Abstract
Background
DPY30 is a common subunit of the human SET1/MLL complex and is an essential protein required for the activity of SET1/MLL methyltransferase. DPY30 regulates the histone H3K4 modification, and dysfunction of DPY30 might contribute to the regulation of cancer immune evasion. However, the functions and regulation of DPY30 in the expression of programmed cell death ligand 1 (PD-L1) is still not completely explored.
Methods
Various online databases were used for data processing and visualization, including UALCAN, Oncomine, cBioPortal, SangerBox, TISIDB, TIMER, and GEPIA databases. The expression of DPY30 and PD-L1 in melanoma tissues were evaluated by IHC. Chromatin Immunoprecipitation (ChIP), RT-PCR and flow cytometry were used to elucidate the underlying molecular mechanism of PD-L1 expression regulation and its function.
Results
The mRNA level of DPY30 in melanoma was higher than in normal tissues. The expression of DPY30 was positively associated with TMB, neoantigens and PD-L1 expression. Furthermore, DPY30 expression showed significant positive correlations with immune suppressor cells and ICP genes involved in T-cell exhaustion. IHC showed that the positive rates of DPY30 and PD-L1 in melanoma tissues were 62% and 58%, respectively. Correlation analysis revealed that DPY30 over-expression was positively associated with PD-L1 expression. Silencing of DPY30 by specific siRNA significantly inhibited PD-L1 expression. ChIP analysis revealed that H3K4me3 levels were enriched in the proximal PD-L1 promoter region in tumor cells. Inhibition of DPY30 still suppressed the PD-L1 level in IFN-γ treated MMAC-SF cells. Furthermore, the apoptosis of PD1+ T-cells in co-culture with MMAC-SF cells by knockdown of DPY30 were markedly reduced.
Conclusion
This study shows the roles of DPY30 in regulating the cancer immune evasion in melanoma. Targeting the DPY30-H3K4me3 axis might be an alternative approach to enhance the efficacy of checkpoint immunotherapy.
Keywords:
Introduction
Melanoma stands out as one highly immunogenic cancer that approaches durable response to checkpoint immunotherapy, leading to the first approval of immune checkpoint inhibitors in the treatment of melanoma.Citation1,Citation2 Immune checkpoint inhibitors targeting the cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1) led to a considerable improvement of survival in patients with metastatic melanoma.Citation3 Unfortunately, about 60% of patients with melanoma showed primary resistance to PD-1 blockade, and 20−30% of initial responders would develop secondary drug resistance.Citation4 It has been suggested that the expression of PD-L1 in tumor cells is considered a predictive biomarker for response to immunotherapy,Citation5 but the mechanism underlying melanoma resistance to PD-1/PD-L1 inhibitors is still elusive.
PD-L1 is expressed on the surface of several cell types, including T-cells, B-cells, antigen-presenting cells (APCs), monocytes, and epithelial cells, and is upregulated in response to IFN-γ.Citation6 IFN-γ is a key cytokine produced by activated T cells, as well as natural killer (NK) and NK T cells, in the tumor microenvironment.Citation7 Notably, tumor cells can adaptively increase the expression of PD-L1 under the control of IFN-γ.Citation8 IFN-γ binds with receptor and subsequently activates JAK/STAT signaling pathway, which leads to the downstream expression and activation of IRF-1, further inducing PD-L1 expression on tumor cells.Citation9 Furthermore, high PD-L1 expression detected in cancer cells and associated with poor prognosis.Citation10 The binding of PD-L1 to PD-1 in T cells induced T cell apoptosis and reducing immunocompetence, which thus helps cancer cells escape immune surveillance and killing.Citation11 Moreover, it has been reported that MYC and Sox2 overexpression,Citation12,Citation13 and the loss of PTENCitation14 regulate constitutive PD-L1 expression in tumor cells. PD-L1 has become a key protein in immunotherapy, and its regulatory mechanisms are being intensively studied.
Histone H3K4 methylation is one of the most prominent epigenetic modifications, and plays critical roles in regulating target gene transcription.Citation15 Especially histone H3K4 trimethylation (H3K4me3) selectively localizes at gene promoters and transcriptional start sites and directly regulates gene transcription.Citation16 As the most notable histone H3K4 methylation enzymes in mammals, the SET1/MLL complexes comprise Set1a or −1b or Mll1, −2, −3, or −4 as the catalytic subunit, and Wdr5, Rbbp5, Ash2l, and Dpy30 as integral core subunits necessary for the full methylation activity.Citation17 DPY30 directly facilitates genome-wide H3K4 methylation and involves in regulating fundamental cellular processes.Citation18 It has been reported that DPY30 significantly promotes cell proliferation, migration, invasion, and epithelial–mesenchymal transition of epithelial ovarian cancer.Citation19 Similarly, DPY30 plays a critical role in the proliferation and appropriate differentiation of hematopoietic progenitor cells.Citation20 Moreover, DPY30 regulates cell senescence through ID protein expression.Citation21 However, the roles and mechanisms of DPY30 in the development of melanoma are still unclearly known.
In this study, we analyzed the expression of DPY30 in melanoma and investigated the correlation between DPY30 and progression, and cancer immune evasion in melanoma on the basis of various databases. Finally, we evaluated DPY30 and PD-L1 levels in melanoma tissues by IHC analysis, and tested the hypothesis that PD-L1 expression is regulated by an epigenetic mechanism in melanoma.
Materials and Methods
Cell Culture and Reagents
Human melanoma MMAC-SF (RRID: CVCL_1420) and WM-115 (RRID: CVCL_0040) cells were propagated in DMEM medium and MEM medium at humid atmosphere (5% CO2, 37°C), respectively. The culture medium was supplemented with 10% FBS, penicillin (100 u/mL) and streptomycin (100 mg/mL). FITC Annexin V Apoptosis Detection Kit I (cat# 556547) was supplied by BD Biosciences (San Jose, CA, USA). Chromatin Immunoprecipitation Kit (17–10086) was obtained from Merck (Millipore, CA, USA). H3K4Me3 (9751S) was purchased from Cell Signaling Technology (Danvers, MA, USA). IFN-γ (300–02) was supplied by Peprotech (Jiangsu China). PE anti-mouse PD-L1 (124307) and PE anti-mouse PD-1 (135205) were purchased from Biolegend (San Diego, CA, USA). Anti PD-L1 antibody (ab205921) was obtained from Abcam (Cambridge, MA, USA). Anti DPY30 antibody (MA5-32900) was acquired Invitrogen (Carlsbad, CA, USA).
Gene Analysis in Public Datasets
The Oncomine database (http://www.oncomine.org) and UALCAN (http://ualcan.path.uab.edu) database were used to search and analyze the expression levels of DPY30 reported by other studies. The TIMER (http://TIMER.cistrome.org) online tool was used to observe the pan-cancer analysis of DPY30 mRNA expression. DNA methylation of DPY30 in skin cutaneous melanoma (SKCM) was estimated and visualized using MEXPRESS (https://mexpress.be) and UALCAN. cBioPortal (https://www.cbioportal.org/) was used to search and download mutation information. SangerBox (https://www.SangerBox.com/) was used to analyze immune checkpoint (ICP), tumor mutational burden (TMB), and neoantigens. TIMER (cistrome.shinyapps.io/timer) was used to investigate the correlations between DPY30 and tumor infiltrating immune cells. TISIDB (http://cis.hku.hk/TISIDB/index.php) and GEPIA (http://gepia.cancer-pku.cn) were used to explore the correlations between DPY30 expression and immune or molecular subtypes of different cancer types.
Tissue Microarray and Evaluation of Immunostaining
The commercial melanoma tissue arrays were constructed by Avilabio Co. Ltd. in Shaanxi, China. Briefly, the tissue microarrays including 50 melanoma patients were prepared from archival formalin-fixed, paraffin embedded tissue blocks. Immunohistochemical (IHC) staining was performed as previously described.Citation22 Protein expression levels were determined using this formula: overall score = positive percentage score × intensity score. The staining intensity was scored as negative (0), weak (1), moderate (2) or strong (3). The percentage of positive cells was scored as 0 (0–10%), 1 (10–25%), 2 (25–50%) or 3 (50–75%) or 4 (>75%).Citation23 DPY30 in nucleus was defined as negative (combined score from 0 to 6) or positive (combined score from 8 to 12). PD-L1 expression was considered positive when ≥5% membranous staining of tumor cells were positive, as described in previous studies.Citation24
Real-Time PCR (RT-PCR) Analysis
Total RNA was isolated using the Trizol reagent (15596–026; Invitrogen, USA). Reverse transcription (RT) was performed using the TransScript First-strand cDNA Synthesis SuperMix (TransGen), according to the manufacturer’s protocol. The primer sequences were as follows: DPY30 forward primer: 5′-AACGCAGGTTGCAGAAAATCCT−3′, Reverse primer, 5′-TCTGATCCAGGTAGGCACGAG−3′; PD-L1 forward primer: 5′-GCTGTTGAAGGACCAGCTCT-3′, Reverse primer, 5′-TGGAGGATGTGCCAGAGGTA-3′; GAPDH forward primer: 5′-AATCCCATCACCATCTTCC-3′, Reverse primer, 5′-CATCACGCCACAGTTTCC-3′.
Chromatin Immunoprecipitation (ChIP)
Interaction of H3K4me3 in the PD-L1 promoters was examined using Chromatin Immunoprecipitation Kits. Briefly, MMAC-SF and WM-115 cells were used for each ChIP. DNA-protein complex was crosslinked with 1% formaldehyde, lysed with IP lysis buffer, digested with micrococcal nuclease, ultrasonicated, and incubated with anti-H3K4me3. After incubation with protein G beads and washing with wash buffers, chromatin fragments were harvested using the elution buffer. Finally, after decrosslinking and protein digestion, the resulting DNA fragments were purified and fold enrichment was examined using qPCR.
Knockdown by siRNA
Small interfering RNAs for gene DPY30 were purchased from Qiagen (validated FlexiTube siRNA). Transfection of MMAC-SF and WM-115 cells were carried out using Lipofectamine® RNAiMAX vehicle according to the manufacturer’s instruction following optimization. As a negative control, a non-targeting sequence siRNA was employed. Knockdown at mRNA level was confirmed by RT-PCR. RT-PCR was carried out on an ABI PRISM 7900HT sequence detection system (Applied Biosystems).
Preparation of Human T Cells and Co-Culture
The human T cells were isolated from the three healthy volunteers using a human T cell isolation kit (Miltenyi Biotec) according to the manufacturer’s protocol. T cells were cultured in Human ImmunoCult-XFT Cell Expansion medium (Stem Cell) with penicillin (100 u/mL) and streptomycin (100 mg/mL) at 37°C with 5% CO2. T cells were prestimulated with IFN-γ (Peprotech) for 48 hours before co-culture with MMAC-SF cells. After 24 hours of co-culture, PD1+ T cells apoptosis was determined by flow cytometry.
Flow Cytometry
Melanoma cells and human T cells were collected and incubated with the appropriate antibodies in room temperature for 30 min. After washing, the appropriate FITC or DyLight649-conjugated antibodies were used to combine with the first antibodies and were analyzed using FlowJo software (v10, BD, USA). For IFN-γ treatment, cells were first transfected with siRNA mimics as described above, they were treated with 1 ng/mL or 5 ng/mL IFN-γ. Then, cells were collected for flow cytometry. Co-culture with MMAC-SF cells and T cells were harvested, and washed twice then resuspended in 1 mL binding buffer as per BD Biosciences manufacturer’s protocol. 5 μL Annexin V-FITC and 5 μL PI were added to the cell suspension for staining in the dark for 15 min. Cells were then analyzed by flow cytometry.
Western Blotting
Total protein was extracted using RIPA lysis buffer supplemented with 1% cocktail and 1% PMSF. The concentration of proteins was measured by the BCA protein assay kit. The quantified proteins were resolved by 10% SDS-PAGE gels electrophoresis and then transferred to a PVDF membrane. Blots were blocked with 5% non-fat dry milk for 1 h at 37°C, and then primary antibodies were probed for 2 h against DPY30 (1:500), PD-L1 (1:2000) and β-actin (1:1000) at a room temperature. Corresponding secondary antibodies were labeled with horseradish peroxidase (1:1000) and incubated for 1.5 h at 37°C. Finally, the blots were placed on film using the enhanced chemiluminescent system (ECL, Bio-Ras Laboratories, Hercules, CA, USA).
Statistical Analysis
Statistical analyses were performed using Student’s t-test comparing 2 groups. Statistical analysis was performed with SPSS 23.0 statistical software (Chicago, IL, USA). Here, the difference was indicated statistically significant when P < 0.01 and P < 0.05.
Results
DPY30 Is Upregulated in Melanoma
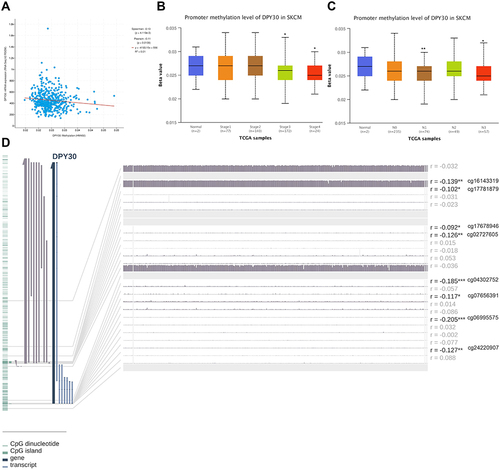
To explore the roles of DPY30 in melanoma progression, we firstly analyzed its expression in Oncomine database. The results showed that the mRNA level of DPY30 in melanoma was higher than in normal tissues (). From the UALCAN database we also identified that the mRNA level of DPY30 was higher in metastatic melanoma compared with primary melanoma (). Meanwhile, we found that DPY30 had a high expression level in almost all cancer cell lines (). In addition, we analyzed the genetic alterations of DPY30 on the outcomes of SKCM patients via the cBioPortal database. Based on TCGA, we found that DPY30 was altered in 5% of SKCM patients (), which included shallow deletion, diploid, gain, and amplification (). However, the genetic alterations of DPY30 represented no significant impact on the overall survival rates of SKCM patients (). Then, we analyzed the methylation status of DPY30 on the progression of SKCM patients through cBioPortal and UALCAN database. The results revealed that expression of DPY30 mRNA was significantly negatively correlated with its methylation status (). From the UALCAN database we also identified that the degree of DPY30 gene methylation was inversely related to the tumor’s stage (). By using the MEXPRESS to explore, we found that among 22 predicted CpG sites of DPY30, 8 CpG sites, including cg16143319 (r = −0.139, P < 0.01), cg17781879 (r = −0.102, P < 0.05), cg17678946 (r = −0.092, P < 0.05), cg02727605 (r = −0.126, P < 0.01), cg04302752 (r = −0.185, P < 0.001), cg07656391 (r = −0.117, P < 0.05), cg06995575 (r = −0.205, p < 0.001), and cg24220907 (r = −0.127, P < 0.01), exhibited negative correlation with DPY30 expression ().
Figure 1 DPY30 expression in melanoma. (A) DPY30 gene copy number in Haqq melanoma vs normal tissues. (B) Comparison of DPY30 expression in primary and metastatic melanoma. (C) DPY30 expression in different cancers and normal tissues in TCGA data. (D) DPY30 gene expression and mutation analysis in SKCM. (E) The distribution of DPY30 genomic alterations in SKCM. (F) The graph of the correlation between DPY30 expression and copy number alterations in SKCM. (G) Kaplan–Meier curve of OS in SKCM patients with altered (red) and unaltered (blue) DPY30 gene. *P< 0.05, **P< 0.01, ***P< 0.001.
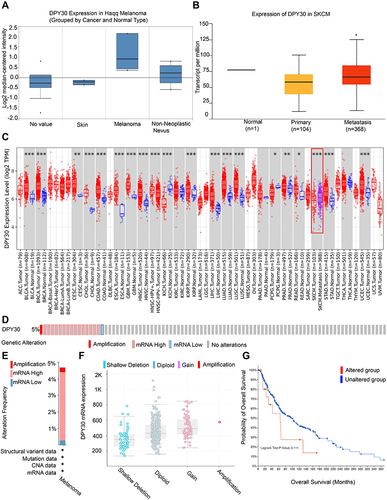
Figure 2 DNA methylation and its correlation with DPY30 expression. (A) Correlation analysis of DPY30 mRNA expression with DPY30 promoter methylation status by the UALCAN database. (B) The methylation levels of DPY30 gene promoter region were compared in TNM stage from UALCAN database. (C) The methylation levels of DPY30 gene promoter region were compared in N stage from UALCAN database. (D) DNA methylation of the DPY30 promoter region in TCGA-SKCM dataset. *P< 0.05, **P< 0.01, ***P< 0.001.
DPY30 Expression Is Related to ICP, TMB and Neoantigen
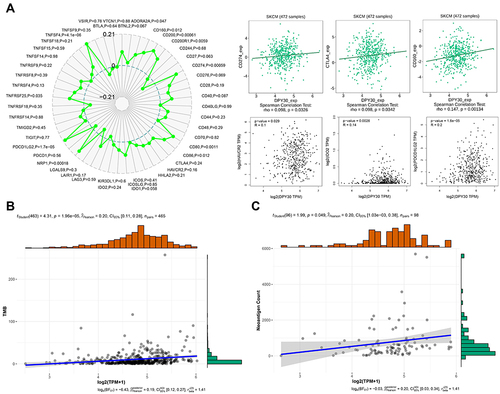
ICP, TMB, and neoantigens in tumor microenvironment were related to antitumor immunity and could predict the therapeutic efficacy of tumor immunotherapy.Citation25–27 Then, we analyzed the correlations between DPY30 expression and ICP, TMB, and neoantigens. We utilized SangerBox and TISIDB to explore the relationships between DPY30 and immune checkpoint genes involved in T-cell exhaustion. Our results indicated that DPY30 expression was positively associated with CD274 (r = 0.489, P = 0.0326), CTLA4 (r = 0.098, P = 0.0342), CD160 (r = 0.147, P = 0.001), HAVCR2 (r = 0.1, P = 0.029), IDO2 (r = 0.14, P = 0.002), and PDCD1LG2 (r = 0.2, P < 0.0001) (). By using the SangerBox to explore, we identified that DPY30 was positively associated with TMB levels (r = 0.20, P < 0.0001) (). By analyzing the relationships between DPY30 expression and neoantigens, we found that DPY30 expression was positively related to neoantigens (r = 0.20, P = 0.049) (). The above results demonstrated that DPY30 might be involved in cancer immune evasion in melanoma.
Figure 3 DPY30 expression is related to ICP, TMB, and neoantigen. (A) Correlations of DPY30 expression with ICP genes (CD274, CTLA4, CD160, HAVCR2, IDO2, and PDCD1LG2) involved in T-cell exhaustion. (B) The relationship between DPY30 expression and TMB. (C) The relationship between DPY30 expression and neoantigen.
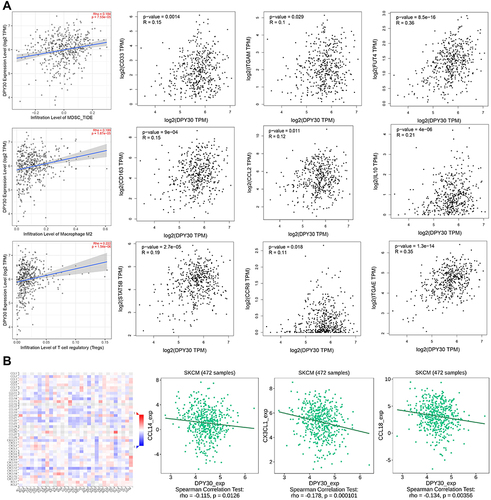
DPY30 Positively Correlated with Immunosuppressive TME in Melanoma
Thus, we further performed correlation analyses to investigate the association between DPY30 and immunosuppressive TME by using TIMER and GEPIA database. The results indicated that DPY30 expression was positively correlated with immunosuppressive cells including myeloid-derived suppressor cells (MDSCs) (CD33: r = 0.315, P = 0.0014; ITGAM: r = 0.1, P = 0.029; FUT4: r = 0.36, P < 0.0001), tumour-associated macrophages (TAMs) (CD163: r = 0.15, P < 0.001; CCL2: r = 0.12, P = 0.011; IL10: r = 0.21, P < 0.001), and regulatory T cells (Tregs) (STAT5B: r = 0.19, P < 0.0001; CCR8: r = 0.11, P = 0.018; ITGAE: r = 0.35, P < 0.0001) (). We further assessed the correlation of DPY30 expression with chemokines using TISIDB database. The result revealed that DPY30 was negatively associated with chemokines, such as CCL14 (r = −0.115, P = 0.012), CX3CL1 (r = −0.178, P = 0.0001), and CCL18 (r = −0.134, P = 0.003) (). Taken together, our findings revealed that DPY30 was closely associated with the immunosuppressive TME in SKCM.
DPY30 Over-Expression Is Positively Associated with PD-L1 Expression
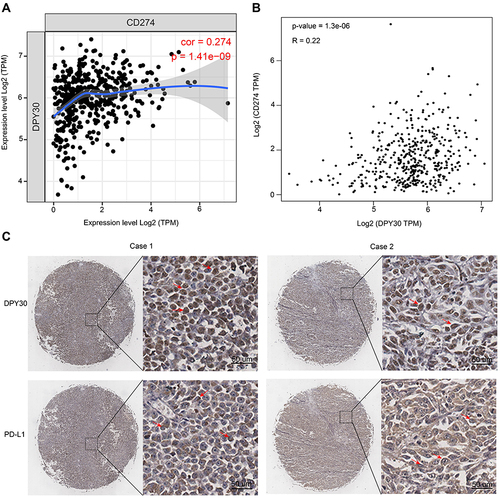
PD-L1 played essential roles in reprogramming the immunosuppressive TME, so we firstly analyzed the association between DPY30 and CD274 in melanoma by using TIMER and GEPIA. We found that the expression of DPY30 and CD274 was positively correlated (, Cor = 0.274, p = 1.41e-9; , R = 0.22, p = 1.3e-6). We further evaluated the levels of DPY30 and PD-L1 in melanoma tissues in human melanoma tissue arrays. The results showed that 31 of 50 of the primary lesions exhibited positive staining for DPY30, and DPY30 protein was localized in the nucleus of melanoma cells. Then, we detected PD-L1 in the same type of tissue arrays. The results demonstrated that the positive rates of PD-L1 was 58% (29/50), and PD-L1 protein was detected on the cell membrane and in the cytoplasm of melanoma cells (). Pearson correlation analysis demonstrated that the expression of DPY30 was closely associated the level of PD-L1, and DPY30 was positively correlated with PD-L1 (r = 0.336, p = 0.017) ().
Table 1 Correlation Between the Expression of DPY30 and PD-L1
Figure 5 The expression of DPY30 correlates with PD-L1 in human melanoma tissues. (A and B) Spearman correlation analysis of DPY30 expression with CD274 expression. (C) Representative IHC staining of DPY30 and PD-L1 in the same melanoma tissue. Red arrows point to the location of DPY30 protein and PD-L1 protein.
The DPY30-H3K4me3 Axis-Mediated PD-L1 Expression
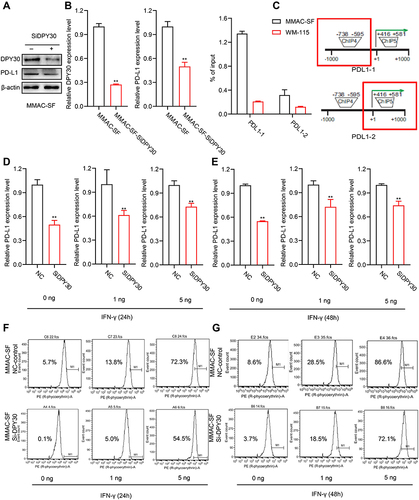
To study the regulation between DPY30 and PD-L1, we used the siRNA targeting DPY30 in MMAC-SF cells. Silencing of DPY30 by specific siRNA significantly inhibited PD-L1 expression in MMAC-SF cells, and this was supported by Western blot and RT-PCR (). To determine whether this decrease in PD-L1 expression was due to DPY30-mediated chromatin modifications. ChIP analysis revealed that H3K4me3 levels were enriched in the proximal PD-L1 promoter region in MMAC-SF cells (). In addition, TISIDB was used to analyze the function of DPY30 and found that DPY30, as part of the MLL1/MLL complex, is involved in the methylation of histone H3K4 especially trimethylation (Supplementary Table 1). These results suggested that DPY30 may mediate PD-L1 expression through H3K4me3.
Figure 6 Function of DPY30 in regulation of PD-L1 expression. (A and B) MMAC-SF cells were transiently transfected with DPY30-specific SiRNA, and the protein levels of DPY30 and PD-L1 were analyzed by Western blotting, and the mRNA levels of DPY30 and PD-L1 were detected by RT-PCR. (C) The H3K4me3 levels in the PD-L1 promoter region in MMAC-SF and WM-115 cell lines were analyzed by ChIP. (D and E) MMAC-SF cells and MMAC-SF Si-DPY30 cells were treated with IFN-γ at the prescribed dose for 24 h or 48h, and analyzed for PD-L1 mRNA level by RT-PCR. (F and G) MMAC-SF cells and MMAC-SF Si-DPY30 cells were treated with IFN-γ at the prescribed dose for 24 h or 48h, stained with PD-L1 specific MAb, and analyzed by flow cytometry. **P< 0.01.
DPY30 Regulates the Expression of PD-L1
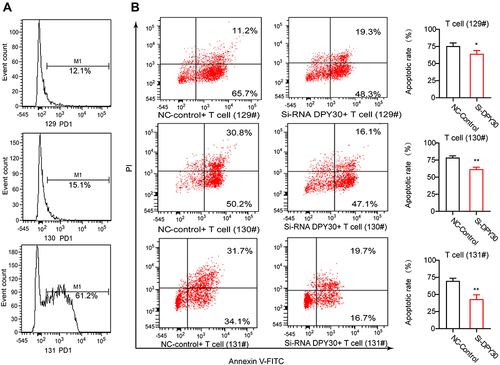
IFN-γ induces PD-L1 expression and disruption of this pathway might sensitize melanoma patients to immune-based therapies. To further study the regulation between DPY30 and PD-L1, we detected the PD-L1 level by IFN-γ treatment and down-regulation of DPY30 in MMAC-SF cells. The results of RT-PCR showed that the level of PD-L1 in MMAC-SF si-DPY30 cells remained unchanged in the 1ng IFN-γ treatment group and slightly increased in the 5ng IFN-γ treatment group compared with the non-IFN -γ treatment group at 24 hours of IFN-γ treatment. The overall PD-L1 level at 48 h after IFN -γ treatment was higher than at 24h, but the trend kept the same as that at 24h (). Moreover, the results of flow cytometry indicated that knockdown of DPY30 gene could significantly suppress PD-L1 levels after IFN-γ (1 or 5 ng) treated MMAC-SF cells for 24 hours. After 48 hours of IFN-γ treatment, Knockdown of DPY30 still suppressed PD-L1 levels (). To further validate the effect of down-regulation of DPY30 in MMAC-SF cells, and we detected the apoptosis of PD1+ T cells by flow cytometry in a co-culture model. We detected PD-1 level in human T cells from 3 normal samples by flow cytometry (), and the apoptosis of PD1+ T-cells in co-culture with MMAC-SF cells by knockdown of DPY30 were markedly reduced ().
Figure 7 Knockdown of DPY30 in MMAC-SF cells inhibited T cell apoptosis. (A) The PD1+ level in human T cells from 3 normal samples were analyzed by flow cytometry. (B) Flow cytometry was used to analyze the apoptosis of T cells in the co-culture model with MMAC-SF cells and PD1+ T cells. *P< 0.05, **P< 0.01. #Indicates the sample number.
Discussion
As the major H3K4 methylation enzymes in mammals, the SET1/MLL complexes are considered potential pharmacologic targets in epigenetic therapeutics. DPY30, a core subunit of SET1/MLL complexes, is important for facilitating genome-wide H3K4 methylation.Citation18 Accumulating evidence have revealed significant upregulation of DPY30 mRNA expression were observed in hepatocellular carcinoma,Citation28 cervical squamous cell carcinoma,Citation29 ovarian cancer,Citation19 and cholangiocarcinoma.Citation23 Our study showed that the mRNA level of DPY30 in melanoma was higher than in normal tissues. In addition, DPY30 could induce epithelial to mesenchymal transition by activating Wnt/β-catenin signaling in the progression of cervical squamous cell carcinoma.Citation29 Similarly, our research also proved that the mRNA level of DPY30 was higher in metastatic melanoma compared with primary melanoma. These findings suggested that DPY30 over-expressed in melanoma, which might play essential roles in the progression of melanoma.
DNA methylation is an important biological process involving in cancer development,Citation30,Citation31 and changes in DNA methylation in cancer have been regarded as diagnostic or prognostic biomarkers.Citation32 A previous study indicated that 36 DNA methylation markers from 304 patients with hepatocellular carcinoma could predict poor survival in patients with hepatocellular carcinoma.Citation33 Furthermore, comprehensive genome-scale analysis of aberrant DNA methylation enabled classification of colorectal carcinoma patients based on their prognosis.Citation34 In this study, we analyzed the methylation status of DPY30 gene through the UALCAN database. The analysis presented that the degree of DPY30 gene methylation was negatively correlated with tumor stage, and the expression of DPY30 mRNA was significantly negatively correlated with its methylation status. Furthermore, among 22 predicted CpG sites of DPY30, 8 CpG sites exhibited negative correlation with DPY30 expression. These findings suggested that the methylation status of DPY30 might be a novel direction for prognosis evaluation of melanoma.
Immune checkpoint play essential roles in regulating the immunosuppressive TME.Citation35 Immune checkpoint inhibitors, including anti-PD-1 and anti-CTLA4 antibodies, have revolutionized the management of melanoma and significantly improved the clinical outcomes for patients with melanoma.Citation36 However, immune checkpoint inhibitors are only effective in a part of patients with melanoma, and two-thirds of patients are resistant and will require further treatment.Citation37 Our study indicated that DPY30 expression had significant positive associations with ICP genes involved in T-cell exhaustion, including CD274, CTLA4, CD160, HAVCR2, IDO2, and PDCD1LG2. Further studies indicated that DPY30 was positively correlated with immunosuppressive cells in SKCM including MDSCs, TAMs, and Tregs, suggesting that DPY30 was closely associated with the immunosuppressive TME in SKCM. IHC results showed that the positive rates of DPY30 and PD-L1 in melanoma tissues were 62% and 58%, respectively. Correlation analysis revealed that DPY30 was positively associated with PD-L1 expression. Furthermore, we found that silencing of DPY30 by specific siRNA significantly inhibited PD-L1 expression. Therefore, we believe that DPY30 might be involved in cancer immune evasion in melanoma.
Histone H3K4 methylation is one of the most significant epigenetic modifications, and plays critical roles in regulating target gene transcription.Citation38,Citation39 The most notable H3K4 methylation enzymes in mammals are the SET1/MLL complexes, which have a profound effect on numerous biological processes including cancer progression.Citation40–42 SET1/MLL family complexes comprise Set1a, Set1b, MLL1, MLL2, MLL3, and MLL4 as the catalytic subunit, and Wdr5, Rbbp5, Ash2l, and DPY30 as the common core subunits.Citation17,Citation39,Citation43 The roles of the MLL/SET1 complexes in transcriptional regulation have been extensively studied. Among these subunits, MLL1 has been shown to bind to the CD274 promoter to catalyze H3K4me3 to activate PD-L1 transcription in tumor cells. Inhibition of MLL1 activity or silencing of MLL1 expression reduced H3K4me3 level in the CD274 promoter region and downregulated PD-L1 expression in tumor cells.Citation44 Previous study showed that MLL3 was located in the enhancer of PD-L1 and regulated the H3K4me1 level in the enhancer regions. Depletion of MLL3 reduced the binding of H3K4me1 in the PD-L1 enhancer regions and Pol II Ser-5p in PD-L1 promoter regions.Citation45 Furthermore, it has been reported that WDR5 was positively correlated with PD-L1 expression, and WDR5 inhibitor inhibited PD-L1-based immune evasion by blocking the WDR5-MLL complex mediating H3K4me3 of target genes.Citation46 DPY30 is a core subunit of SET1/MLL complexes, and it has been reported that DPY30 promoted the expression of endogenous MYC by regulating chromatin accessibility.Citation47 Previous study also showed that DPY30 regulated angiogenesis and tumor growth in an H3K4me3-dependent manner, and PDE4B was a key downstream effector of DPY30-H3K4me3 axis. Targeting DPY30-PDE4B axis was an effective approach for treatment of glioblastoma.Citation48 DPY30 promoted the expression of Hif-1α and its targeted glycolytic genes by regulating H3K4me3 modification.Citation28 In this study, we found that H3K4me3 levels were enriched in the proximal PD-L1 promoter region in melanoma cells. Inhibition of DPY30 suppressed the PD-L1 level in IFN-γ treated melanoma cells. Our study demonstrated that DPY30 might regulate PD-L1 expression through H3K4me3, which might be a promising therapeutic target in melanoma.
Conclusion
In summary, our study firstly demonstrated that the elevated DPY30 levels in melanoma tissues, and the expression of DPY30 was closely related to the level of PD-L1. DPY30 was associated with cancer immune evasion by regulating PD-L1 expression through epigenetic mechanism in melanoma.
Abbreviations
APCs, antigen-presenting cells; ChIP, Chromatin Immunoprecipitation; CTLA-4, cytotoxic T lymphocyte antigen 4; H3K4me3, histone H3K4 trimethylation; IHC, Immunohistochemical; ICP, immune checkpoint; MDSCs, myeloid-derived suppressor cells; NK, natural killer; PD-1, programmed cell death protein 1; PD-L1, programmed cell death ligand 1; TAMs, tumour-associated macrophages; TMB, tumor mutational burden; Tregs, regulatory T cells.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics Approval and Informed Consent
The study’s protocol was approved by the ethics committee of Cancer Hospital, Chinese Academy of Medical Sciences, Peking Union Medical College, and informed consent was obtained from the study participants. Our experiments followed the guidelines outlined in the Declaration of Helsinki.
Disclosure
The authors declare no conflicts of interest for this work.
Acknowledgments
We acknowledge all the public databases we used in the study for providing their platforms and contributors for uploading their meaningful datasets.
Additional information
Funding
References
- Larkin J, Chiarion-Sileni V, Gonzalez R., et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(1):23–34. doi:10.1056/NEJMoa1504030
- Robert C, Schachter J, Long GV, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372(26):2521–2532. doi:10.1056/NEJMoa1503093
- Ugurel S, Rohmel J, Ascierto PA, et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies-update 2017. Eur J Cancer. 2017;83:247–257. doi:10.1016/j.ejca.2017.06.028
- Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet. 2018;392(10151):971–984. doi:10.1016/s0140-6736(18)31559-9
- Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi:10.1158/1535-7163.MCT-14-0983
- Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–416. doi:10.1093/annonc/mdv615
- Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. doi:10.1016/s1359-6101(01)00038-7
- Benci JL, Xu B, Qiu Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–54 e12. doi:10.1016/j.cell.2016.11.022
- Mimura K, Teh JL, Okayama H, et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109(1):43–53. doi:10.1111/cas.13424
- Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76(3):359–370. doi:10.1016/j.molcel.2019.09.030
- Wu M, Huang Q, Xie Y, et al. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J Hematol Oncol. 2022;15(1):24. doi:10.1186/s13045-022-01242-2
- Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–231. doi:10.1126/science.aac9935
- Wu R, Wang C, Li Z, et al. SOX2 promotes resistance of melanoma with PD-L1 high expression to T-cell-mediated cytotoxicity that can be reversed by SAHA. J Immunother Cancer. 2020;8(2):e001037. doi:10.1136/jitc-2020-001037
- Parsa AT, Waldron JS, Panner A, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–88. doi:10.1038/nm1517
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–849. doi:10.1038/nrm1761
- Soares LM, He PC, Chun Y, Suh H, Kim T, Buratowski S. Determinants of histone H3K4 methylation patterns. Mol Cell. 2017;68(4):773–85 e6. doi:10.1016/j.molcel.2017.10.013
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi:10.1146/annurev-biochem-051710-134100
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144(4):513–525. doi:10.1016/j.cell.2011.01.020
- Zhang L, Zhang S, Li A, Zhang A, Zhang S. DPY30 is required for the enhanced proliferation, motility and epithelial-mesenchymal transition of epithelial ovarian cancer cells. Int J Mol Med. 2018;42(6):3065–3072. doi:10.3892/ijmm.2018.3869
- Yang Z, Augustin J, Chang C, et al. The DPY30 subunit in SET1/MLL complexes regulates the proliferation and differentiation of hematopoietic progenitor cells. Blood. 2014;124(13):2025–2033. doi:10.1182/blood-2014-01-549220
- Simboeck E, Gutierrez A, Cozzuto L, et al. DPY30 regulates pathways in cellular senescence through ID protein expression. EMBO J. 2013;32(16):2217–2230. doi:10.1038/emboj.2013.159
- Sun L, Guo C, Cao J, et al. Over-expression of alpha-enolase as a prognostic biomarker in patients with pancreatic cancer. Int J Med Sci. 2017;14(7):655–661. doi:10.7150/ijms.18736
- Hong ZF, Zhang WQ, Wang SJ, et al. Upregulation of DPY30 promotes cell proliferation and predicts a poor prognosis in cholangiocarcinoma. Biomed Pharmacother. 2020;123:109766. doi:10.1016/j.biopha.2019.109766
- Li T, Zhang C, Zhao G, et al. IGFBP2 regulates PD-L1 expression by activating the EGFR-STAT3 signaling pathway in malignant melanoma. Cancer Lett. 2020;477:19–30. doi:10.1016/j.canlet.2020.02.036
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi:10.1016/j.ccell.2015.03.001
- Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–2501. doi:10.1056/NEJMc1713444
- Peng M, Mo Y, Wang Y, et al. Neoantigen vaccine: an emerging tumor immunotherapy. Mol Cancer. 2019;18(1):128. doi:10.1186/s12943-019-1055-6
- Liu B. DPY30 functions in glucose homeostasis via integrating activated histone epigenetic modifications. Biochem Biophys Res Commun. 2018;507(1–4):286–290. doi:10.1016/j.bbrc.2018.11.023
- He FX, Zhang LL, Jin PF, Liu DD, Li AH. DPY30 regulates cervical squamous cell carcinoma by mediating epithelial-mesenchymal transition (EMT). Onco Targets Ther. 2019;12:7139–7147. doi:10.2147/OTT.S209315
- Flausino CS, Daniel FI, Modolo F. DNA methylation in oral squamous cell carcinoma: from its role in carcinogenesis to potential inhibitor drugs. Crit Rev Oncol Hematol. 2021;164:103399. doi:10.1016/j.critrevonc.2021.103399
- Zhang J, Yin J, Luo L, et al. Integrative analysis of DNA methylation and transcriptome identifies a predictive epigenetic signature associated with immune infiltration in gliomas. Front Cell Dev Biol. 2021;9:670854. doi:10.3389/fcell.2021.670854
- Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15(7):459–466. doi:10.1038/s41571-018-0004-4
- Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–1956. doi:10.1002/hep.27732
- Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22(2):271–282. doi:10.1101/gr.117523.110
- Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):1482–1492. doi:10.1093/annonc/mdw168
- Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet. 2021;398(10304):1002–1014. doi:10.1016/s0140-6736(21)01206-x
- Pires da Silva I, Ahmed T, Reijers ILM, et al. Ipilimumab alone or ipilimumab plus anti-PD-1 therapy in patients with metastatic melanoma resistant to anti-PD-(L)1 monotherapy: a multicentre, retrospective, cohort study. Lancet Oncol. 2021;22(6):836–847. doi:10.1016/s1470-2045(21)00097-8
- Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20(3):341–348. doi:10.1016/j.ceb.2008.03.019
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi:10.1016/j.molcel.2006.12.014
- Vedadi M, Blazer L, Eram MS, Barsyte-Lovejoy D, Arrowsmith CH, Hajian T. Targeting human SET1/MLL family of proteins. Protein Sci. 2017;26(4):662–676. doi:10.1002/pro.3129
- Lu K, Tao H, Si X, Chen Q. The histone H3 lysine 4 presenter WDR5 as an oncogenic protein and novel epigenetic target in cancer. Front Oncol. 2018;8:502. doi:10.3389/fonc.2018.00502
- Jiang H. The complex activities of the SET1/MLL complex core subunits in development and disease. Biochim Biophys Acta Gene Regul Mech. 2020;1863(7):194560. doi:10.1016/j.bbagrm.2020.194560
- Dou Y, Milne TA, Ruthenburg AJ, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi:10.1038/nsmb1128
- Lu C, Paschall AV, Shi H, et al. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst. 2017;109(6):djw283. doi:10.1093/jnci/djw283
- Xiong W, Deng H, Huang C, et al. MLL3 enhances the transcription of PD-L1 and regulates anti-tumor immunity. Biochim Biophys Acta Mol Basis Dis. 2019;1865(2):454–463. doi:10.1016/j.bbadis.2018.10.027
- Zhang J, Zhou Q, Xie K, et al. Targeting WD repeat domain 5 enhances chemosensitivity and inhibits proliferation and programmed death-ligand 1 expression in bladder cancer. J Exp Clin Cancer Res. 2021;40(1):203. doi:10.1186/s13046-021-01989-5
- Yang Z, Shah K, Busby T, et al. Hijacking a key chromatin modulator creates epigenetic vulnerability for MYC-driven cancer. J Clin Invest. 2018;128(8):3605–3618. doi:10.1172/JCI97072
- Dixit D, Prager BC, Gimple RC, et al. Glioblastoma stem cells reprogram chromatin in vivo to generate selective therapeutic dependencies on DPY30 and phosphodiesterases. Sci Transl Med. 2022;14(626):eabf3917. doi:10.1126/scitranslmed.abf3917