Abstract
Purpose
To explore the mechanisms by which circRNA/miRNA/mRNA competitive endogenous RNAs (ceRNA) networks regulate CRSwNP.
Methods
The expression profiles of circRNAs, miRNAs, and mRNAs from patients with CRSwNP and control subjects were acquired from the Gene Expression Omnibus database. The circRNA/miRNA/mRNA ceRNA network was constructed based on the predicted circRNA–miRNA interactions and miRNA–mRNA interactions. Hub-mRNAs were screened by protein–protein interaction network analysis and Cytoscape molecular complex detection. The expression of factors in tissue and in hsa_circ_0031594 siRNA transfection cells was verified by RT-qPCR and the association between them was revealed by Spearman correlation analysis. Receiver operating characteristic curve analysis was performed with the pROC R package.
Results
The differential expression of 5423 circRNAs, 415 miRNAs, and 3673 mRNAs was identified in CRSwNP subjects compared to control subjects. Among these, 9 circRNAs, 39 miRNAs, and 78 mRNAs were screened to construct a ceRNA network. Ultimately, a subnetwork including circRNA hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, RACGAP1, CHEK1 and PRC1 was screened out. RT-qPCR validated that the expression of hsa_circ_0031594, NCAPG2, PRC1 was significantly increased, and hsa-miR-1260b and hsa-miR-6507-5p were expressed significantly less in patients with CRSwNP than in control subjects. In addition, the AUCs of hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, and PRC1 to discriminate CRSwNP patients were 0.995, 0.842, 0.862, 0.765, and 0.816. Spearman correlation showed that the expression of hsa_circ_0031594 was negatively correlated with hsa-miR-1260b and hsa-miR-6507-5p, and positively correlated with NCAPG2 and PRC1. In human nasal epithelial cell (HNEpC) line, knocking down hsa_circ_0031594 could increase the expression of hsa-miR-1260b and hsa-miR-6507-5p, and reduce the expression of NCAPG2 and PRC1.
Conclusion
CeRNA networks including hsa_circ_0031594, hsa-miR-1260b, and NCAPG2, and hsa_circ_0031594, hsa-miR-6507-5p, and PRC1 may be key regulators for CRSwNP occurrence, and may be potential targets for the pathogenesis and treatment development of CRSwNP.
Keywords:
Introduction
Chronic sinusitis with nasal polyps (CRSwNP) is characterized by chronic inflammation of the nasal cavity and paranasal sinus mucosa.Citation1 Patients with CRSwNP experience a high burden of symptoms, including stuffy nose, runny nose, and loss of the sense of smell, which have negative impacts on physical and mental health. Despite advances in medical therapy and surgical intervention for CRSwNP, the recurrence rate remains highCitation2 and the pathophysiological mechanisms underlying CRSwNP remain unclear.
Circular RNAs (circRNAs) are a type of ncRNA deriving from exon, intron, or intergenic regions, and are characterized by having a covalently closed continuous loop.Citation3 It is widely reported that circRNA plays an important role in cell and tissue proliferation.Citation4,Citation5 For example, circGCN1L1 promotes cell proliferation and apoptosis by targeting miR-330-3p and TNF-α.Citation6 CircRNAs play important roles in CRSwNP occurrence.Citation7 Yu et al found that 1794 circRNAs were downregulated and 1081 circRNAs were upregulated in CRSwNP patients relative to control subjects.Citation7 MicroRNAs (miRNAs) are a class of small noncoding RNAs that regulate the expression of target genes by preventing the translation or inducing the degradation of target mRNAs.Citation8 Zhang et al identified differential miRNA expression profiles between patients with CRSsNP and those with eosinophilic CRSwNP.Citation9 They reported that up-regulated expression of miR-125b contributes to mucosal eosinophilia in eosinophilic CRSwNP.Citation9 Silveira et al reported that miR-205-5p was directly implicated with cell cycle regulation, and related to T2-polarity in CRSwNP.Citation10 Liu et al found that upregulated miR-21 can inhibit activation of NF-κB P65 and increase IL-10 expression through targeting to PDCD4 in nasal epithelial cells, which could further limit the expression of pro-inflammatory cytokines in CRSwNP.Citation11 Although many studies have indicated differential expressions of circRNAs and miRNAs between patients with CRSwNP and other patients or controls, the specific regulatory mechanisms through regulation of circRNA/miRNA/mRNA networks contributing to CRSwNP remain unclear.
CircRNAs can act as miRNA ‘sponges’ or protein scaffolds to regulate gene transcription.Citation12 CircRNAs can regulate mRNA expression by competitively associating with miRNAs, instead of their target mRNAs, thus forming competitive endogenous RNA (ceRNA) network.Citation6,Citation13–15 However, whether the differential expression of circRNAs can regulate CRSwNP through forming circRNA/miRNA/mRNA ceRNA networks remains unclear. Our study explores the mechanisms through circRNA/miRNA/mRNA ceRNA networks may regulate CRSwNP pathogenesis, providing rationale for these RNAs to be considered as potential targets for the diagnosis and treatment of CRSwNP.
Materials and Methods
Microarray Data Information
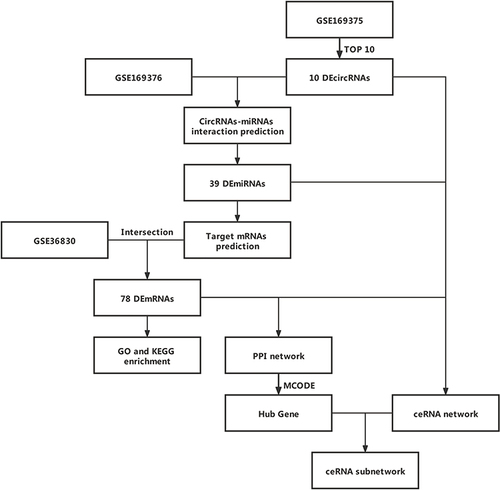
The raw expression data for circRNA, miRNA, and mRNA was collected from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Genomic microarray analysis of circRNA expression (GSE169375) and miRNA expression (GSE169376) was obtained for 9 nasal mucosa samples, including samples from 6 CRSwNP patients and 3 control subjects.Citation7 Significantly differentially expressed (DE) circRNAs between the 6 CRSwNP samples and the 3 control samples were screened by a cutoff of P < 0.05 and |log2fold change (FC)|>1.5,Citation16,Citation17 using the Limma R package. Significantly DEmiRNAs were identified by a cutoff of P < 0.05 using the Limma R package. MRNA expression data was obtained from nasal polyp tissue samples from 6 CRSwNP subjects and from uncinate tissue from 6 control subjects in GSE36830.Citation18 DEmRNAs were identified with a p-value of P < 0.05 using the Limma R package. The basic information for the samples in the three GEO datasets used in this study are shown in .
Table 1 Basic Information of the Three Microarray Datasets from GEO
Prediction of circRNA–miRNA Interaction
The circBase website (http://www.circbase.org/)Citation19 was used to evaluate the characteristics of circRNAs. The top ten DEcircRNAs (), including the top 5 up-regulated and the top 5 down-regulated DEcircRNAs, were identified based on the absolute value of log2FC. The interactions between DEmiRNAs and the top ten DEcircRNAs were predicted using the the miRanda (http://www.microrna.org)Citation20 and the RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) databases.Citation21
Table 2 Basic Characteristics of the Top Ten DEcircRNAs
Prediction of miRNA–mRNA Interaction
The mRNAs that were potentially related to the differentially expressed miRNAs were screened based on the intersection predicted from three online miRNA target prediction tools, including TargetScan (http://www.targetscan.org/),Citation22 miRDB (http://www.mirdb.org/),Citation23 and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php).Citation24 The miRNA-related mRNAs that were not differentially expressed between CRSwNP subjects and control subjects in GSE36830 were excluded.
Construction of a circRNA–miRNA–mRNA ceRNA Network
The circRNA/miRNA/mRNA regulatory network was constructed by integrating circRNA-miRNA pairs and miRNA-mRNA pairsCitation25 and was visualized using Cytoscape software (http://cytoscape.org/; version 3.8.0).
Functional Annotation of mRNAs in the ceRNA Network
The biological processes, cellular components, and molecular functions related to the mRNAs in the ceRNA network were analyzed by Gene Ontology (GO) (http://geneontology.org/). Signalling pathways related to the mRNAs in the ceRNA network were analyzed by Kyoto Encyclopedia of Gene and Genome (KEGG) (https://www.kegg.jp/). The results were depicted using clusterProfiler R package.
Construction of PPI Network and Identification of Hub Genes
A PPI network related to mRNAs in the ceRNA network was established by the string database (minimum required interaction score = 0.400 and hide disconnected nodes in the network) (http://string-db.org/). The resulting PPI network was visualized using Cytoscape software. Subsequently, the hub-mRNAs were evaluated using the Cytoscape Molecular Complex Detection (MCODE) (module with the following criteria: degree = 2; node score = 0.2; k-core = 2; and max. depth = 100),Citation26 according to the topology.
Patient Inclusion and Exclusion Criteria
A total of 28 subjects including 14 patients with CRSwNP and 14 control subjects were enrolled in this study. Patient inclusion criteria: diagnosed with CRSwNP according to the European Position Paper on Rhinosinusitis and Nasal Polyps (EPOS);Citation27 no treatment with corticosteroids, immunomodulatory agents, or antibiotics within 4 weeks before enrollment. Patients with acute infections, acetylsalicylic acid-intolerance, fungal sinusitis, immunodeficiency, coagulation disorder, or cystic fibrosis and pregnant women were excluded. Nasal polyp tissues from patients with CRSwNP and nasal mucosal tissues from control subjects were collected. This study was approved by the Institution Ethics Committee of Yantai Yuhuangding Hospital and all patients provided informed consent before enrollment. Our experiments were performed in accordance with the Declaration of Helsinki.
RNA Extraction and RT-qPCR
Total RNA was extracted from tissue samples using Trizol reagent (Sparkjade, China), according to the manufacturer’s instructions. Then 1 µg RNA was reverse-transcribed using AG reverse transcription kit (Accurate Biology, China). The RT-qPCR reactions were performed using SYBR Green qPCR Mix kit (Sparkjade, China) according to the manufacturer’s instructions. The extraction and amplification of miRNAs was performed according to the manufacturer’s instructions (Vazyme, China). The primers for RT-qPCR are shown in .
Table 3 Primer Sequences of circRNA, miRNAs, mRNAs for RT-qPCR
ROC Curve and Correlation Analysis
ROC curve analysis was carried out to calculate the areas under curve (AUC) and estimate the functional utility of the selected the hub gene in discriminating CRSwNP patients from normal subjects. The pROC R package was used for ROC analyses. The correlation between hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2 and PRC1 was assessed by Spearman correlation analysis.
Nasal Epithelial Cell Line Culture and Small Interference RNA Transfection
The human nasal epithelial cell (HNEpC) line was obtained from BeNa Culture Collection (BNCC, China). The HNEpC cells were cultured by Minimum Essential Medium containing 10% fetal bovine serum at 37°C with 5% CO2. Three small interference RNAs (siRNAs) for hsa_circ_0031594 were generated by GenePharma (GenePharma Corporation, Shanghai, China). The sequences: hsa_circ_0031594-1 siRNA, 5’-ACGCAACCAGGCAAUGGUGTT-3’; hsa_circ_0031594-2 siRNA, 5’-UUACGCAACCAGGCAAUGGTT-3’; hsa_circ_0031594-3 siRNA, 5’-ACCAGGCAAUGGUGGCUUGCU-3’. The siRNA sequences of negative control (NC) were 5’-UUCUCCGAACGUGUCACGUTT-3’; All siRNAs were transfected by Lipofectamine® 3000 transfection reagent (Thermo Fisher Scientific, America) according to the instruction. After transfection for 48 h, the expression of hsa_circ_0031594 was tested by real-time PCR analysis to assess the transfection efficiency.
Nuclear Cytoplasm Separation and Nucleic Acid Electrophoresis
3*106 HNEpC cells were collected for nuclear cytoplasm separation and RNA extraction by using the PARIS™ kit (Thermo Fisher Scientific, America) according to the manufacturer’s instructions. The RNA is reversely transcribed using HiScript Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). The hsa_circ_0031594 was amplified by using 2x EasyTaq PCR SuperMix+ Dye (TransGen Biotech, China) PCR reagent according to the instructions. The amplified nucleic acid was tested by agarose gel electrophoresis. The expression of hsa_circ_0031594, U6 (control expressed in the nuclear), and GAPDH (control expressed in the cytoplasm) in nuclear and cytoplasm were analyzed by PCR.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism (GraphPad Software, CA). The significance of differences between groups was tested by Student’s t-test. The relative expression of genes was calculated using the 2−ΔΔCT method. A p-value of P < 0.05 was considered as statistically significant.
Results
Identification of Differentially Expressed circRNAs, miRNAs, and mRNAs Between CRSwNP Patients and Controls
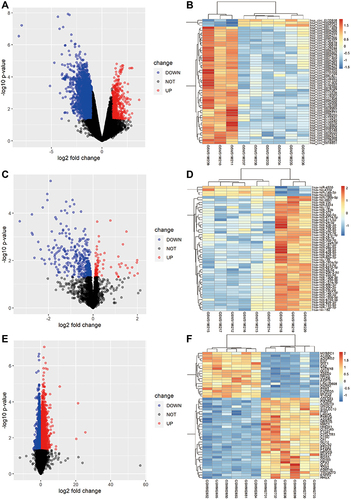
A flowchart detailing the analysis steps is shown in . In brief, 5423 DEcircRNAs were found to be differentially expressed in patients with CRSwNP compared to controls in GSE169375. The DEcircRNAs are shown in a volcano plot (), and the top 50 DEcircRNAs visualized in a heatmap (). Additionally, 415 DEmiRNAs in patients with CRSwNP compared to controls () were identified from GSE169376, and the top 50 DEmiRNAs were visualized in a heatmap (). Meanwhile, 3673 DEmRNAs in patients with CRSwNP compared to controls were identified from GSE36830 (), and the top 50 DEmRNAs were visualized in a heatmap ().
Figure 2 Differentially expressed circRNAs, miRNAs and mRNAs in CRSwNP subjects compared with control subjects. (A) Volcano map for all DEcircRNAs from GSE169375. (B) Heatmap of the top 50 DEcircRNAs from GSE169375. (C) Volcano map for all DEmiRNAs from GSE169376. (D) Heatmap of the top 50 DEmiRNAs from GSE169376. (E) Volcano map for all DEmRNAs from GSE36830. (F) Heatmap of the top 50 DEmRNAs in GSE36830.
Construction of the ceRNA Network
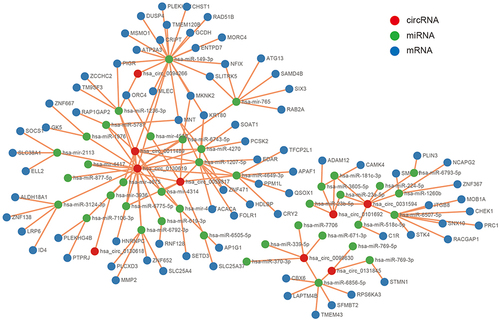
To better comprehend the role of circRNAs in regulating CRSwNP, a circRNA/miRNA/mRNA regulatory network was generated. The top ten DEcircRNAs () were selected as initial candidates for the ceRNA network construction. Of the 415 DEmiRNAs, 39 miRNAs were predicted to be related to the top ten DEcircRNAs by the miRanda and RNAhybrid databases. Of the DEcircRNAs, hsa_circ_0092542 had no potential interaction with miRNAs, so it was excluded from subsequent analyses. We chose 78 mRNAs predicted to interact with the 39 miRNAs. Finally, the 9 DEcircRNAs, the 39 miRNAs, and the 78 mRNAs (Tables S1 and S2) were used to construct the ceRNA network (). 62 circRNA-miRNA interaction pairs and 88 miRNA-mRNA interaction pairs were found within the ceRNA network ().
GO and KEGG Enrichment Analyses
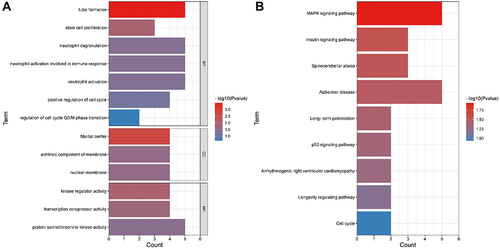
In order to understand the biological functions and pathways impacted by the 78 DEmRNAs in CRSwNP, GO annotation and KEGG pathway analysis were performed. GO enrichment analysis consists of biological processes, molecular function, and cellular components (). The 78 DEmRNAs were primarily enriched in biological processes such as neutrophil degranulation, neutrophil activation, positive regulation of cell cycle, and tube formation. The 78 DEmRNAs were mainly enriched in cellular components including fibrillar center, extrinsic component of membrane, and nuclear membrane, and were enriched in molecular functions including kinase regulator activity, transcription corepressor activity, and protein serine/threonine kinase activity ( and Table S3). The 78 DEmRNAs in CRSwNP were significantly enriched in KEGG pathways including the MAPK signaling pathway, insulin signaling pathway, p53 signaling pathway, and the cell cycle ( and Table S4).
Hub Genes and ceRNA Subnetwork Screen
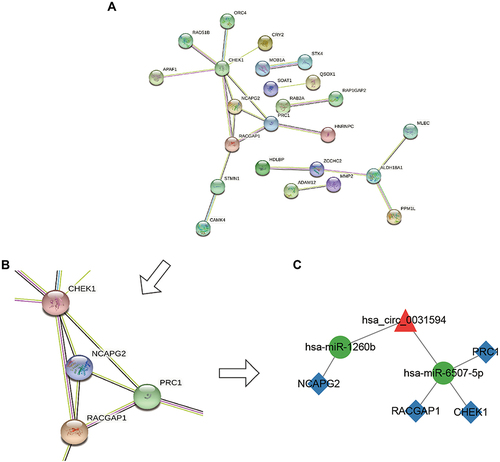
The PPI network was established by the string database. The PPI network of the 78 DEmRNAs contained 24 proteins and 21 interaction edges (). The hub interaction cluster consisted of four genes: polycomb inhibitory complex 1 (PRC1), checkpoint kinase (CHEK1), Rac GTPase Activating Protein 1 (RACGAP1), and Non-SMC lectin II complex subunit G2 (NCAPG2), as identified by the Cytoscape MCODE module (). By mapping the 4 hub genes into the preliminary circRNA/miRNA/mRNA network, a circRNA/miRNA/mRNA subnetwork consisting of hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, PRC1, CHEK1, RACGAP1, NCAPG2 was proposed ().
Verification of Expression of the ceRNA Subnetwork Factors in CRSwNP
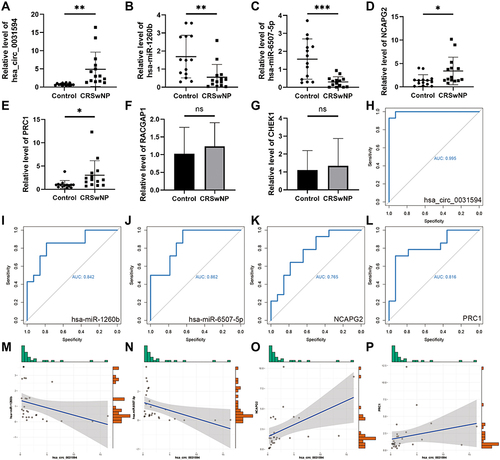
In order to verify the ceRNA subnetwork analysis, we tested the expression of ceRNA subnetwork factors in nasal polyp samples from CRSwNP patients and nasal mucosa samples from control subjects. We found that expression of hsa_circ_0031594 was significantly higher in nasal polyps than in control nasal mucosa (). Expression of hsa-miR-1260b and hsa-miR-6507-5p was significantly lower in nasal polyps than in control nasal mucosa ( and ). The expression of NCAPG2 and PRC1 in nasal polyps was significantly higher than in control nasal mucosa ( and ). However, the expression of RACGAP1 and CHEK1 was not significantly different between CRSwNP and control samples ( and ). ROC analysis was performed to verify the correlation between gene expression and CRSwNP. The AUCs of hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, and PRC1 were 0.995, 0.842, 0.862, 0.765, and 0.816, respectively (). Spearman correlation analysis results () showed that the expression of hsa_circ_0031594 was negatively correlated with the expression of hsa-miR-1260b (Spearman correlation coefficient = −0.55, p = 0.003) and hsa-miR-6507-5p (Spearman correlation coefficient = −0.58, p = 0.001), and positively correlated with the expression of NCAPG2 (Spearman correlation coefficient = 0.60, p = 0.001), and PRC1 (Spearman correlation coefficient = 0.47, p = 0.011).
Figure 6 The factor expression levels and diagnostic values of core subnetwork, and correlation analysis in CRSwNP. (A–E) Confirmation of the expression of hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, and PRC1 in the subnetwork using RT-qPCR (CRSwNP (n=14); control subjects (n=14). *P < 0.05, **P < 0.01, ***P<0.001. (F and G) Confirmation of the expression of RACGAP1 and CHEK1 in the subnetwork using RT-qPCR (CRSwNP (n=7); control subjects (n=7)). (H–L) ROC curves for testing the hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2 and PRC1 by RT-qPCR. (M) The correlation between hsa_circ_0031594 and hsa-miR-1260b (Spearman correlation coefficient = −0.55, p = 0.003). (N) The correlation between hsa_circ_0031594 and hsa-miR-6507-5p (Spearman correlation coefficient = −0.58, p = 0.001). (O) The correlation between hsa_circ_0031594 and NCAPG2 (Spearman correlation coefficient = 0.60, p = 0.001). (P) The correlation between hsa_circ_0031594 and PRC1 (Spearman correlation coefficient = 0.47, p = 0.011).
Knocking Down hsa_circ_0031594 Upregulated hsa-miR-1260b and hsa-miR-6507-5p and Downregulated NCAPG2 and PRC1
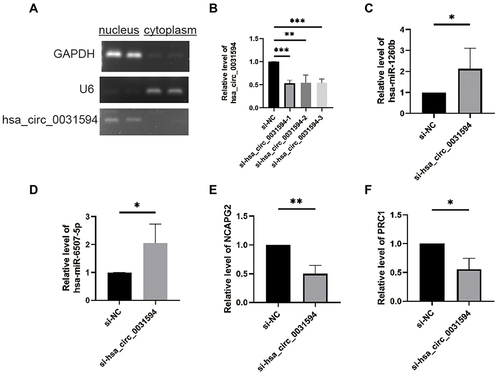
Through nuclear and cytoplasm separation, hsa_circ_0031594 was localized in cytoplasm of HNEpC cells (). When knocking down hsa_circ_0031594 expression by siRNAs in HNEpC cells, the expression of hsa_circ_0031594 were obviously downregulated (). Meanwhile, the expression levels of hsa-miR-1260b and hsa-miR-6507-5p were increased and the level of NCAPG2, and PRC1 was reduced in the hsa_circ_0031594 siRNA group than in the NC group (). These results showed knocking down hsa_circ_0031594 could upregulate hsa-miR-1260b and hsa-miR-6507-5p and downregulate NCAPG2 and PRC1.
Figure 7 Knocking down hsa_circ_0031594 affected the expression of hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, and PRC1. (A) The subcellular localization of hsa_circ_0031594 in human nasal epithelial cells HNEpC. (B) SiRNAs obviously knocked down hsa_circ_0031594 expression. **P < 0.01, ***P < 0.001. (C and D) Knocking down hsa_circ_0031594 obviously increased the hsa-miR-1260b and hsa-miR-6507-5p expression. *P < 0.05. (E and F) Knocking down hsa_circ_0031594 obviously reduced the NCAPG2 and PRC1 expression. *P < 0.05, **P < 0.01.
Discussion
CRSwNP is heterogeneous disease that is associated with significant public health problems and causes heavy socioeconomic burden.Citation28,Citation29 Abnormal gene expression is a key factor leading to the high incidence of CRSwNP,Citation30,Citation31 but it is unclear how these genes are regulated. In our study, we explored the function of a circRNA/miRNA/mRNA ceRNA network in regulating CRSwNP through bioinformatics analysis and RT-qPCR verification. We found that hsa_circ_0031594/hsa-miR-1260b/NCAPG2 and hsa_circ_0031594/hsa-miR-6507-5p/PRC1 ceRNA subnetworks might be primary regulators of CRSwNP, and our study provides potential targets for the diagnosis and treatment of CRSwNP.
Research by Yu et al identified the differential expression of 2875 circRNAs and 192 miRNAs between CRSwNP subjects and control subjects.Citation7 This indicated that circRNAs and miRNAs may play important roles in the occurrence and development of CRSwNP. However, how these circRNAs and miRNAs interact to regulate CRSwNP remained unknown. Functionally, circRNAs can act as ‘miRNA sponges’ and competitively bind miRNAs to regulate the target mRNA post-transcriptional activity,Citation32 thus forming a ceRNA network to regulate disease occurrence or development. For example, hsa_circ_0000520/miR-556-5p/NLRP3 could mediate cell pyroptosis and inflammation to regulate ovalbumin-induced allergic rhinitis in mice.Citation33 However, no ceRNA networks have yet been reported to regulate CRSwNP. In our study, differentially expressed genes from GSE169375, GSE169376, and GSE36830 were extracted to construct a ceRNA network in order to explore circRNA/miRNA/mRNA functions in CRSwNP. We found that the hsa_circ_0031594/ hsa-miR-1260b/ NCAPG2 and hsa_circ_0031594/ hsa-miR-6507-5p/PRC1 ceRNA subnetworks might be hub regulators of CRSwNP. We used RT-qPCR to validate the differential expression of hsa_circ_0031594, hsa-miR-1260b, hsa-miR-6507-5p, NCAPG2, and PRC1 between CRSwNP patients and control subjects. In HNEpC, knocking down hsa_circ_0031594 could increase the expression of hsa-miR-1260b and hsa-miR-6507-5p, and reduce the expression of NCAPG2 and PRC1. These data suggests that ceRNA networks play important roles in CRSwNP.
Neutrophils are predominant in approximately 50% of patients with CRS without nasal polyps, and also play a role in patients with severe type 2 CRS with nasal polyp disease.Citation34 Neutrophils promote tissue remodeling and impair the epithelial barrier by producing TGF-β2 and oncostatin M.Citation35,Citation36 In addition, activated neutrophils can induce an accumulation of eosinophils,Citation37 and enhance the progression of nasal polyps through a Th2 immune response,Citation38,Citation39 thereby establishing a more persistent and severe pathogenesis of CRSwNP. Neutrophils also induce exacerbation of type 2 immune responses by releasing neutrophil extracellular traps into extracellular space following rhinovirus infection.Citation40 In our study, GO analysis revealed that the 78 DEmRNAs in the ceRNA network were enriched in biological processes related to neutrophil activation, indicating that circRNA/miRNA/mRNA ceRNA network may regulate CRSwNP occurrence and development by activating neutrophils. Moreover, we found through KEGG analysis that the MAPK pathway is involved in CRSwNP. This is consistent with a previous report in which TGF-β1 activated MAPK signaling pathways to promote the development of CRSwNP by initiating EMT.Citation41 These results demonstrate that the genes involved in the ceRNA network play important roles in CRSwNP occurrence and development.
Through PPI network and Cytoscape MCODE analysis of hub genes, PRC1, RACGAP1, CHEK1, and NCAPG2 were screened out. These genes are enriched in biological processes involved in the cell cycle, according to GO analysis. NCAPG2 can promote cell proliferation by regulating G2/MCitation42 or by activating STAT3 and NF-κB/ miR-188-3p pathways.Citation43,Citation44 PRC1 regulates cell proliferation and cell cycle transition through P53.Citation45 In addition, PCR1 reinforces Wnt signaling to promote cell proliferation.Citation46 CHEK1, a serine/threonine specific protein kinase, phosphorylates many downstream effectors to initiate cell cycle checkpoints, cell cycle arrest, DNA repair, and cell death.Citation47 RACGAP1 promotes cell proliferation and restricts apoptosis.Citation48,Citation49 The depletion of RACGAP1 leads to the formation of multinucleated cells, the failure of cytokinesis, and apoptosis.Citation50–52
Abnormal epithelial proliferation is one of the typical features of CRSwNP,Citation53 which leads to abnormal function and remodeling of the nasal epithelial cell barrier, especially squamous cell metaplasia and secretory hyperplasia of epithelial cells.Citation54,Citation55 Abnormal epithelial proliferation can also change the local inflammatory environment and lead to chronic inflammation by producing the epithelial-derived cytokines IL33 and thymic stromal lymphopoietin (TSLP).Citation54–58 These data indicates that cell cycle-associated factors play important roles in CRSwNP pathogenesis. In our study, cell cycle-related factors NCAPG2, PRC1, RACGAP1, and CHEK1 were identified as hub genes for CRSwNP. Through RT-qPCR, we verified increased expression of NCAPG2 and PRC1 in CRSwNP subjects compared to control subjects, showing that NCAPG2 and PRC1 may regulate CRSwNP pathogenesis by mediating cell cycle and cell proliferation.
In the circRNA/miRNA/mRNA subnetwork, hsa_circ_0031594 may act as a ceRNA to capture hsa-miR-1260b and positively regulate the expression of NCAPG2. In addition, hsa_circ_0031594 can also act as a hsa-miR-6507-5p sponge and positively regulate the expression of PRC1/RACGAP1/CHEK1. Hsa_circ_0031594 is upregulated in CRSwNP, which is consistent with a previous report.Citation7 Hsa-miR-6507-5p has been reported to be upregulated in patients with achalasia, however its mechanism has not been studied.Citation59 We found that hsa-miR-6507-5p and hsa-miR-1260b were downregulated in CRSwNP. Hsa_circ_0031594 (Circ-EGLN3) has been reported to promote renal cell carcinoma (RCC) cell proliferation and aggressiveness by enhancing the IRF7 level via sponging miR-1299,Citation60 or by upregulating HMGXB3 via sponging miR-1224-3p.Citation61 Hsa-miR-1260b, mediated by YY1, activates KIT signaling by targeting SOCS6 to regulate NSCLC cell proliferation and apoptosis.Citation62 Therefore, we speculate that the circRNA/miRNA/mRNA network related to hsa_circ_0031594 is a new candidate target related to cell proliferation in CRSwNP. Verification of the differential expression by RT-qPCR revealed that the hsa_circ_0031594/ hsa-miR-1260b/ NCAPG2 axis and hsa_circ_0031594/ hsa-miR-6507-5p/ PRC1 axis may be key pathways in regulating CRSwNP, and ROC analysis results also showed that these subnetworks may have important clinical diagnostic significance. The hsa_circ_0031594/hsa-miR-6507-5p/RACGAP1/CHEK1 axis was not verified in our study, and whether this axis plays a more important role in CRSwNP requires further evaluation.
Conclusion
In conclusion, this study identified a comprehensive circRNA/miRNA/mRNA network related to CRSwNP, which included the 9 DEcircRNAs, the 39 miRNAs, and the 78 mRNAs. Furthermore, the hub-ceRNA network, which included hsa_circ_0031594, hsa-miR-1260b, and NCAPG2 axis, and hsa_circ_0031594, hsa-miR-6507-5p, and PRC1 axis was identified and verified by clinical CRSwNP samples expression analysis and in-vitro cell experiments. Our study will help to explore the mechanisms underlying the occurrence and development of CRSwNP and further develop potential treatment strategies for the disease. However, since these results are mainly based on computational biology and RT-qPCR experimental verification, further biological and molecular experiments will be needed to verify our hypothesis.
Abbreviations
CRSwNP, chronic sinusitis with nasal polyps; CeRNA, competitive endogenous RNA; MiRNA, microRNA; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Gene and Genome; PPI, protein–protein interaction; MCODE, molecular complex detection; ROC, receiver operating characteristic; AUC, area under curve.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Approval and Informed Consent
All study surgical procedures and experiment protocols were performed in accordance with standard guidelines approved by the Institution Ethics Committee of Yantai Yuhuangding Hospital.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82071021), and the Major Scientific and Technological Innovation Project of Shandong Province (2020CXGC011302).
Additional information
Funding
References
- Zhang Y, Lou H, Wang Y, Li Y, Zhang L, Wang C. Comparison of corticosteroids by 3 approaches to the treatment of chronic rhinosinusitis with nasal polyps. Allergy Asthma Immunol Res. 2019;11(4):482–497. doi:10.4168/aair.2019.11.4.482
- Chalermwatanachai T, Vilchez-Vargas R, Holtappels G, et al. Chronic rhinosinusitis with nasal polyps is characterized by dysbacteriosis of the nasal microbiota. Sci Rep. 2018;8(1):7926. doi:10.1038/s41598-018-26327-2
- Tsai CY, Shen CY, Liu CW, et al. Aberrant non-coding RNA expression in patients with systemic lupus erythematosus: consequences for immune dysfunctions and tissue damage. Biomolecules. 2020;10(12):1641. doi:10.3390/biom10121641
- Xu Q, Cheng D, Li G, et al. CircHIPK3 regulates pulmonary fibrosis by facilitating glycolysis in miR-30a-3p/FOXK2-dependent manner. Int J Biol Sci. 2021;17(9):2294–2307. doi:10.7150/ijbs.57915
- Ji N, Wang Y, Gong X, Ni S, Zhang H. CircMTO1 inhibits ox-LDL-stimulated vascular smooth muscle cell proliferation and migration via regulating the miR-182-5p/RASA1 axis. Mol Med. 2021;27(1):73. doi:10.1186/s10020-021-00330-2
- Zhu H, Hu Y, Wang C, Zhang X, He D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death Dis. 2020;11(4):284. doi:10.1038/s41419-020-2447-7
- Yu J, Kang X, Xiong Y, Luo Q, Dai D, Ye J. Gene expression profiles of circular RNAs and MicroRNAs in chronic rhinosinusitis with nasal polyps. Front Mol Biosci. 2021;8:643504. doi:10.3389/fmolb.2021.643504
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
- Zhang XH, Zhang YN, Li HB, et al. Overexpression of miR-125b, a novel regulator of innate immunity, in eosinophilic chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2012;185(2):140–151. doi:10.1164/rccm.201103-0456OC
- Silveira MLC, Tamashiro E, Santos ARD, et al. miRNA-205-5p can be related to T2-polarity in chronic rhinosinusitis with nasal polyps. Rhinology. 2021;59(6):567–576. doi:10.4193/Rhin21.109
- Liu R, Du J, Zhou J, et al. Elevated microRNA-21 is a brake of inflammation involved in the development of nasal polyps. Front Immunol. 2021;12:530488. doi:10.3389/fimmu.2021.530488
- Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842. doi:10.1261/rna.047126.114
- Li B, Huang N, Wei S, et al. lncRNA TUG1 as a ceRNA promotes PM exposure-induced airway hyper-reactivity. J Hazard Mater. 2021;416:125878. doi:10.1016/j.jhazmat.2021.125878
- Liu Y, Xue M, Du S, et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat Commun. 2019;10(1):1637. doi:10.1038/s41467-019-09649-1
- Qian W, Cai X, Qian Q, et al. lncRNA ZEB1-AS1 promotes pulmonary fibrosis through ZEB1-mediated epithelial-mesenchymal transition by competitively binding miR-141-3p. Cell Death Dis. 2019;10(2):129. doi:10.1038/s41419-019-1339-1
- Chen Y, Huang X, Zhu K, et al. LIMD2 is a prognostic and predictive marker in patients with esophageal cancer based on a cerna network analysis. Front Genet. 2021;12:774432. doi:10.3389/fgene.2021.774432
- Zhu M, Li M, Zhou W, et al. Qianggan extract improved nonalcoholic steatohepatitis by modulating lncRNA/circRNA immune ceRNA networks. BMC Complement Altern Med. 2019;19(1):156. doi:10.1186/s12906-019-2577-6
- Stevens WW, Ocampo CJ, Berdnikovs S, et al. Cytokines in chronic rhinosinusitis. Role in eosinophilia and aspirin-exacerbated respiratory disease. Am J Respir Crit Care Med. 2015;192(6):682–694. doi:10.1164/rccm.201412-2278OC
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi:10.1261/rna.043687.113
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi:10.1371/journal.pbio.0020363
- Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–454. doi:10.1093/nar/gkl243
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi:10.7554/eLife.05005
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–d131. doi:10.1093/nar/gkz757
- Huang HY, Lin YC, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48(D1):D148–D154. doi:10.1093/nar/gkz896
- Zhao H, Chen L, Shan Y, et al. Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure. Aging. 2021;13(4):6076–6090. doi:10.18632/aging.202590
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi:10.1186/1471-2105-4-2
- Fokkens WJ, Lund VJ, Hopkins C, et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58(Suppl S29):1–464. doi:10.4193/Rhin20.401
- McCormick JP, Thompson HM, Cho DY, Woodworth BA, Grayson JW. Phenotypes in chronic rhinosinusitis. Curr Allergy Asthma Rep. 2020;20(7):20. doi:10.1007/s11882-020-00916-6
- Wang SB, Chen SM, Zhu KS, Zhou B, Chen L, Zou XY. Increased lipopolysaccharide content is positively correlated with glucocorticoid receptor-beta expression in chronic rhinosinusitis with nasal polyps. Immun Inflamm Dis. 2020;8(4):605–614. doi:10.1002/iid3.346
- Peng Y, Zi XX, Tian TF, et al. Whole-transcriptome sequencing reveals heightened inflammation and defective host defence responses in chronic rhinosinusitis with nasal polyps. Eur Respir J. 2019;54(5):1900732. doi:10.1183/13993003.00732-2019
- Böscke R, Vladar EK, Könnecke M, et al. Wnt signaling in chronic rhinosinusitis with nasal polyps. Am J Respir Cell Mol Biol. 2017;56(5):575–584. doi:10.1165/rcmb.2016-0024OC
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
- Yu X, Wang M, Zhao H, Cao Z. Targeting a novel hsa_circ_0000520/miR-556-5p/NLRP3 pathway-mediated cell pyroptosis and inflammation attenuates ovalbumin (OVA)-induced allergic rhinitis (AR) in mice models. Inflamm Res. 2021;70(6):719–729. doi:10.1007/s00011-021-01472-z
- Delemarre T, Bochner BS, Simon HU, Bachert C. Rethinking neutrophils and eosinophils in chronic rhinosinusitis. J Allergy Clin Immunol. 2021;148(2):327–335. doi:10.1016/j.jaci.2021.03.024
- Shi LL, Xiong P, Zhang L, et al. Features of airway remodeling in different types of Chinese chronic rhinosinusitis are associated with inflammation patterns. Allergy. 2013;68(1):101–109. doi:10.1111/all.12064
- Wang H, Li ZY, Jiang WX, et al. The activation and function of IL-36γ in neutrophilic inflammation in chronic rhinosinusitis. J Allergy Clin Immunol. 2018;141(5):1646–1658. doi:10.1016/j.jaci.2017.12.972
- Gevaert E, Zhang N, Krysko O, et al. Extracellular eosinophilic traps in association with Staphylococcus aureus at the site of epithelial barrier defects in patients with severe airway inflammation. J Allergy Clin Immunol. 2017;139(6):1849–1860.e1846. doi:10.1016/j.jaci.2017.01.019
- Ryu G, Kim DW. Th2 inflammatory responses in the development of nasal polyps and chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2020;20(1):1–8. doi:10.1097/ACI.0000000000000588
- Yu S, Cao C, Li Q, et al. Local IL-17 positive T cells are functionally associated with neutrophil infiltration and their development is regulated by mucosal microenvironment in nasal polyps. Inflamm Res. 2021;70(1):139–149. doi:10.1007/s00011-020-01424-z
- Toussaint M, Jackson DJ, Swieboda D, et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med. 2017;23(6):681–691. doi:10.1038/nm.4332
- Yang HW, Lee SA, Shin JM, Park IH, Lee HM. Glucocorticoids ameliorate TGF-β1-mediated epithelial-to-mesenchymal transition of airway epithelium through MAPK and Snail/Slug signaling pathways. Sci Rep. 2017;7(1):3486. doi:10.1038/s41598-017-02358-z
- Zhan P, Xi GM, Zhang B, et al. NCAPG2 promotes tumour proliferation by regulating G2/M phase and associates with poor prognosis in lung adenocarcinoma. J Cell Mol Med. 2017;21(4):665–676. doi:10.1111/jcmm.13010
- Meng F, Zhang S, Song R, et al. NCAPG2 overexpression promotes hepatocellular carcinoma proliferation and metastasis through activating the STAT3 and NF-κB/miR-188-3p pathways. EBioMedicine. 2019;44:237–249. doi:10.1016/j.ebiom.2019.05.053
- Wu J, Li L, Jiang G, Zhan H, Zhu X, Yang W. NCAPG2 facilitates glioblastoma cells’ malignancy and xenograft tumor growth via HBO1 activation by phosphorylation. Cell Tissue Res. 2021;383(2):693–706. doi:10.1007/s00441-020-03281-y
- Wu F, Shi X, Zhang R, et al. Regulation of proliferation and cell cycle by protein regulator of cytokinesis 1 in oral squamous cell carcinoma. Cell Death Dis. 2018;9(5):564. doi:10.1038/s41419-018-0618-6
- Chen J, Rajasekaran M, Xia H, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65(9):1522–1534. doi:10.1136/gutjnl-2015-310625
- Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134(5):1013–1023. doi:10.1002/ijc.28226
- Wang C, Wang W, Liu Y, Yong M, Yang Y, Zhou H. Rac GTPase activating protein 1 promotes oncogenic progression of epithelial ovarian cancer. Cancer Sci. 2018;109(1):84–93. doi:10.1111/cas.13434
- Mi S, Lin M, Brouwer-Visser J, et al. RNA-seq identification of RACGAP1 as a metastatic driver in uterine carcinosarcoma. Clin Cancer Res. 2016;22(18):4676–4686. doi:10.1158/1078-0432.CCR-15-2116
- Niiya F, Xie X, Lee KS, Inoue H, Miki T. Inhibition of cyclin-dependent kinase 1 induces cytokinesis without chromosome segregation in an ECT2 and MgcRacGAP-dependent manner. J Biol Chem. 2005;280(43):36502–36509. doi:10.1074/jbc.M508007200
- Kamijo K, Ohara N, Abe M, et al. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17(1):43–55. doi:10.1091/mbc.e05-06-0569
- Yang XM, Cao XY, He P, et al. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155(4):1233–1249.e1222. doi:10.1053/j.gastro.2018.07.010
- Deng H, Li M, Zheng R, et al. YAP promotes cell proliferation and epithelium-derived cytokine expression via NF-κB pathway in nasal polyps. J Asthma Allergy. 2021;14:839–850. doi:10.2147/JAA.S315707
- Meng J, Zhou P, Liu Y, et al. The development of nasal polyp disease involves early nasal mucosal inflammation and remodelling. PLoS One. 2013;8(12):e82373. doi:10.1371/journal.pone.0082373
- Okada N, Nakayama T, Asaka D, et al. Distinct gene expression profiles and regulation networks of nasal polyps in eosinophilic and non-eosinophilic chronic rhinosinusitis. Int Forum Allergy Rhinol. 2018;8(5):592–604. doi:10.1002/alr.22083
- Ishinaga H, Kitano M, Toda M, et al. Interleukin-33 induces mucin gene expression and goblet cell hyperplasia in human nasal epithelial cells. Cytokine. 2017;90:60–65. doi:10.1016/j.cyto.2016.10.010
- Liao B, Cao PP, Zeng M, et al. Interaction of thymic stromal lymphopoietin, IL-33, and their receptors in epithelial cells in eosinophilic chronic rhinosinusitis with nasal polyps. Allergy. 2015;70(9):1169–1180. doi:10.1111/all.12667
- Nagarkar DR, Poposki JA, Tan BK, et al. Thymic stromal lymphopoietin activity is increased in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2013;132(3):593–600.e512. doi:10.1016/j.jaci.2013.04.005
- Gholipour M, Mikaeli J, Mowla SJ, et al. Identification of differentially expressed microRNAs in primary esophageal achalasia by next-generation sequencing. Turk J Biol. 2021;45(3):262–274. doi:10.3906/biy-2101-61
- Lin L, Cai J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. J Cell Biochem. 2020;121(11):4377–4385. doi:10.1002/jcb.29620
- Zhang G, Wang J, Tan W, et al. Circular RNA EGLN3 silencing represses renal cell carcinoma progression through the miR-1224-3p/HMGXB3 axis. Acta Histochem. 2021;123(6):151752. doi:10.1016/j.acthis.2021.151752
- Xia Y, Wei K, Yang FM, et al. miR-1260b, mediated by YY1, activates KIT signaling by targeting SOCS6 to regulate cell proliferation and apoptosis in NSCLC. Cell Death Dis. 2019;10(2):112. doi:10.1038/s41419-019-1390-y