the probes and anti-digoxigenin-fluorescein-Fab fragments (Roche Diagnostics, Indianapolis, IN, USA) sequentially. The samples were treated with ProLong Gold Antifade Reagent and 40, 6-diamidino-2-phenySlindole (DAPI, Invitrogen, Carlsbad, CA, USA). The samples were imaged with the Olympus FluoView™ FV1000 confocal microscope (Olympus, Hamburg, Germany).
RNA Interference and Cell Transfection
β-catenin expression was inhibited using small interfering RNA (siRNA). siRNAs targeting β-catenin (F: 5′-UUGUUAUCAGAGGACUAAAUA-3′; R: 5′-UAUUUAGUCCUCUGAUAAACAA-3′) were synthesized by Tsingke (Beijing, China). The transfection of siRNAs was accomplished based on the instructions of the jetPRIME kit (Polyplus, France).
Construction of Plasmids and the Adenovirus Package and Cell Infection
The TGFB2-OT1 sequence was synthesized and inserted into a pcDNA 3.1 plasmid to construct a TGFB2-OT1 overexpression plasmid (Sangon Biotech, Shanghai, China; called TGFB2-OT1 below). The empty vector was used as a control (GeneCopoeia, Rockville, USA; called Vector below). The pcDNA-β-catenin, pcDNA-GSK3β, and pcDNA-HA-Ub plasmids were synthesized and provided by Genecreate (Wuhan, China). Cells were transfected using the jetPRIME kit (Polyplus, France) according to the given instructions. Stable expression constructs for TGFB2-OT1 (AV TGFB2-OT1), short-hairpin β-catenin (AV sh-β-catenin), and Vector (AV Vector) were packaged into an adenovirus by Genecreate (Wuhan, China), with a virus titer of 2×1010 PFU/mL.
Tube-Formation Assay
Standard Matrigel with high concentration was added to the lower chamber of a μ-Slide Angiogenesis (ibidi, Germany) glass well (10 μL per well) and then placed at 37°C for 30 min for solidification. Then, 2×104 human umbilical vein endothelial cells (HUVECs) were resuspended in indicated condition mediums (CMs) obtained from different cells and added to the upper chamber (50 μL per well). The cells were incubated for 6 h and then observed and recorded using an inverted microscope (Olympus, Japan). Wimasis (https://www.wimasis.com/en/) was used for analysis.
In vivo Experiments
Four-week-old male athymic BALB/C-nude mice were purchased from SPF Biotechnology (Beijing, China). All animals were housed in the Center for Animal Experiments, Wuhan University, and all experiments were also performed at this center. Nude mice were randomized into different groups (4 mice in each group) and a subcutaneous tumor model was constructed. For modeling, 5×106 hepatoma cells in 200 μL serum-free Dulbecco’s Modified Eagle Medium (DMEM) were subcutaneously injected into the axilla of the right forelimb in model mice. Starting from 1 week post-injection, subcutaneous tumor volumes were measured every 2 days. Subcutaneous tumors were injected with different adenoviruses (0.2 mL per mouse) or saline on the fourth day since the start of measurement. The tumors were injected every 2 days for a week. After 18 days, all mice were sacrificed. Its subcutaneous tumors, lung and liver tissues were isolated and preserved in formaldehyde for hematoxylin-eosin (HE) and immunohistochemical (IHC) staining. For survival curve, when the volume of mice reached the ethical standard, the mice were sacrificed.
RNA Pull-Down Assay
The biotin-labeled RNA TGFB2-OT1 was incubated with MyOne Streptavidin C1 dynabeads (Thermo fisher, IL, USA) at 4°C for 2 hours. The cell lysates added RNase inhibitor were mixed into the above mixture rotated at 4°C overnight. The RNA-protein mixture was boiled in Elution buffer and sodium dodecyl sulfate (SDS) buffer. Subsequently, mass spectrometry (MS) and Western blot (WB) analyses were used to identify the eluted proteins. The full-length transcript of TGFB2-OT1 was 1370 bp in length, with sense 1, sense 2, and sense 3 corresponding to its 1–430 bp, 431–1040 bp, and 1041–1370 bp fragments, respectively.
RNA Immunoprecipitation (RIP)
RIP assays were carried out using the Magna RIP RNA binding protein immunoprecipitation kit (Millipore, MA, USA) according to the manufacturer’s instructions. In brief, Cell lysis added RNase inhibitor and protease inhibitor cocktail was centrifuged at 10,000 ×g at 4°C for 10 min. Magnetic beads were blended with 5 µg immunoprecipitation (IP)-grade antibodies at 28°C for 30 min with gentle rotation. Add the bead-antibody complexes to the cell lysate supernatant and incubate at 4°C for 3 hours with gentle rotation. After purification, RNA was analyzed using quantitative real-time PCR (RT-qPCR). The production of RT-qPCR was assessed by agarose gel electrophoresis (AGE).
Immunofluorescence (IF)
After properly treating, 4% paraformaldehyde-fixed cells were permeabilized with 0.5% Triton X-100 and then incubated sequentially with the primary and secondary antibodies (detail in the Supplementary Table S2) according to the manufacturers’ instructions. Slides were counterstained with DAPI and imaged using the Olympus FluoView™ FV1000 confocal microscope.
Co-Immunoprecipitation (Co-IP)
Cells were cultured in 10-cm2 culture dishes and transfected with plasmids to perform Co-IP. Cell precipitates were collected and added into 2-mL centrifuge tubes. Then, 1 mL IP buffer (20 mM Tris-HCl, pH7.4; 150 mM NaCl; 1 mM EDTA, pH8.0; 1% NP-40; 1× Protease and Phosphatase Inhibitor Cocktail) was added per tube to lyse cells for 30 min. Pre-washed Protein A/G Magnetic beads (MCE) were added into the centrifuge tubes and rotated on a disc rotating mixer for 30 min. Then, antibodies were added and mixtures were incubated for more than 3 h at 4°C. Finally, the specific proteins interacting with the target protein were pulled down using magnetic beads. These proteins were separated from the magnetic beads after mixing with 5× SDS loading buffer and heated to 96°C for 10 min. The samples were sent to Singleronbio (Nanjing, China) for MS analysis.
Endogenous and Exogenous Ubiquitination Assays
For the endogenous ubiquitination assays, Huh-7 and HCCLM3 cells were co-transfected with β-catenin and TGFB2-OT1 or vector plasmids. For the exogenous ubiquitination assays, Huh-7 and HCCLM3 cells were co-transfected with HA-Ub, β-catenin, and TGFB2-OT1 plasmids or vector plasmids. Before IP assays, 100 nM MG132 was added to the CMs for 6 h. Cells were lysed using an IP lysis buffer. The subsequent steps were the same as those used for Co-IP.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software) and SPSS 22.0 software (SPSS, USA). Results were presented as the mean ± standard deviation (SD). Student’s t-tests were applied for the analysis of differences between two groups. Two-way analysis of variance (ANOVA) was used for the comparison of more than two groups. The correlation between TGFB2-OT1 and β-catenin expression was analyzed using the Pearson correlation test. Two‐sided P values, indicating the correlation between TGFB2-OT1 expression and clinical variables, were calculated using the χ2 test. In this study, the statistical significance of all experiments was standardized at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Other methods and materials list in the Supplementary File.
Results
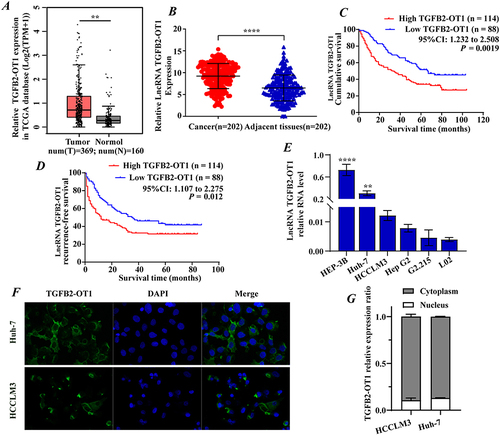
lncRNA TGFB2-OT1 is Highly Expressed in HCC and Indicates a Poor Prognosis
lncRNA TGFB2-OT1 was predicted to have non-coding potential using several prediction tools (Figure S1A) and was confirmed to be conserved among different species (Figure S1B). Data analysis of TCGA database suggested that TGFB2-OT1 was highly expressed in various tumor tissues (Figure S1C), including liver cancer tissues (). RT-qPCR analysis of 202 pairs of HCC samples further showed that TGFB2-OT1 expression was higher in tumor tissues than in the adjacent normal liver tissues (). The prognosis of these 202 patients was examined. Kaplan–Meier survival analysis suggested that high expression of TGFB2-OT1 was associated with a poor overall survival (OS, ) and recurrence-free survival (RFS, ) rate. RT-qPCR was performed to detect the expression of TGFB2-OT1 in different hepatoma cell lines. We found that TGFB2-OT1 was overexpressed in hepatoma cells when compared with normal hepatocytes (). FISH experiments showed that TGFB2-OT1 was mainly localized in the cytoplasm (). RT-qPCR analysis of cytoplasmic and nuclear RNA further proved that TGFB2-OT1 mainly expressed in the cytoplasm (). Further analysis of the relationship between TGFB2-OT1 expression and the clinicopathological features of HCC patients () revealed that TGFB2-OT1 expression was significantly associated with age (P = 0.001), cirrhosis (P = 0.003), tumor size (P < 0.001), tumor encapsulation (P = 0.029), tumor protruding from the liver surface (P = 0.040), and alpha fetoprotein (AFP, P < 0.001) levels.
Table 1 The Relationship Between TGFB2-OT1 Expression and the Clinicopathological Features of HCC Patients
Figure 1 lncRNA TGFB2-OT1 is highly expressed in hepatocellular carcinoma (HCC) and is associated with a poor prognosis in HCC patients. (A) Data analysis from TCGA database showed that TGFB2-OT1 was highly expressed in the tumor tissues. (B) TGFB2-OT1 was upregulated in 202 HCC tissues when compared with 202 adjacent tissues. (C–D) Kaplan–Meier curves showing the overall survival (OS) and recurrence-free survival (RFS) rates of the TGFB2-OT1 high and TGFB2-OT1 low groups. (E) RT-qPCR for TGFB2-OT1 in HCC cell lines. (F) FISH analysis demonstrating the subcellular localization of TGFB2-OT1 in HCC cells. (G) RT-qPCR analysis for TGFB2-OT1 in the cytoplasmic and nuclear. **P < 0.01; ****P < 0.0001.
lncRNA TGFB2-OT1 Facilitates the Progression of HCC in vitro and in vivo
To further investigate the biological function of TGFB2-OT1, we constructed TGFB2-OT1 overexpression plasmids and selected Huh-7 and HCCLM3 cells as overexpression models. Overexpression efficiency was verified using RT-qPCR (Figure S2A). Subsequent Cell Counting Kit-8 (CCK-8), EdU, colony formation, wound healing, and transwell experiments demonstrated that TGFB2-OT1 overexpression could promote the proliferation (; representative image in Figure S2B–D), invasion (; representative image in ), and migration ( and ; representative image in Figure S2F) of HCC cells in vitro.
Figure 2 lncRNA TGFB2-OT1 facilitates proliferation, invasion, and migration in vitro and in vivo. (A) EdU assay indicating that the upregulation of TGFB2-OT1 significantly increased the number of EdU-positive hepatoma cells. (B and C) The number of hepatoma cell colonies was increased after TGFB2-OT1 overexpression. (D) Transwell assay demonstrating that TGFB2-OT1 overexpression facilitated tumor cell invasion. (E and F) Wound-healing assay revealing that TGFB2-OT1 overexpression facilitated cell migration. (G) Representative images of xenografts with AV-TGFB2-OT1 overexpression (n=4), AV-Vector (n=4) expression, and saline treatment (n=4) removed from nude mice. (H) Tumor volume curves indicating that TGFB2-OT1 significantly increased the tumor volume of xenografts. (I) Kaplan–Meier curves of survival in nude mice. (J) Tumor weight histogram of the xenografts. (K and L) Representative images of hematoxylin and eosin (HE)-stained lung metastatic tumors. Magnification 200× and 400×. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
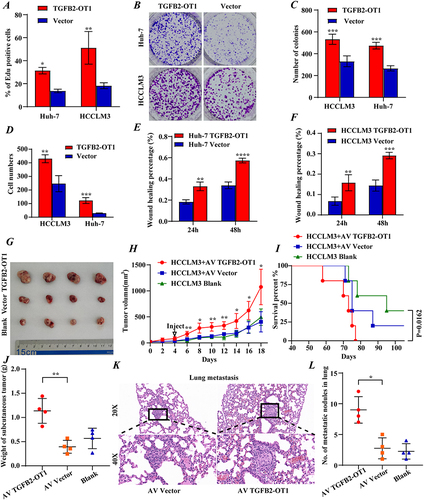
To further confirm the function of TGFB2-OT1 in vivo, subcutaneous tumors were treated with adenoviruses. Tumors injected with the TGFB2-OT1-overexpressing adenovirus had a faster growth rate than tumors from the control group ( and ). Kaplan–Meier survival analysis confirmed the poor OS of mice in the TGFB2-OT1 overexpression group (). The weight of subcutaneous tumors was higher in the TGFB2-OT1 overexpression group (), and the number of metastatic nodules in the lungs was also significantly higher in this group ( and ). Further, the overexpression of TGFB2-OT1 upregulated the percentage of positively Ki67 stained cells. (Figure S2G and H).
lncRNA TGFB2-OT1 Affects HCC Angiogenesis in vitro and in vivo
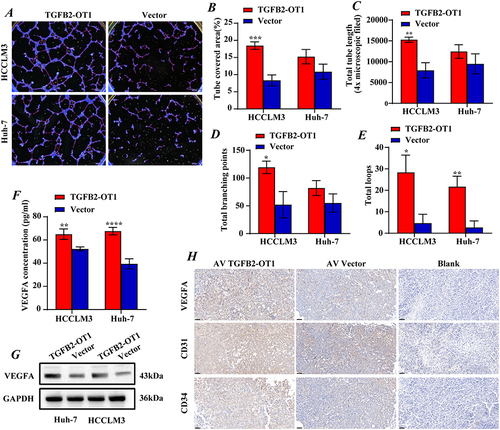
TGFB2-OT1 has been verified to promote inflammation and autophagy in VECs.Citation11 To explore whether TGFB2-OT1 affects angiogenesis in HCC, we conducted a tube-formation assay. We found that TGFB2-OT1 overexpression significantly promoted the number of vasculature nodes, vasculature length, and vasculature area in HUVECs (). We also measured vascular endothelial growth factor A (VEGFA) concentrations in condition mediums and found obviously higher VEGFA levels in the overexpression group (). W analysis in hepatoma cells also indicated that TGFB2-OT1 overexpression enhanced the expression of VEGFA (). We performed IHC on stably TGFB2-OT1 overexpressing subcutaneous tumor tissues and found that the expression levels of VEGFA, CD31, and CD47 were significantly increased ().
Figure 3 lncRNA TGFB2-OT1 affects angiogenesis in vitro and in vivo in HCC. (A–E) TGFB2-OT1 significantly increased the tube-covered area, total tube length, total branching points, and total loops in HUVECs incubated with CMs. (F) ELISA demonstrated that the concentration of exocrine VEGFA in CMs was significantly increased after TGFB2-OT1 overexpression. (G) Western blot (WB) analysis of VEGFA protein expression in Huh-7 and HCCLM3 cells with or without TGFB2-OT1 overexpression. (H) Xenografts were fixed, embedded in paraffin, and stained for VEGFA, CD31, and CD34. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
lncRNA TGFB2-OT1 Binds to β-Catenin
MS analysis of RNA pulldown products indicated that lncRNA binds to β-catenin (Figure S3A). We further verified the presence of this interaction using the RIP assay ( and ) and cell co-localization assay (). Then, we used the RNAfold web server website (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/ RNAfold.cgi) to predict the secondary structure of TGFB2-OT1. TGFB2-OT1 was accordingly truncated into three segments (, agarose electrophoresis analysis of fragments in Figure S3B). Using these three sequences, additional RNA pull-down experiments were performed. All three segments were found to bind to the β-catenin protein ().
Figure 4 lncRNA TGFB2-OT1 binds to β-catenin and increases intracellular β-catenin protein expression. (A) Agarose electrophoresis of RNA immunoprecipitation (RIP) products after PCR. (B) PCR analysis of RIP products. (C) Representative immunofluorescence images showing the co-localization of TGFB2-OT1 and β-catenin. (D) Representative images of different fragments of TGFB2-OT1. (E) WB analysis of β-catenin in HCCLM3 cells after treatment with RNAs corresponding to different fragments of TGFB2-OT1. (F) WB analysis of β-catenin in Huh-7 and HCCLM3 cells with or without TGFB2-OT1 overexpression. (G) Representative images of immunohistochemical staining (IHC) for β-catenin in xenografts. ****P < 0.0001.
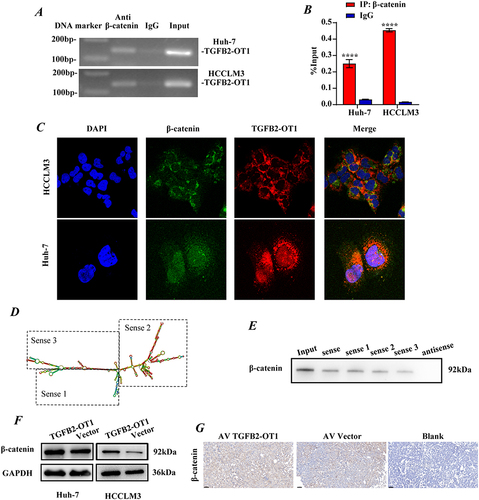
lncRNA TGFB2-OT1 Promotes Intracellular β-Catenin Protein Expression
To further investigate the relationship between the lncRNA TGFB2-OT1 and β-catenin, the relative expression of TGFB2-OT1 and the mRNA levels of β-catenin in tissue samples were measured. No significant correlation was observed at the RNA expression level (Figure S3C), and this was further corroborated by similar findings from the GEPIA website (http://gepia2.cancer-pku.cn/#index, Figure S3D). Surprisingly, we observed an obvious difference in β-catenin expression across samples with varying TGFB2-OT1 levels (Figure S4E). Thus, we further analyzed the mRNA and protein expression of β-catenin in HCC cell lines after the overexpression of TGFB2-OT1. We found that while β-catenin mRNA levels did not change significantly (Figure S4F), its protein levels were significantly upregulated (). Meanwhile, IHC analysis of subcutaneous tumor tissues revealed that the expression of β-catenin was elevated in the TGFB2-OT1 overexpression group ().
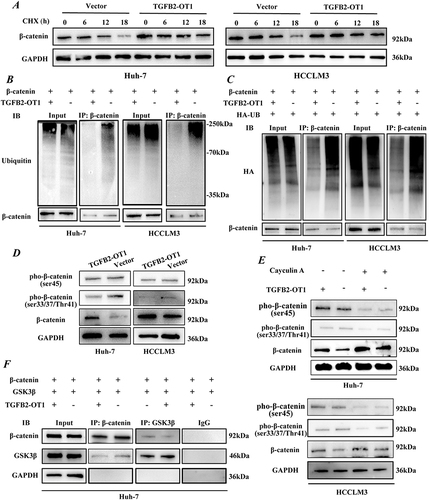
TGFB2-OT1 Stabilizes β-Catenin by Antagonizing the Binding Between GSK3β and β-Catenin
To further investigate the specific molecular mechanism through which TGFB2-OT1 modulates β-catenin expression, we incubated hepatoma cells in CMs containing 500 nM cycloheximide (CHX) for 0, 6, 12, and 18 h. The degradation of β-catenin was slower in the TGFB2-OT1 overexpression group (, S4A and B). Therefore, we performed endogenous () and exogenous ubiquitination assays (). The results revealed that TGFB2-OT1 overexpression inhibited the levels of ubiquitinated β-catenin. In the classic Wnt/β-catenin signaling pathway, β-catenin is sequentially phosphorylated at Ser45 by casein kinase I isoform-α (CK1α) and at Ser33, Ser37, and Thr41 by glycogen synthase kinase 3β (GSK3β). These changes lead to the ubiquitination and degradation of β-catenin. Thus, we examined the levels of phosphorylated β-catenin (pho-β-catenin) after TGFB2-OT1 overexpression. Ser33, Ser37, and Thr41 pho-β-catenin were found to be distinctly downregulated after TGFB2-OT1 overexpression, although Ser45 pho-β-catenin was largely unaffected (). Coincidentally, the addition of 100 nM MG132 for 6 h significantly attenuated Ser33, Ser37, and Thr41 pho-β-catenin levels when was TGFB2-OT1 overexpressed (Figure S4C). Hence, we incubated hepatoma cells in CMs containing 100 nM calyculin A (potent and cell-permeable protein phosphatase 1 and protein phosphatase 2A inhibitor, MCE; Shanghai, China) for 30 min. We found that TGFB2-OT1 overexpression inhibited the phosphorylation levels of β-catenin at Ser33, Ser37, and Thr41 but did not affect phosphorylation at Ser45 (). Competitive binding assays illustrated that the binding of β-catenin to GSK3β was significantly reduced after TGFB2-OT1 overexpression ( and S4D).
Figure 5 TGFB2-OT1 stabilizes β-catenin by antagonizing the binding between GSK3β and β-catenin. (A) WB analysis of β-catenin expression in Huh-7 and HCCLM3 cells treated with cycloheximide (CHX) for 0, 6, 12, and 18 h. (B) Endogenous ubiquitination assay of β-catenin in Huh-7 and HCCLM3 cells with or without TGFB2-OT1 overexpression. (C) Exogenous ubiquitination assay of β-catenin in Huh-7 and HCCLM3 cells with or without TGFB2-OT1 overexpression. (D) WB analysis of phosphorylated β-catenin expression levels in Huh-7 and HCCLM3 cells with or without TGFB2-OT1 overexpression. (E) WB analysis of phosphorylated β-catenin expression levels in Huh-7 and HCCLM3 cells treated with Calyculin A for 30 min. (F) Co-immunoprecipitation (Co-IP) and WB analysis in Huh-7 cells illustrated that lncRNA TGFB2-OT1 competes with GSK3β for binding to β-catenin.
β-Catenin Knockdown Reverses the Phenotypic Effects of lncRNA TGFB2-OT1 on HCC
To verify whether TGFB2-OT1 promotes the progression of HCC by stabilizing β-catenin, we knocked down β-catenin in TGFB2-OT1-overexpressing cell lines. We found that a low expression of β-catenin (knock-down efficiency was verified by RT-qPCR, Figure S5A) reversed the effects of TGFB2-OT1 on tumor cell proliferation (, S5B and S6A–B), invasion, and migration ( and , the representative image in Figure S6C) in vivo. Enzyme-linked immunosorbent assay (ELISA) and WB analyses indicated that the knockdown of β-catenin could also reverse the protein expression and secretion of VEGFA ( and ) induced by TGFB2-OT1 overexpression. Similarly, the knockdown of β-catenin rescued the increase in tumor angiogenesis caused by TGFB2-OT1 overexpression ( and S6D–F).
Figure 6 Knockdown of β-catenin reverses the phenotypic effects of lncRNA TGFB2-OT1 on HCC in vitro and in vivo. (A) EdU assays in Huh-7 and HCCLM3 cells transfected with different plasmids. (B and C) Transwell assays indicated that the knockdown of β-catenin reverses the effects of TGFB2-OT1 on hepatoma cells. (D) WB analysis of VEGFA, β-catenin, and phosphorylated β-catenin expression levels in Huh-7 and HCCLM3 cells transfected with different plasmids. (E) ELISA for examining exocrine VEGFA levels in CMs from hepatoma cells. (F and G) Representative images of angiogenesis after incubation with different CMs and related statistical analysis. (H) Representative images of xenografts from nude mice following rescue experiments. (I and J) Tumor volume curves and tumor weight histograms of xenografts from nude mice with tumors treated with different adenovirus constructs. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
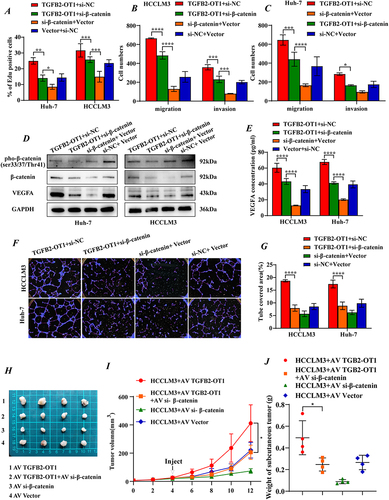
We constructed a subcutaneous tumor model in Balb/c-nude mice and found that the knockdown of β-catenin reverses the increase in tumor proliferation caused by TGFB2-OT1 overexpression (). IHC results suggested that the knockdown of β-catenin also reverses the effect of TGFB2-OT1 overexpression on Ki67, CD31, CD47, and VEGFA expression (Figure S6G).
In summary, the findings showed that lncRNA TGFB2-OT1 interferes with the binding between β-catenin and GSK3β, thus inhibiting the phosphorylation and degradation of β-catenin. This results in an enhanced capacity for proliferation, invasion, migration, and tumor angiogenesis in HCC ().
Discussion
lncRNA TGFB2-OT1 was first discovered as a mediator of inflammation and autophagy in VECs. Huang et al revealed that lncRNA TGFB2-OT1 could bind to MIR3960, MIR4488, and MIR4459 to upregulate the expression of the miRNAs targets ceramide synthase 1 (CERS1); N-acetyltransferase 8-like (NAT8L); and La ribonucleoprotein domain family, member 1 (LARP1), which participate in autophagy by affecting mitochondrial function and producing IL6, IL8, and IL1B in VECs.Citation11 Based on these findings, we examined whether TGFB2-OT1 functions analogously in the process of tumor angiogenesis in HCC. Our results first indicated the function of TGFB2-OT1 in HCC. They demonstrated that TGFB2-OT1 is highly expressed in HCC and is associated with poor OS and RFS in HCC patients. Moreover, TGFB2-OT1 promotes the proliferation, migration, invasion, and angiogenesis of HCC in vivo and in vitro. However, the microRNAs which were found to bind TGFB2-OT1 in previous study, were not examined to express in HCC.
In this study, the concentration of VEGFA in the CMs of hepatoma cells was found to be increased when TGFB2-OT1 was overexpressed. Consequently, IHC for VEGFA and the vascular markers CD31 and CD34 pointed to active angiogenesis in the TGFB2-OT1 overexpression group. Interestingly, we found that the knockdown of β-catenin could reverse the effects of TGFB2-OT1 on angiogenesis in HCC. Our results showed that TGFB2-OT1 promoted the accumulation of β-catenin and stabilized it, potentially allowing it to translocate to the nucleus and activate downstream genesCitation14 such as nuclear DNA-binding T cell factor/lymphoid enhancer-binding factor (TCF/LEF)15 and hypoxia-inducible factor 1α (HIF1α).Citation16 Notably, the xenograft model in our study tended to show a classical capillary vascular pattern instead of a sinusoid-like vascular pattern, called the vessels that encapsulate tumor clusters (VETCs).Citation17 This may be because our xenograft model was based on subcutaneous tumors, whose angiogenesis differs from that of liver tumors. Whether TGFB2-OT1 could promote VETCs in HCC is not clear. Tumor angiogenesis has become a key target of cancer treatment strategies in recent years.Citation18,Citation19 VEGF signaling inhibitors, such as bevacizumab and sorafenib, have been approved for cancer systemic therapy.Citation20 TGFB2-OT1 which conducted as an activator of β-catenin to upregulate VEGFA had not applied to the therapeutic experiment of HCC in vivo and vitro, while it was expected to combine existing drugs to achieve extraordinary effect.
The Wnt/β-catenin pathway, which is highly conserved and tightly regulated, plays an essential role in hepatobiliary development and liver homeostasis.Citation21 As the core protein of this signaling pathway,Citation22 β-catenin has received widespread attention in the field of cancer research.Citation23,Citation24 In our study, we found that the ubiquitination and degradation of β-catenin were reduced when TGFB2-OT1 overexpressed. The expression of the β-catenin protein is finely controlled by the β-catenin destruction complex, which contains the scaffold protein Axin, adenomatous polyposis coli protein (APC), glycogen synthase kinase 3β (GSK3β), and casein kinase I isoform-α (CK1α).Citation25 Excess β-catenin is sequentially phosphorylated—first at Ser45 by CK1α, and then at Ser33, Ser37, and Thr41 by GSK3β.Citation26 Thus, we examined the levels of pho-β-catenin. Surprisingly, Ser33, Ser37, and Thr41 pho-β-catenin were downregulated, while Ser45 pho-β-catenin showed no difference in expression. This indicated that TGFB2-OT1 may mediate GSK3β-induced pho-β-catenin expression. In contrast to findings from Wu et al,Citation27 we found that TGFB2-OT1 did not affect the phosphorylation of GSK3β. Interestingly, Yue et al discovered that in colon cancer, the binding of lncRNA CYTOR to β-catenin impedes CK1-induced β-catenin phosphorylation at Ser45 rather than at Ser33, Ser37, and Thr41.Citation28 Our study confirmed that lncRNA TGFB2-OT1 could act independently to regulate the phosphorylation of β-catenin and inhibit its degradation instead of depending on the Wnt ligand in HCC.Citation29
Though lncRNAs were widely known as no protein-coding potential in the eukaryotic genome. A small number of non-coding RNAs, including circRNAs, miRNAs and lncRNAs, have been verified the protein-coding potentialCitation30 and functioned in cancer progression.Citation31 Huang et al disclosed that lncRNA HOXB-AS3 was bound by the ribosome and encoded a highly conserved peptide.Citation32 It was the peptide rather than lncRNA HOXB-AS3 that mediated the glucose metabolism reprogramming in colon cancer and suppressed the colon cancer growth. Interestingly, Liang et al discovered the similar result in HCC. CircRNA circβ-catenin was derived from the CTTNB1. Due to the active internal ribosome entry site (IRES)Citation33 and open reading frame (ORF),Citation34,Citation35 circβ-catenin encoded the β-catenin isomer β-catenin-370aa, which repressed the GSK3β-induced β-catenin degradation and promoted the progression of HCC.Citation36 Functional peptides are usually encoded by short ORTs in ncRNAs. To further explore the potential peptide of lncRNA TGFB2-OT1, we predicted the ORF, IRES and adenosine methylation in the website. However, the ORF5 which was predicted most likely to translate, had no active IRES or adenosine methylation. Wang et al published RNA-sequencing documents that contained the translating mRNAs (mRNAs bound to ribosome-nascent chain) in lung cancer cells. TGFB2-OT1 was not found to bind to ribosomes in their research.Citation37 In summary, lncRNA TGFB2-OT1 itself functioned in the Wnt/β-catenin pathway.
In this study, we investigated the effect of TGFB2-OT1 on the angiogenesis and development of HCC in detail and examined the related mechanisms. TGFB2-OT1 was overexpressed in HCC and was associated with poor OS and RFS in HCC patients. TGFB2-OT1 affected the proliferation, migration, and angiogenesis of HCC in vitro and in vivo. Further experiments revealed that TGFB2-OT1 acted as a competing element and antagonized the binding between GSK3β and β-catenin. It impeded the GSK3β-induced expression of pho-β-catenin and stabilized β-catenin, promoting progression and angiogenesis in HCC. However, the mechanism via which TGFB2-OT1-mediated β-catenin upregulated VEGFA expression was not clarified. The value of TGFB2-OT1 as a molecular target in the treatment of HCC needs to be explored further.
Conclusion
TGFB2-OT1 is overexpressed in HCC and predicts poor OS and RFS in HCC patients. In patients with HCC, TGFB2-OT1 expression is associated with age, cirrhosis, tumor size, tumor encapsulation, tumor protrusion from the liver surface, and AFP levels. TGFB2-OT1 affects the proliferation, migration, and angiogenesis of HCC both in vitro and in vivo. Mechanistically, TGFB2-OT1 acts as a competing element antagonizing the binding of GSK3β to β-catenin. Thus, TGFB2-OT1 impedes the GSK3β-induced expression of pho-β-catenin and stabilizes β-catenin, promoting HCC progression and angiogenesis. TGFB2-OT1 may act as a new target combined with other chemotherapy drugs applicated in the treatment of HCC in the future.
Abbreviations
HCC, Hepatocellular carcinoma; lncRNA, long non-coding RNA; VECs, Vascular endothelial cells; MS, Mass spectrometry; RIP, RNA immunoprecipitation; Co-IP, Co-immunoprecipitation; WB, Western blot; HUVEC, Human umbilical vein endothelial cell; RT-qPCR, quantitative real-time PCR; FISH, Fluorescence in situ hybridization; IF, Immunofluorescence; siRNA, small interfering RNA; CMs, Condition mediums; ELISA, Enzyme linked immunosorbent assay; VEGFA, Secretory vascular endothelial growth factor A;IHC, Immunohistochemical; HE, Hematoxylin-eosin; AGE, Agarose gel electrophoresis; RIPA, Radio-Immunoprecipitation Assay; SDS-PAGE, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis; DAB, Diaminobenzidine; OS, Overall survival rate; PFS, Progression-free survival rate; AFP, Alpha fetoprotein; CHX, Cycloheximide; Pho-β-catenin, phosphorylated β-catenin; GSK3β, Glycogen synthase kinase 3β; CK1α, Casein kinase I isoform-α; CERS1, Ceramide synthase 1;NAT8L, N-acetyltransferase 8-like; LARP1, La ribonucleoprotein domain family, member 1; TCF, nuclear DNA-binding T cell factor; LEF, lymphoid enhancer-binding factor; HIF1α, hypoxia-inducible factor 1α; APC, Adenomatous polyposis coli protein; IRES, internal ribosome entry site; ORF, open reading frame.
Data Accessibility
The data of this study are included in this article and its online Supplementary Material Files. Please contact the corresponding author if any further information is required.
Ethics Statement
The study was approved by the Ethics Committee of the Zhongnan Hospital of Wuhan University, and the study enrollment was carried out after a written informed consent was obtained from each patient. Our research has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). The animal experiments were conducted at the Institution of Model Animal of Wuhan University and in accordance with the ARRIVE guidelines and the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. Our experiment was approved by the Animal Care and Use Committee of the Zhongnan Hospital of Wuhan University (WQ20210088).
Clinical Perspectives
Previous studies have shown that TGFB2-OT1 acts as a competing endogenous RNA and binds to microRNAs, thereby promoting inflammation and autophagy in vascular endothelial cells (VECs) by enhancing the production of inflammatory factors. However, TGFB2-OT1 has not been well-studied in the context of cancer, and its biological function in HCC is not clear. Our findings showed that lncRNA TGFB2-OT1 interferes with the binding between β-catenin and GSK3β, thus inhibiting the phosphorylation and degradation of β-catenin. This results in an enhanced capacity for proliferation, invasion, migration, and tumor angiogenesis in HCC. As a molecule mediating VEGFA and β-catenin which play essential part in tumor targeting therapy, TGFB2-OT1 may act as a new target combined with other chemotherapy drugs applicated in the treatment of HCC in the future.
Disclosure
The authors declare no conflicts of interest.
Acknowledgments
The authors acknowledge the resources and support provided by the medical research center, Zhongnan Hospital of Wuhan University.
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi:10.3322/caac.21660
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
- Mattiuzzi C, Lippi G. Current cancer epidemiology. J Epidemiol Glob Health. 2019;9:217–222. doi:10.2991/jegh.k.191008.001
- Hepatocellular carcinoma. % J nature reviews. Dis Primers. 2021;7:7. doi:10.1038/s41572-021-00245-6
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi:10.1002/hep.29270
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi:10.1016/j.cell.2018.01.011
- Sedaghati M, Kebebew E. Long noncoding RNAs in thyroid cancer. Curr Opin Endocrinol Diabetes Obes. 2019;26:275–281. doi:10.1097/MED.0000000000000497
- Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol Cancer. 2019;18:28. doi:10.1186/s12943-019-0957-7
- Bian Z, Zhang J, Li M, et al. LncRNA–FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24:4808–4819. doi:10.1158/1078-0432.CCR-17-2967
- Jiang X, Wang G, Liu Y, et al. A novel long non-coding RNA RP11-286H15.1 represses hepatocellular carcinoma progression by promoting ubiquitination of PABPC4. Cancer Lett. 2021;499:109–121. doi:10.1016/j.canlet.2020.11.038
- Huang S, Lu W, Ge D, et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi:10.1080/15548627.2015.1106663
- Zhao X, Su L, He X, Zhao B, Miao J. Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy. 2020;16:70–85. doi:10.1080/15548627.2019.1598750
- Ma H, Su L, Zhang S, Kung H, Miao J. Inhibition of ANXA7 GTPase activity by a small molecule promotes HMBOX1 translation of vascular endothelial cells in vitro and in vivo. Int J Biochem Cell Biol. 2016;79:33–40. doi:10.1016/j.biocel.2016.08.010
- Vlad A, Röhrs S, Klein-Hitpass L, Müller O. The first five years of the Wnt targetome. Cell Signal. 2008;20:795–802. doi:10.1016/j.cellsig.2007.10.031
- Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4(11):a007906–a007906. doi:10.1101/cshperspect.a007906
- Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–217. doi:10.1038/ncb1534
- Fang JH, Zhou HC, Zhang C, et al. A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology. 2015;62:452–465. doi:10.1002/hep.27760
- Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20:409–426. doi:10.1007/s10456-017-9562-9
- Song D, Lan J, Chen Y, et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene. 2021;40:2952–2967. doi:10.1038/s41388-021-01747-z
- Crawford Y, Ferrara N. VEGF inhibition: insights from preclinical and clinical studies. Cell Tissue Res. 2009;335:261–269. doi:10.1007/s00441-008-0675-8
- Perugorria MJ, Olaizola P, Labiano I, et al. Wnt-beta-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16:121–136. doi:10.1038/s41575-018-0075-9
- Brembeck FH, Rosário M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16:51–59. doi:10.1016/j.gde.2005.12.007
- Yu F, Yu C, Li F, et al. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther. 2021;6:307. doi:10.1038/s41392-021-00701-5
- Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi:10.1002/hep.27893
- Stamos JL, Weis WI. The β-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi:10.1101/cshperspect.a007898
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi:10.1016/j.devcel.2009.06.016
- Wu H, Lu XX, Wang JR, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15:1506–1522. doi:10.1080/15548627.2019.1586250
- Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/beta-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26:1287–1298. doi:10.1016/j.ymthe.2018.02.024
- Cong F, Schweizer L, Varmus H. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development. 2004;131:5103–5115. doi:10.1242/dev.01318
- Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi:10.1186/s12943-020-1147-3
- Zhou B, Yang H, Yang C, et al. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89–99. doi:10.1016/j.canlet.2020.10.002
- Huang JZ, Chen M, Chen D, et al. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68:171–84 e6. doi:10.1016/j.molcel.2017.09.015
- Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi:10.1038/nature10398
- Kondo T, Plaza S, Zanet J, et al. Small peptides switch the transcriptional activity of shavenbaby during drosophila embryogenesis. Science. 2010;329:336–339. doi:10.1126/science.1188158
- Anderson DM, Anderson KM, Chang CL, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi:10.1016/j.cell.2015.01.009
- Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi:10.1186/s13059-019-1685-4
- Wang T, Cui Y, Jin J, et al. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 2013;41:4743–4754. doi:10.1093/nar/gkt178