Abstract
Background
Plasmacytoid dendritic cells (pDCs) are the main producers of type I interferon (IFN-I), and the excessive production of IFN-I is a hallmark of systemic lupus erythematosus (SLE). Both SLC15A4 and miR-31-5p are SLE susceptibility-related genes, and SLC15A4 has been implicated an important role in endolysosomal toll-like receptor (TLR) activation in pDCs. However, whether miR-31-5p exerts a regulating effect on SLC15A4 expression in pDCs is unclear.
Methods
The expression of SLC15A4 and miR-31-5p in peripheral blood mononuclear cells (PBMCs) of SLE patients was measured by RT-qPCR analyses. The quantitative analysis of IFN-α secretion in the patients’ serum was performed by ELISA assay. Luciferase-reporter assay was applied to confirm the interaction between miR-31-5p and SLC15A4. The expression of miR-31-5p, SLC15A4 and IFN-stimulated genes (ISGs, such as MX1, OAS1 and IFIT3) was detected by Western blot and RT-qPCR assays and further IRF5 phosphorylation was evaluated by immunofluorescence after transfected with miR-31-5p mimics or inhibitor in THP-1 and CAL-1 cells.
Results
The expression of miR-31-5p was downregulated and negatively correlated with the overexpression of SLC15A4 in PBMCs of SLE patients. In addition to this, the secretion of IFN-α was overexpressed in sera of SLE and positively correlated with SLC15A4 level. We found that miR-31-5p directly targeted SLC15A4 and negatively regulated the expression of SLC15A4 in THP-1 and CAL-1 cells. In vitro inhibition of miR-31-5p increased the phosphorylation of IRF5 and the induction of ISGs stimulated by R848, overexpression of miR-31-5p get the reverse results.
Conclusion
miR-31-5p might involve in SLE pathogenesis through regulating IFN-I expression by negatively regulating SLC15A4 to increase the levels of IFN-α and ISGs in pDCs.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with a broad spectrum of clinical presentations and primarily affects women of childbearing age.Citation1 As we know, type I IFN (IFN-I) is mostly produced by pDCs and plays a significant role in the pathogenesis of SLE. Solute carrier family 15 member 4 (SLC15A4) regulates lysosome antigen processing and endosomal toll-like receptors (TLRs) TLR7/9-mediated inflammatory responses.Citation2 In pDCs, SLC15A4 is essential for TLR to trigger the production of IFN-I, especially IFNα/β, which is involved in the pathogenesis of SLE.Citation3 Genetic, clinical and experimental data have strongly suggested that IFN-I plays an important role in SLE. Studies have also shown that SLC15A4 mutant C57BL/6-Fas(lpr) mice, in which pDCs were present but showed incapacitation to produce IFN-I in response to endosomal TLR ligands, which led to an absence of autoantibodies, reduced lymphadenopathy and splenomegaly, and extended survival.Citation4–6
Previous studies have demonstrated that both the proliferation and acquisition of activation markers were normal in SLC15A4-deficient B cells.Citation3 However, SLC15A4 or TASL (TLR adaptor interacting with SLC15A4 on the lysosome) deficiency caused defects in the activation of IRF5 but not in the MAPK or NF-kB pathway in pDC cell line CAL-1.Citation7 Recently, Kobayashi et al have reported that knocking down SLC15A4 expression impaired TLR7/8 or TLR9 triggered IFN-I production and decreased mTORC1 activity in CAL-1 cells.Citation8 These findings revealed that SLC15A4 plays a key role in pDCs and can be used as a potential therapeutic target.
MicroRNAs (miRNAs) critically contribute to the progress of autoimmune diseases,Citation9 and the dysregulation of miRNA activity has been associated with the pathogenesis of SLE.Citation10 miRNAs composed of 22 nucleotides are regulatory RNAs and act on target mRNAs in a sequence-specific manner to either promote their mRNA cleavage and degradation or reduce their translational efficiency. Dysregulated expression of miRNAs has been reported in SLE patients, including miR-16-5p, miR-374a-5p, miR-34a-5p, miR-31-5p, and miR-1-3p, which have been the miRNAs most frequently associated with susceptibility-related genes.Citation11 Certain down-regulated miRNAs showed negative correlation with the SLE disease activity index (SLEDAI) and IFN score.Citation12–14 Among them, miR-31 was dysregulated in both PBMCs and splenocytes of MRL/lpr mice compared in MRL control mice.Citation15 However, whether there is a direct link between the dysregulated miR-31-5p and increased IFN-I secretion in the pDCs of SLE patients is unclear.
The present study showed that miR-31-5p was decreased in the PBMCs of SLE patients and it was a negative regulator of the TLR signaling pathway. Our data indicated that downregulation of miR-31-5p contributed to the overactivation of the IRF5 signaling pathway and increased the expression of ISGs by targeting SLC15A4 in CAL-1 cells, while overexpression of miR-31-5p downregulating IFN-I production by reducing SLC15A4 level.
Materials and Methods
Human Samples
In accordance with the Declaration of Helsinki, all patients and control participants provided written informed consent. All experimental protocols were approved by the Ethics Committee of Southwest Hospital of Third Military Medical University, Chongqing, China (approval no. KY2020317). Peripheral blood samples were obtained from patients diagnosed with SLE (n = 21), and control samples were obtained from healthy volunteers (n = 21), who were recruited from the Department of Dermatology, Southwest Hospital. Disease activity was assessed using SLEDAI (9.8 ± 4.3).Citation16 The PBMCs were separated from peripheral blood by Ficoll-Paque density gradient separation, and the PBMCs and serum were cryopreserved for later use.
Cell Culture
The THP-1 cells were obtained from ATCC and propagated according to ATCC instructions. The cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). The CAL-1 cells were from NIH (provided by Dr. Liu Yuanyuan, School of Public Health, Tianjin Medical University, Tianjin, China), which were cultured in RPMI-1640 medium supplemented with 10% FBS, 1% P/S, 2 mM glutamine, 25 mM HEPES and 50 mM β-mercaptoethanol. All cells were maintained at 37°C in 5% CO2.
Cytokine Quantification
Human IFN-α was detected in the serum of SLE patients (n = 21) and healthy controls (n = 21) by using an ELISA kit for detecting human IFN-α (Elabscience, Wuhan, China). All samples were analyzed in triplicate in independent experiments. The correlation between IFN-α and SLC15A4 was calculated.
Real Time-Quantitative PCR (RT-qPCR)
Total RNA was extracted using RNAiso Plus reagent (Takara, Shiga, Japan). The mRNA first-strand cDNA was obtained with a PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara). miRNA first-strand cDNA was synthesized with a miR-X miRNA First-Strand Synthesis Kit (Takara). RT-qPCR analysis was performed using TB Green Premix Ex Taq II (Takara) in a real-time PCR machine (Bio-Rad, California, USA). To normalize the expression data, primers for the housekeeping genes GAPDH and U6 (Sangon Biotech, Shanghai, China) were used. miR-31-5p expression was normalized to that of the small nuclear RNA U6. The relative expression of genes and miRNAs was analyzed by relative quantification (2−ΔΔCT). The primer sequences and miR-31-5p sequence are summarized in .
Table 1 Sequence of PCR Primers and miR-31-5p
Cell Transfection
THP-1 and CAL-1 cells were seeded in 24-well plates or 96-well plates at densities of 2.5 × 105 cells or 5 × 104 cells, respectively, and then transfected with negative controls (mimics control, inhibitor control), miR-31-5p mimics or miR-31-5p inhibitor (40µM final concentration, GenePharma, Shanghai, China) by using Lipofectamine 3000 reagent (Invitrogen, CA, USA). The transfected cells were cultured for 24h before analysis and then stimulated with R848 (5µg/mL, Invivogen, France) for 2h, 4h or 24h.
Dual-Luciferase Reporter Assay
HEK-293 cells were transfected with a dual-luciferase plasmid containing the wild-type (SLC15A4-WT) or mutated full-length 3′ UTR (SLC15A4-MT) from human SLC15A4 (RefSeq NM_145648.4) and miR-31-5p mimics (10pmol/well, GenePharma) at 10,000 cells per well using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) for 24h. Dual-luciferase reporter expression was evaluated with a Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) following the manufacturer’s protocol. The ratio of firefly luciferase to Renilla luciferase was established and compared to the transfection control. The mutations were introduced in the predicted miR-31-5p-binding sites in the SLC15A4 3′ UTR (position 541–548) using site-directed mutagenesis and RT-qPCR. The SLC15A4-WT sequence was cttcctgaga acaatcttgc TCTTGCCatg ttctttgatt taggctggta, and SLC15A4-MT sequence was cttcctgaga acaatcttgc AGAACGGatg ttctttgatt taggctggta.
Western Blot Assay
Total protein was extracted by cell lysis with RIPA buffer and quantified with BCA assay. Next, equal amounts of protein (20µg) were separated via SDS-PAGE, and a PVDF membrane was used for protein transfer. The membrane was incubated with primary antibodies (4°C, overnight) against SLC15A4 (1:1000, ab64429, Abcam, Cambridge, MA), TASL (1:1000, ab69152, Abcam), IRF5 (1:1000, ab181553, Abcam), phospho-IRF5 (1:2000, AF8382, Affinity Biosciences, USA) and GAPDH (1:5000, bs1009R, Bioss, Bejing, China). Then, the membrane was washed with TBST three times (10 min each) and subsequently incubated for 1h with an anti-rabbit IgG HRP-conjugated secondary antibody. The greyscale value of the protein bands was determined with ImageJ software.
Immunofluorescence
CAL-1 cells were transfected with negative controls, miR-31-5p mimics or miR-31-5p inhibitor for 24h and stimulated with R848 for 4h. Then, the cells were washed with PBS and fixed with cold 100% methanol for 10 min, blocked with a BSA/PBS solution, washed with PBS, stained with an anti-phospho-IRF5 antibody (1:200, AF8382, Affinity Biosciences) overnight at 4°C, and incubated with an Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (1:400, ab150077, Abcam, Shanghai, China). The samples were mounted with DAPI containing diamond Prolong mounting solution (AR1176, Boster, Wuhan, China) and were analyzed on a Leica confocal microscope.
Statistical Analyses
Data were analyzed and plotted with GraphPad Prism (version 8, GraphPad Software, La Jolla, CA). For normally distributed data, Student’s t-test for unpaired or paired values was performed. Spearman correlation test was performed only for correlation studies. P values less than 0.05 were considered statistically significant.
Results
Decreased miR-31-5p Expression and Enhanced SLC15A4 Expression in SLE Patients
The results of RT-qPCR assays showed that the mRNA expression of SLC15A4 was greatly increased in PBMCs of SLE patients compared to that of the healthy controls (). However, miR-31-5p mRNA expression in PBMCs showed a significantly lower level in SLE patients than in healthy controls (), and it was significantly and negatively correlated with the SLC15A4 level ().
Figure 1 SLC15A4 expression was negatively correlated with the decreased expression of miR-31-5p, and positively correlated with the increased IFN-α in SLE. (A and B) The mRNA expression of SLC15A4 and miR-31-5p in PBMCs from SLE patients and healthy controls was detected by RT-qPCR normalized to that of GAPDH and U6. The data are reported as the mean ± SD (n=21 for each) of individual experiments. Student’s t-test, ***p < 0.001 and ****p< 0.001. (C) Correlation between the levels of SLC15A4 and miR-31-5p in SLE patient PBMCs. Each dot represents an individual SLE patient. P values were determined by Spearman correlation test. (D) The secretion of IFN-α in SLE serum, as measured by ELISA. Student’s t-test, ****p < 0.0001. (E) Correlation between the levels of SLC15A4 in PBMCs and IFN-α in the sera of SLE patients. Each dot represents an individual SLE patient. P values were calculated by Spearman correlation test.
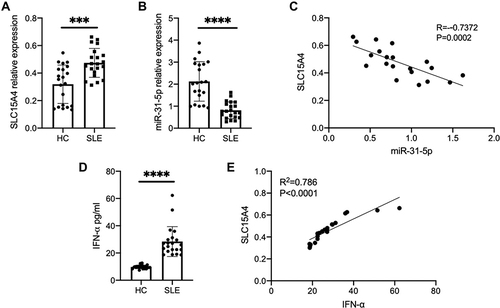
SLC15A4 is critical for TLR7/8/9-mediated inflammatory signaling. Our data showed that SLC15A4 was markedly upregulated () and the IFN-α protein was significantly increased in SLE patients (), and these changes were significantly positively correlated (), suggesting that SLC15A4 might be a potent regulator of the IFN-I pathway.
miR-31-5p is a Negative Regulator of SLC15A4 Gene Expression
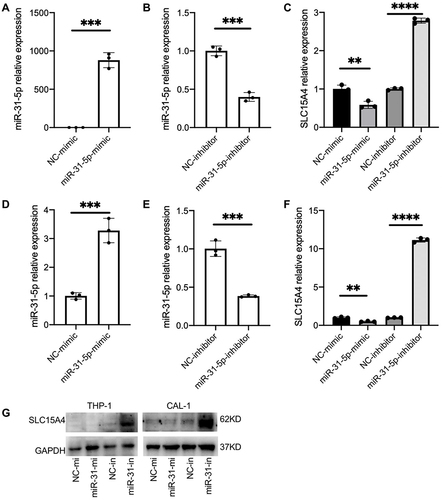
To determine whether decreased expression of miR-31-5p is correlated with an increased expression of SLC15A4 in SLE, we verified the regulatory function of miR-31-5p and studied the underlying molecular mechanisms using THP-1 and CAL-1 cells. The results demonstrated that overexpression of miR-31-5p by transfection with miR-31-5p mimics inhibited SLC15A4 expression compared with negative control. In contrast, inhibition of miR-31-5p expression by transfection with miR-31-5p inhibitor significantly elevated SLC15A4 expression in THP-1 () and CAL-1 cells (). Western blot assays showed that overexpression of miR-31-5p significantly reduced the protein levels of SLC15A4 compared to those of the negative controls, while the miR-31-5p inhibitor significantly increased SLC15A4 production in THP-1 and CAL-1 cells ().
Figure 2 miR-31-5p is a negative regulator of SLC15A4. (A–F) Quantitative analysis was determined by RT-qPCR assay of the mRNA levels of miR-31-5p and SLC15A4 in THP-1 (A–C) and CAL-1 cells (D–F) transfected with negative controls (NC-mimic and NC-inhibitor), miR-31-5p mimics (miR-31-5p mimic) or miR-31-5p inhibitor as indicated and then stimulated with R848 (5µg/mL) for 24h. Student’s t-test, **p < 0.01, ***p < 0.001 and ****p < 0.0001. (G) THP-1 and CAL-1 cells were transfected with negative controls (NC-mi and NC-in), miR-31-5p mimics (miR-31-mi) or miR-31-5p inhibitor (miR-31-in) and then stimulated with R848 (5µg/mL) for 24h. Whole-cell lysates were prepared, and Western blot was performed for SLC15A4 and GAPDH.
miR-31-5p Directly Targets the SLC15A4 Gene to Regulate Its Function
miR-31-5p negatively regulated the expression of SLC15A4, suggesting that miR-31-5p might target the SLC15A4 gene directly. The TargetScan online database predicted that miR-31-5p have complementary SLC15A4 mRNA-binding sites (). The luciferase assay revealed that miR-31-5p mimics inhibited the expression of luciferase reporters fused with the wild-type SLC15A4 3′UTR, while mutation of the target sequence in the SLC15A4 3′UTR abolished this inhibition (), indicating that miR-31-5p directly targeted SLC15A4.
Figure 3 miR-31-5p directly targets SLC15A4 and negatively regulates the IRF5 pathway. (A) Sequence alignment of miR-31-5p and its putative binding sites in the 3′UTR of SLC15A4. Mutations of the miR-31-5p target sites are indicated on top. (B) Luciferase activity in HEK-293 cells transfected with SLC15A4-WT or SLC15A4-MT, cotransfected with mimics control (NC-mimic) or miR-31-5p-mimics (miR-31-5p-mimic). Error bars represent the SEM. Student’s t-test, ns p > 0.05 and ***p < 0.001. (C) THP-1 and CAL-1 cells were transfected with negative controls (NC-mi and NC-in), miR-31-5p mimics (miR-31-mi) or miR-31-5p inhibitor (miR-31-in), respectively, and then stimulated with R848 (5µg/mL) for 24h. Whole-cell lysates were prepared, and Western blot was performed for TASL, IRF5 and GAPDH. (D) CAL-1 cells were transfected with negative controls (NC-mimics and NC-inhibitor, NM and NI), miR-31-5p mimics (31-mi) or miR-31-5p inhibitor (31-in) and stimulated with R848 (5µg/mL) for 2h and 4h respectively. Whole-cell lysates were prepared and Western blot was performed for p-IRF5 and GAPDH.
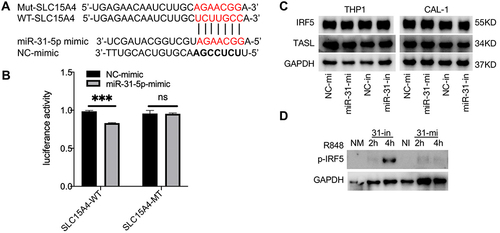
To verify the regulatory effect of miR-31-5p on SLC15A4 function, the expression levels of TASL and IRF5 were detected in THP-1 and CAL-1 cells. As expected, we found that inhibition or overexpression of miR-31-5p did not alter the expressions of TASL or total IRF5 (), which was consistent with the published literature.Citation7 While CAL-1 cells transfected with negative controls showed minimal p-IRF5 levels, cells transfected with miR-31-5p inhibitor stimulated with R848 for 4h significantly enhanced p-IRF5 levels ().
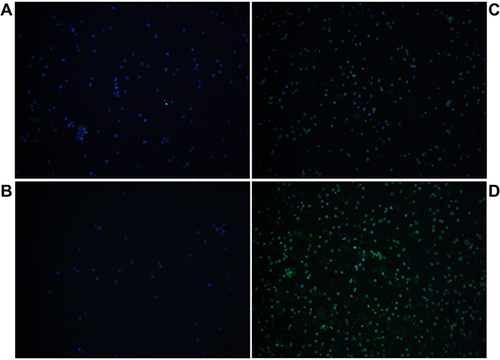
Under the laser confocal microscopy, the miR-31-5p inhibitor significantly increased the p-IRF5 level, while miR-31-5p mimics did not markedly alter the p-IRF5 level compared with that in the controls (), consistent with the data shown in .
Figure 4 miR-31-5p negatively regulates IRF5 phosphorylation. CAL-1 cells were transfected with mimics control (A), miR-31-5p mimics (C), inhibitor control (B) or miR-31-5p inhibitor (D), and then stimulated with R848 (5µg/mL) for 4h. Then, the cells were stained with anti-phospho-IRF5 antibody and incubated with Alexa Fluor 488 goat anti-rabbit IgG (green). DAPI was used to stain the nuclei (blue). The images were captured with a laser confocal microscope.
miR-31-5p is a Negative Regulator and SLC15A4 is a Positive Regulator of the IFN-I Pathway
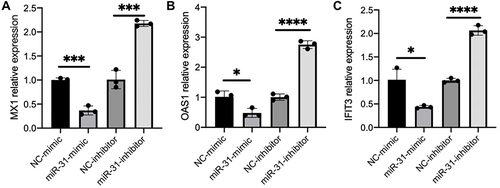
To validate miR-31-5p functions as a cell intrinsic negative regulator of proinflammatory cytokines in pDC cells, we performed RT-qPCR assay to measure the expression level of ISGs in CAL-1 cells after transfection with negative controls, miR-31-5p mimics or miR-31-5p inhibitor. The results demonstrated that miR-31-5p mimics decreased the induction of ISGs, including MX1, OAS1 and IFIT3, while miR-31-5p inhibitor exerted reverse effects ().
Figure 5 miR-31-5p is a negative regulator and SLC15A4 is a positive regulator of the IFN-I pathway. (A–C) Normalized mRNA expression of MX1, OAS1, and IFIT3 relative to that of GAPDH in CAL-1 cells transfected with negative controls (NC-mimics and NC-inhibitor), miR-31-5p-mimics (miR-31-mimic) or miR-31-5p-inhibitor (miR-31-inhibitor), and stimulated with R848 (5µg/mL) for 24h. Student’s t-test, *p < 0.05, ***p < 0.001 and ****p < 0.0001.
Discussion
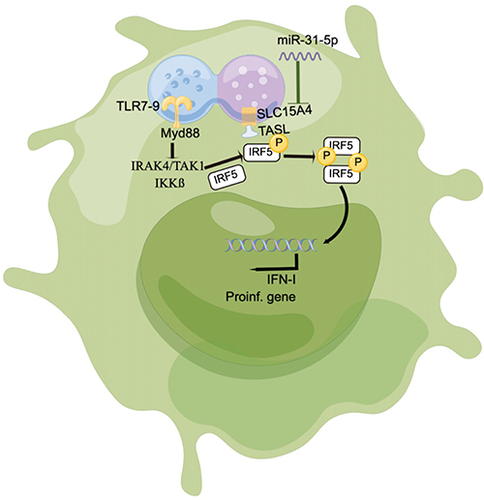
SLC15A4 is involved in disease progression mainly through the production of IFN-I and proinflammatory cytokines in pDCs, which play critical roles in the pathogenesis of SLE and murine models.Citation2,Citation5 miRNAs have been shown to correlate with inflammatory cytokines in immune cells that play an important role in SLE pathogenesis.Citation15 In our study, we confirmed that miR-31-5p was negatively correlated with SLC15A4 expression in the PBMCs of SLE patients, and that miR-31-5p also negatively regulated SLC15A4 expression and IRF5 activation, subsequently IFN-I production was modulated in pDC cells ().
Figure 6 Schematic showing that miR-31-5p targets SLC15A4 in pDCs of SLE, negatively regulating IFN-I production.
SLC15A4 is crucial for pDC to produce proinflammatory cytokines and activate the endosomal TLR signaling pathway.Citation17 Knocking out SLC15A4 has been previously shown to abrogate TLR7 signaling and amelioration of murine lupusCitation16 and deletion of SLC15A4 or TASL, which both contain risk alleles for SLE, specifically impaired the activation of the IRF pathway in previous studies.Citation7,Citation16 The important functions of SLC15A4 have been identified in TLR7 and TLR9-mediated signaling pathways in pDC and B cells.Citation3 In mouse SLC15A4−/− mast cells limited mast cell functions and inflammatory responses were induced by controlling the mTORC1-TFEB signaling axis.Citation18 In C57BL/6, NZB/W F1 mice and BXSB male mice with TLR7 gene duplication, a function-impairing mutation or deletion of the SLC15A4 gene diminished IFN-I production by TLR-activated pDCs.Citation5,Citation19 Thus, SLC15A4 may be analyzed to identify important targets for therapeutic intervention in human lupus.Citation4 However, the molecular mechanisms underlying the repression of SLC15A4 expression are still largely unknown. In this study, we demonstrated that miR-31-5p directly targeted the SLC15A4 3′UTR and that miR-31-5p negatively regulated SLC15A4 expression.
pDCs specialize in secretion of IFN-I in response to pathogens, and evidence suggests that pDCs are intimately involved in the pathogenesis of lupus.Citation4 Previous study indicated that dysregulation of miR-31 in immune cells resulted in impaired activation, survival, Th17 cell differentiation, inflammatory factors and glycolytic metabolism.Citation20–22 The TGF-β/miR-31/CEACAM1-S axis has been shown to inhibit CD4 + CD25 + Treg differentiation in SLE.Citation23 In this study, we were the first to reveal the effect of miR-31-5p on pDCs in SLE. Overexpression of miR-31-5p significantly decreased the expression of ISGs in CAL-1 cells but was significantly increased by transfection with miR-31-5p inhibitor. Moreover, the expression of miR-31-5p regulated the activation of the IRF5 signaling pathway in CAL-1 cells.
In conclusion, our data revealed that the expression of the miR-31-5p/SLC15A4 showed a strong correlation with inflammatory cytokines and a potential interplay between miR-31-5p and SLC15A4, which contributes to SLE pathogenesis by enhancing SLC15A4 expression and activating the TLR7 signaling pathway in pDCs. Therefore, this study suggests the potential diagnostic or therapeutic value of miR-31-5p/SLC15A4 for SLE.
Data Sharing Statement
The raw data supporting the conclusions of this article will be made available by the corresponding author, without undue reservation.
Ethics Approval and Consent to Participate
This study was approved by the Ethical Committee of Southwest Hospital of Third Military Medical University and conducted according to Declaration of Helsinki principles. Written informed consent was obtained from all participants. The reference number provided by the ethics committee was KY2020317.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study received the support of the National Natural Science Foundation of China (No.81773316) and Senior Medical Talents Program of Chongqing for Young and Middle-aged. We thank Dr. Yuanyuan Liu at School of Public Health, Tianjin Medical University for providing the pDC cell line CAL-1. We thank the patients, nurses, and doctors from the Department of Dermatology and the Traditional Chinese Medicine department of Southwest Hospital for participating in this study.
References
- Strickland FM, Hewagama A, Lu Q, et al. Environmental exposure, estrogen and two X chromosomes are required for disease development in an epigenetic model of lupus. J Autoimmun. 2012;38(2–3):J135–J143. doi:10.1016/j.jaut.2011.11.001
- Kobayashi T, Nguyen-Tien D, Sorimachi Y, et al. SLC15A4 mediates M1-prone metabolic shifts in macrophages and guards immune cells from metabolic stress. Proc Natl Acad Sci USA. 2021;118(33):e2100295118. doi:10.1073/pnas.2100295118
- Kobayashi T, Shimabukuro-Demoto S, Yoshida-Sugitani R, et al. The histidine transporter SLC15A4 coordinates mTOR-dependent inflammatory responses and pathogenic antibody production. Immunity. 2014;41(3):375–388. doi:10.1016/j.immuni.2014.08.011
- Baccala R, Gonzalez-Quintial R, Blasius AL, et al. Essential requirement for IRF8 and SLC15A4 implicates plasmacytoid dendritic cells in the pathogenesis of lupus. Proc Natl Acad Sci USA. 2013;110(8):2940–2945. doi:10.1073/pnas.1222798110
- Katewa A, Suto E, Hui J, et al. The peptide symporter SLC15a4 is essential for the development of systemic lupus erythematosus in murine models. PLoS One. 2021;16(1):e0244439. doi:10.1371/journal.pone.0244439
- Blasius AL, Arnold CN, Georgel P, et al. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107(46):19973–19978. doi:10.1073/pnas.1014051107
- Heinz LX, Lee J, Kapoor U, et al. TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7-9. Nature. 2020;581(7808):316–322. doi:10.1038/s41586-020-2282-0
- Kobayashi T, Nguyen-Tien D, Ohshima D, et al. Human SLC15A4 is crucial for TLR-mediated type I interferon production and mitochondrial integrity. Int Immunol. 2021;33(7):399–406. doi:10.1093/intimm/dxab006
- Snowhite IV, Allende G, Sosenko J, Pastori RL, Messinger Cayetano S, Pugliese A. Association of serum microRNAs with islet autoimmunity, disease progression and metabolic impairment in relatives at risk of type 1 diabetes. Diabetologia. 2017;60(8):1409–1422. doi:10.1007/s00125-017-4294-3
- Omidi F, Hosseini SA, Ahmadi A, et al. Discovering the signature of a lupus-related microRNA profile in the gene expression omnibus repository. Lupus. 2020;29(11):1321–1335. doi:10.1177/0961203320944473
- Navarro Quiroz E, Navarro Quiroz R, Pacheco Lugo L, et al. Integrated analysis of microRNA regulation and its interaction with mechanisms of epigenetic regulation in the etiology of systemic lupus erythematosus. PLoS One. 2019;14(6):e0218116. doi:10.1371/journal.pone.0218116
- Tang Y, Luo X, Cui H, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60(4):1065–1075. doi:10.1002/art.24436
- Tu Y, Guo R, Li J, et al. MiRNA regulation of MIF in SLE and attenuation of murine lupus nephritis with miR-654. Front Immunol. 2019;10:2229. doi:10.3389/fimmu.2019.02229
- Zhu Y, Xue Z, Di L. Regulation of MiR-146a and TRAF6 in the diagnose of lupus nephritis. Med Sci Monit. 2017;23:2550–2557. doi:10.12659/MSM.900667
- Wang Z, Heid B, Dai R, Ahmed SA. Similar dysregulation of lupus-associated miRNAs in peripheral blood mononuclear cells and splenic lymphocytes in MRL/lpr mice. Lupus Sci Med. 2018;5(1):e000290. doi:10.1136/lupus-2018-000290
- Harris VM, Harley ITW, Kurien BT, Koelsch KA, Scofield RH. Lysosomal pH is regulated in a sex dependent manner in immune cells expressing CXorf21. Front Immunol. 2019;10:578. doi:10.3389/fimmu.2019.00578
- McCoy RC, Wakefield J, Akey JM. Impacts of neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168(5):916–927 e912. doi:10.1016/j.cell.2017.01.038
- Kobayashi T, Tsutsui H, Shimabukuro-Demoto S, et al. Lysosome biogenesis regulated by the amino-acid transporter SLC15A4 is critical for functional integrity of mast cells. Int Immunol. 2017;29(12):551–566. doi:10.1093/intimm/dxx063
- Rimann I, Gonzalez-Quintial R, Baccala R, et al. The solute carrier SLC15A4 is required for optimal trafficking of nucleic acid-sensing TLRs and ligands to endolysosomes. Proc Natl Acad Sci USA. 2022;119(14):e2200544119. doi:10.1073/pnas.2200544119
- Fan W, Liang D, Tang Y, et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. 2012;64(11):3715–3725. doi:10.1002/art.34596
- Wu Y, Mealer C, Schutt S, et al. MicroRNA-31 regulates T-cell metabolism via HIF1alpha and promotes chronic GVHD pathogenesis in mice. Blood Adv. 2022;6(10):3036–3052. doi:10.1182/bloodadvances.2021005103
- Johansson A, Nyberg WA, Sjostrand M, et al. miR-31 regulates energy metabolism and is suppressed in T cells from patients with Sjogren’s syndrome. Eur J Immunol. 2019;49(2):313–322. doi:10.1002/eji.201747416
- Liu Y, Li C, Yang Y, et al. The TGF-beta/miR-31/CEACAM1-S axis inhibits CD4(+) CD25(+) treg differentiation in systemic lupus erythematosus. Immunol Cell Biol. 2021;99(7):697–710. doi:10.1111/imcb.12449
