Abstract
Objective
The aim of this study was to identify prognostic significance of microRNA-100 (miR-100) in solid tumor.
Methods
Literature search was conducted in databases such as PubMed, Embase, and Web of Science, using the following words “(microRNA-100 OR miR-100 OR mir100) AND (tumor OR neoplasm OR cancer OR carcinoma OR malignancy).” The search was updated up until July 10, 2016. Newcastle–Ottawa scale was used to evaluate the quality of studies. Pooled hazard ratio (HR) with 95% confidence interval (CI) for patients’ survival was calculated by using a fixed-effects or a random-effects model on the basis of heterogeneity. Subgroup analysis, sensitive analysis, and meta-regression were used to investigate the sources of heterogeneity. Publication bias was evaluated by using Begg’s and Egger’s tests.
Results
A total of 16 articles with 1,501 patients were included in the present meta-analysis. It was demonstrated that a lower expression of miR-100 plays a negative role in the overall survival (OS) of patients with solid tumor (HR =1.92; 95% CI =1.25–2.94). In addition, the association between miR-100 and prognosis was also revealed in the following subgroups: non-small-cell lung cancer (NSCLC; HR =2.46; 95% CI =1.98–3.06), epithelial ovarian cancer (EOC; HR =2.29, 95% CI =1.72–3.04), and bladder cancer (BC; HR =4.14, 95% CI =1.85–9.27).
Conclusion
This meta-analysis indicates that lower expression of miR-100 is related to poorer OS in patients with solid tumor, especially in those with NSCLC, EOC, and BC. MiR-100 is a promising prognosis predictor and may be a potential target for therapy in the future.
Keywords:
Introduction
MicroRNAs, noncoding small RNAs, regulate mRNA at the posttranscriptional level, leading to degradation or inhibition of the target mRNA. Research on microRNAs began in 1993.Citation1,Citation2 Since then, an increasing number of microRNAs were discovered, and it was found that play an important role in a variety of biological activities including proliferation, differentiation, invasion, and apoptosis.Citation3–Citation5 The association between microRNAs and cancer was first reported in 2002 by Calin et al, who observed that miR-15 and miR-16 acted as negative regulators of Bcl-2 in cancer, especially chronic lymphocytic leukemia.Citation6 It is now clear that microRNAs play a crucial role in cancer cells. Moreover, the clinical use of microRNA is being widely investigated. As a result, currently, multiple research studies have been exploring the prognostic function of microRNA-100 (miR-100) in cancer patients in order to find a reliable biomarker to guide for cancer treatment.Citation7,Citation8
As a member of the miR-99 family, miR-100 is located on Chromosome 11 at 11q24.1.Citation9 Its aberrant expression was reported in various cancers including nasopharyngeal cancer (NPC),Citation10 oral squamous cell carcinoma,Citation11 esophageal adenocarcinoma (EA),Citation12 small cell carcinoma of the cervix (SCCC),Citation13 non-small-cell lung cancer (NSCLC),Citation14,Citation15 endometrioid endometrial carcinoma (EEC),Citation16 epithelial ovarian cancer (EOC),Citation17,Citation18 bladder cancer (BC),Citation19,Citation20 renal cell carcinoma (RCC),Citation21 esophageal squamous cell carcinoma (ESCC),Citation22,Citation23 hepatocellular carcinoma (HCC),Citation24 colorectal cancer (CRC),Citation25,Citation26 and pancreatic adenocarcinoma.Citation27 This aberrant expression of miR-100 not only has diagnostic implications but also can predict cancer patient survival.Citation28 From this perspective, it may be feasible to predict prognosis based on miR-100 expression level. However, the correlation between them remains controversial to some extent. In some studies, miR-100 was reported as a tumor suppressor, whose low expression level was a negative prognostic factor.Citation16,Citation17 In contrast, in some articles, miR-100 was regarded as an oncogene.Citation21,Citation27 Therefore, this meta-analysis was conducted to identify the relationship between miR-100 expression level and the survival of cancer patients, by pooling the hazard ratio (HR) from studies addressing the correlation between miR-100 and overall survival (OS) of patients with solid tumor.
Materials and methods
Search strategy
A literature search was conducted in databases such as PubMed, Embase, and Web of Science, using the following words “(microRNA-100 OR miR-100 OR mir100) AND (tumor OR neoplasm OR cancer OR carcinoma OR malignancy).” The search was updated up until July 10, 2016. In order not to miss the potentially related articles, reference lists were also screened.
Inclusion and exclusion criteria
Inclusion criteria were as follows: 1) studies exploring any of the following solid tumor (eg, head and neck, thyroid, non-small-cell or small cell lung, breast, esophageal, gastric, pancreatic, hepatobiliary, colorectal, anal, prostate, bladder, renal, cervical, endometrial and ovarian carcinoma, melanoma, and sarcomas); 2) studies dealing with miR-100 expression and OS; 3) studies that categorized patients into low- and high-expression groups based on the miR-100 expression; 4) studies providing HR directly or key information to calculate HR indirectly, such as Kaplan–Meier curves and original survival data; and 5) studies assessing miR-100 expression in tissue or blood. The following were the exclusion criteria: 1) studies on myelomas, lymphomas, or leukemia; 2) nonoriginal articles, such as reviews, articles, or letters; 3) laboratory studies on cell lines or animals level; 4) studies on a set of microRNAs but not miR-100 alone; and 5) studies on nondichotomous miR-100.
Qualitative assessment
Newcastle–Ottawa scale was used to evaluate the quality of the studies based on the following three aspects: selection, comparability, and outcome.Citation29 A study can be given a maximum of one star for each item within the selection and outcome categories, and a maximum of two stars can be given for comparability category. The total stars that can be given for a study ranges from 0 to 9. Studies with ≥6 stars were considered as of high quality.Citation30 Otherwise, studies were excluded from the final meta-analysis.
Data extraction
Data extraction was carried out by Wang and Cheng. The following details of each article were recorded: publication year, first author’s last name, cancer type, treatment, sample size, stage of disease, miR-100 assay, the cutoff value to discriminate high or low expression of miR-100, sample sources, follow-up time, and HR (low versus high expression). HR >1 indicates that lower expression of miR-100 is an unfavorable prognostic factor, and HR <1 suggests that lower expression of miR-100 confers survival advantage to the patients. If the HR value was calculated by a multivariate analysis, then it was extracted directly; if multivariate analysis was not available, then results from univariate analysis were also accepted in the meta-analysis. If both multivariate analysis and univariate analysis were not offered, Kaplan–Meier curves were used to estimate HR value by using the described method.Citation31 In addition, original survival data obtained, if possible, can also be used to calculate HR by conducting survival analysis.
Statistical analysis
In order to test the heterogeneity of pooled HR, Cochran’s Q-test and Higgins I2 statistics were performed. P<0.05 was considered statistically significant. Random-effects model was used to calculate pooled HR when between-study heterogeneity was revealed (P<0.05), and fixed-effects model was conducted when between-study heterogeneity did not reach the statistical significance (P>0.05). Subgroup analysis, sensitive analysis, and meta-regression were used to investigate the sources of heterogeneity. Publication bias was assessed by using Begg’s test and Egger’s test.Citation32,Citation33 STATA version 12.0 (Stata Corporation, College Station, TX, USA) was used to perform all the analyses.
Results
Study selection
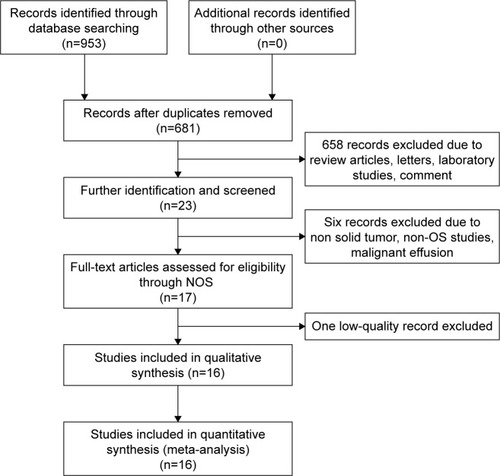
In total, 179, 482, and 292 records were identified from PubMed, Embase, and Web of Science, respectively (). After excluding the replicate records, 272 articles were left. Reviewers Wang and Huang carefully read the abstract or full text as required and removed 658 records because they were review articles, laboratory studies, and so on. Then, 23 publications were retrieved for further analysis; of these studies, two studies focused on biochemical-free survival and recurrence-free survival of prostate cancer instead of OS;Citation34,Citation35 three articles did not address solid tumor.Citation36–Citation38 In addition, a study by Wang et al used malignant effusion, rather than tissue or blood, to assess miR-100 expression in NSCLC patients.Citation39 Therefore, according to the inclusion and exclusion criteria in the present study, 17 articles were accepted for Newcastle–Ottawa scale.
Qualitative assessment
Results from Newcastle–Ottawa scale demonstrated that two studies received eight stars, eight articles received seven stars, six articles received six stars, and one article received one star. Thus, a study by Zhang et al was excluded as it was a low-quality article ().Citation40
Table 1 Newcastle–Ottawa scale
Summary of the analyzed studies
After filtering, 16 studies published between 2011 and 2016 were considered for the final meta-analysis (). The cancer types included EA, SCCC, EEC, human EOC, BC, NSCLC, RCC, ESCC, HCC, CRC, and pancreatic ductal adenocarcinoma. Only four studies clearly stated the research-related treatment. As revealed in , most studies (11 of 16) did not clarify whether patient received adjuvant therapy after surgery. Study sample sizes ranged from 42 to 172 with a total of 1,501 patients from the People’s Republic of China, the USA, Poland, Iran, and Germany. Nine studies enrolled patients with stages I–IV, whereas a study by Dhayat only explored miR-100 in patients with Stage II pancreatic ductal adenocarcinoma.Citation27 Four studies did not specify the stage of disease in the study population. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to assess miR-100 expression in all the studies, and the cutoff value varied among studies with median expression of miR-100 the most widely used. All the authors used tissues as the source of miR-100. The majority of HRs (13 of 16) were reported in the present analysis, all of which were calculated by using a multivariate analysis, whereas four HRs were estimated by analyzing Kaplan–Meier curves.
Table 2 Characteristics of the included articles
Meta-analysis results
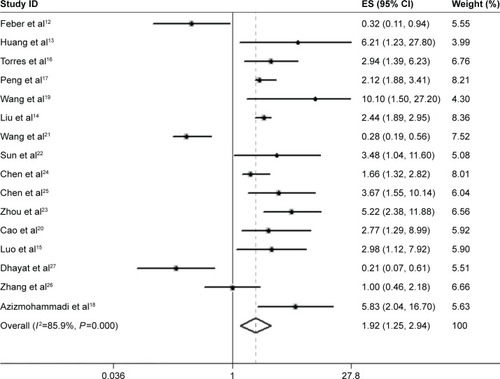
Due to obvious heterogeneity (I2=85.9%; P<0.001), random-effects model was used to calculate pooled HR (HR =1.92; 95% confidence interval [CI] =1.25–2.94), suggesting that lower expression level of miR-100 significantly predicted poorer OS in patients with solid tumor ().
Figure 2 Forest plot of the relationship between miR-100 and overall survival in solid tumor. Note: Weights are from random-effects analysis.
In order to identify the potential source of heterogeneity, subgroups analysis was conducted based on the patient origin, cancer type, and the methods used to calculate HR (). The association between miR-100 and OS was also revealed in the following subgroups: NSCLC (HR =2.46; 95% CI =1.98–3.06; fixed-effects model), EOC (HR =2.29; 95% CI =1.72–3.04; fixed-effects model), and BC (HR =4.14; 95% CI =1.85–9.27; fixed-effects model). Notably, significant heterogeneity was not found among these studies, with P-value of heterogeneity being 0.696, 0.07, and 0.146, respectively. However, no statistic difference was reported in esophageal carcinoma (HR =1.82; 95% CI =0.32–10.32; random-effects model) and CRC (HR =1.70; 95% CI =0.93–3.09; random-effects model). The prognostic role of miR-100 was also significant in the following subgroups: HR by report or multivariate analysis (HR =1.91; 95% CI =1.91–3.06; random-effects model) and Asian patients (HR =2.38; 95% CI =1.56–3.66; random-effects model).
Table 3 Subgroup analysis
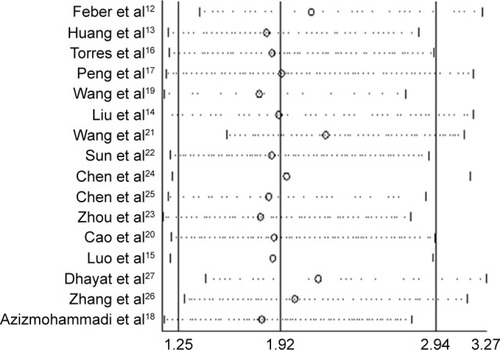
Sensitivity analysis was conducted by sequentially omitting individual studies, indicating that there was not a single study that significantly contributed to heterogeneity (). Furthermore, a meta-regression was also conducted to explore the potential factors that are responsible for heterogeneity. The results showed that the following factors could partly explain the heterogeneity but did not reach statistical significance: patient origin (I2=85.68%; adjusted R2=16.54%; P=0.06), cancer type (I2=83.65%; adjusted R2=10.95%; P=0.59), and multivariate analysis (I2=86.84%, adjusted R2=−8.60%; P=0.97).
Figure 3 Sensitivity analysis: meta-analysis of random-effects estimates (exponential form) with studies omitted.
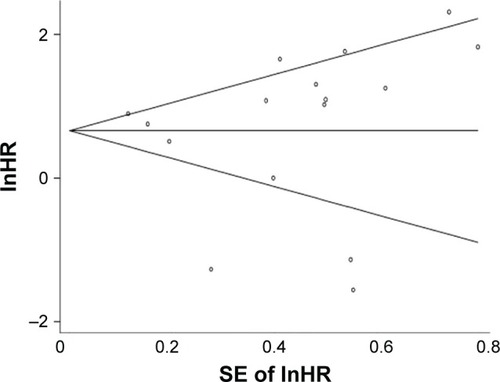
Publication bias was evaluated by using Begg’s funnel plot and Egger’s test. Although the funnel plot revealed slight publication bias, P-values of Begg’s and Egger’s tests were 0.753 and 0.056, respectively, showing no evidence for significant publication bias ().
Discussion
Lack of a reliable biomarker to predict prognosis is a main hurdle to realize individualized treatment. Fortunately, emerging data favor the potential use of miR-100 as a cancer prognostic predictor. In recent years, downstream targets of miR-100 have widely been investigated in several signal pathways, which may provide evidence for its potential clinical use at a molecule level. First, miR-100 facilitated cancer cell proliferation and growth by targeting the mammalian target of rapamycin (mTOR) and fibroblast growth factor receptors (FGFRs). By using GeneChip® miRNA Array and qRT-PCR, Xu et alCitation41 identified decreased expression of miR-100 in bladder tumor tissues and found that ectopic restoration of miR-100 expression suppressed tumor cell proliferation. Furthermore, bioinformatic analysis indicated that mTOR was the direct target of miR-100.Citation41 Consistently, another study on esophageal squamous cell cancer also supported this report. Sun et al transfected esophageal squamous cell cancer cell lines with miR-100 precursor molecules and reported the subsequent inhibition of cell proliferation, which was attributed to the fact that miR-100 downregulated the expression of mTOR by directly targeting its 3′UTR in a post-transcriptional manner.Citation22 In gynecologic cancer, this interaction exists. Torres et al observed that decreased miR-100 in EEC tissues coexisted with increased mTOR kinase.Citation16 With respect to cell growth, another potential target of miR-100 may be FGFRs. Bi et al transfected U2OS cells (human osteo-sarcoma line) with a miR-100 construct or with an antisense of miR-100.Citation42 It was observed that overexpression of miR-100 led to decreased expression of FGFR3 protein. Conversely, inhibition of miR-100 induced FGFR3 protein expression. Bioinformatics analysis and luciferase reporter assay implied that 3′UTR of FGFR3 mRNA was the direct target of miR-100. Based on these experimental results, Bi et alCitation42 pointed out that miR-100 served as a tumor suppressor by preventing translation of FGFR3s. Another study on BC also revealed that the reduction of miR-100 expression induced sixfold upregulation of FGFR3 protein through an interaction with FGFR3 mRNA.Citation43 Similar result was also reported in non-small-cell lung.Citation15 Second, miR-100 participated in cancer cell apoptosis, which also determines the development of the tumor. Chen et al observed that miR-100 precursor elevated expression of cleaved caspase-3 and pro-caspase-3 protein, which are two key apoptosis proteins.Citation24 In order to find the mechanism, flow cytometry and Western blot analysis were used to assess the apoptosis in siRNA/plk 1-transfected human hepatocellular cells. The outcome showed that siRNA/plk 1 could mimic the effect of miR-100, indicating plk1 as a potential target of miR-100. In another digestive cancer, gastric cancer, Notch pathway was reported to participate in the miR-100-mediated apoptosis, which is initiated by the interaction between miR-100 and HS3ST2.Citation44 Third, miR-100 regulated several components of Wnt/β-catenin pathway, including β-catenin, metalloproteinase-7, T-cell factor-4, and lymphoid enhancing factor-1, which were closely related to cell invasion and tumor metastasis.Citation45 Transwell migration and invasion assay by Jiang showed that transfection of miR-100 mimic suppressed migration and invasion of breast cancer compared to the control group. The direct target of miR-100 in Wnt/β-catenin pathway may be FZD-8.Citation46 Bhushan et al also found that downregulation of miR-100 may account for less invasion of breast cancer cells.Citation47 In addition, Fu et al also consolidated the role miR-100 played in invasion and metastasis of esophageal squamous cell cancer by using bioinformatics analyses.Citation48 Fourth, cell cycle arrest, which is responsible for radio-resistance or chemo-resistance, was mediated by miR-100. Xiao et al discovered that the introduction of miR-100 mimic in H69 cells (human small cell lung cancer cell line) led to downregulation of HOXA1 protein, inducing G0-G1 and G1-M cell cycle arrest, which was associated with drug resistance.Citation49 A study by Shi et al documented that miR-100 can target plk1 and mediated G2/M arrest in NPC.Citation10 Together, miR-100 might be a promising prognostic predictor.
Despite the sort of inconsistency, clinical investigations favor predictive use of miR-100 for the survival of cancer patients. Peng et al retrospectively collected fresh surgical tissues from 98 patients with EOC and used qRT-PCR assay to assess the expression level of miR-100.Citation17 It is revealed that the expression of miR-100 was significantly lower in epithelial ovarian tumor tissue than in adjacent normal tissue. Multivariate analysis demonstrated that lower expression of miR-100 was accompanied by shorter survival (HR =2.12; 95% CI =1.88–3.41). Further evidence on endometrial carcinoma also supported the finding that decreased expression level of miR-100 worsens prognosis, with a HR of 2.94 (low/high).Citation16 Similar results were obtained for digestive system cancers. Zhou investigated 120 patients with esophageal squamous cell cancer who received radiotherapy alone and concluded that HRs for locoregional progression-free survival (PFS), distant PFS, and OS (low/high) were 8.9, 8.34, and 8.02, respectively, suggesting that downregulation of miR-100 indicated poor prognosis.Citation23 In BC, Wang et al reported that 3-year OS and 3-year PFS for high expression level of miR-100 were 33 and 26 months, respectively; on the other hand, for low expression level of miR-100, they were merely 23 and 16 months, respectively.Citation19 However, it should be noted that literature on the prognostic value of miR-100 reveals conflicting findings. Wang et al found increased miR-100 expression in cancer tissues compared with normal tissues in patients with renal cell cancer.Citation21 HRs for OS and tumor-specific survival were 0.28 and 0.41, respectively, both of which were statistically significant, suggesting that lower expression of miR-100 confers survival advantage. Dhayat et al focused on pancreatic ductal adenocarcinoma Union for International Cancer Control stage II and found that lower level of miR-100 is a favorable factor to predict treatment response and OS.Citation27 Apart from studies revealing the survival difference, a study by Zhang showed that the difference in miR-100 expression level did not lead to statistic difference in the survival of patients with CRC.Citation26 In order to clarify the relationship between miR-100 and survival of patients with solid tumor, this meta-analysis was conducted.
In order to ensure reliable outcome, the inclusion criteria were strictly limited. First, source of RNA was restricted to tissue or blood. As a result, a study by Wang et al was not accepted, because it explored miR-100 in malignant effusion in NSCLC.Citation39 Second, Newcastle–Ottawa scale was used. As shown in , most studies (16 of 17) were evaluated as of high quality; however, only two studies received nine stars, the highest score. The most common reasons why the research was deprived of stars lay in the comparability and outcome aspects. In terms of comparability, majority of the studies (15 of 17) did not control the most important factor such as TNM stage; some studies (6 of 17) did not control other less important factors, such as age, gender, and smoking status of the patients, which would bring potential bias to the survival outcome. In addition, it is necessary for prognosis studies to describe whether the survival outcome was attained by self-report or blind assessment, which was stated in Newcastle–Ottawa scale. However, only two studies mentioned this category.Citation23,Citation27 Lastly, follow-up rate is not fully recorded. Only a minority of studies described the number of lost patients and offered the reason for loss. Considering these reasons, more stars have been provided in outcome category. It is noteworthy that Zhang et al analyzed survival data from the TCGA database and did not provide sufficient related information to get at least six stars according to Newcastle–Ottawa scale.Citation40 Hence, although it investigated the association between miR-100 expression and OS, it was excluded as a study of low quality.
Finally, 16 studies were included. According to the present study, decreased miR-100 expression level confers survival disadvantage in patients with solid tumor (HR =1.92; 95% CI =1.25–2.94). This relationship was also found in the following subgroups: NSCLC (HR =2.46; 95% CI =1.98–3.06; fixed-effects model), EOC (HR =2.29; 95% CI =1.72–3.04; fixed-effects model), and BC (HR =4.14; 95% CI =1.85–9.27; fixed-effects model). In order to find the potential source of heterogeneity, first, subgroup analysis was conducted, which revealed that heterogeneity declined dramatically in NSCLC group (I2=0; P=0.70), EOC group (I2=69.6%; P=0.07), and BC group (I2=52.7%; P=0.15). Therefore, cancer type might partly account for heterogeneity. Previously, several meta-analyses classified subgroups according to the method used to assess RNA (qRT-PCR versus in situ hybridization) and the source of RNA (blood versus tissue).Citation30 Nonetheless, these variants were similar between the studies included in the present meta-analysis. As a result, the possibility of these variants as the source of heterogeneity was eliminated, and there was no requirement to perform further subgroup analysis. Owing to the results that sensitivity analysis did not reveal the source of heterogeneity, meta-regression was further conducted, which revealed that P-value of patient origin, the method used to calculate HR, and cancer type were all >0.05, with adjusted R2 being 16.54%, −8.6%, and −10.95%, respectively. Meta-regression results showed that although these factors might explain part of heterogeneity, but it did not reach statistical significance. Meta-regression could have been used to analyze whether other factors such as follow-up time, related treatment methods, and cutoff value had an impact on heterogeneity, but, due to insufficient information on or remarkably different definition of cutoff value, the necessity and scientific implication to carry out this meta-regression in these variants were uncertain.
As it is well known, meta-analysis results are more robust than the result of single research. However, it is noteworthy that strict inclusion and exclusion criteria are a prerequisite for pooled data, which made the present results more reliable than the previous ones.Citation50,Citation51 Despite careful assessment of study quality, there are limitations in the present study. First, heterogeneity among the studies included is relatively high. According to subgroup analysis, tumor type could partly explain the source of heterogeneity. Nonetheless, other potential factors may also exist. For instance, pathological type may be a potential source. The pooled HR from ESCC subgroup was 4.61 (95% CI =2.36–8.99; I2=0; fixed-effects model) compared to esophageal cancer (HR =1.82; 95% CI =0.32–10.32; I2=88.5%; random-effects model).Citation22,Citation23 In addition, because of insufficient data, it was difficult to further assess the effects of other variants on heterogeneity. For instance, TNM stage, postoperative treatment, and follow-up method were not clearly stated in all the studies. Second, the power of miR-100 as a stand-alone biomarker is presumably weak and varies for different tumor types, given the fact that interaction between microRNA and its target is complicated. Undoubtedly, microRNA signature may be a better option that has been investigated in other microRNAs.Citation52 However, to date, such microRNA signature comprising miR-100 was not reported. Third, the definition of cutoff value varied among the studies. Some research studies chose median value to define the expression level of miR-100, yet some used mean value or value calculated by receiver operating characteristic curve. Finally, although publication bias was not revealed by Begg’s and Egger’s tests, the funnel plot is slightly asymmetric. The limitations emphasize the need for well-designed and well-conducted prognostic studies on miR-100 in the future.
Conclusion
This meta-analysis indicated that lower expression level of miR-100 is related to poorer OS in solid tumor, especially in patients with NSCLC, EOC, and BC.
Acknowledgments
This work was supported by Youth Science Found of Qilu Hospital of Shandong University, Science and Technology plan project of Shandong province (2012GSF11852) and National Natural Science Foundation of China (81572958).
Disclosure
The authors report no conflicts of interest in this work.
References
- LeeRCFeinbaumRLAmbrosVThe C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14Cell19937558438548252621
- WightmanBHaIRuvkunGPosttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegansCell19937558558628252622
- YoshimuraANumakawaTOdakaHAdachiNTamaiYKunugiHNegative regulation of microRNA-132 in expression of synaptic proteins in neuronal differentiation of embryonic neural stem cellsNeurochem Int201697263327131735
- GuanBLiQShenLMicroRNA-205 directly targets Kruppel-like factor 12 and is involved in invasion and apoptosis in basal-like breast carcinomaInt J Oncol201649272073427278159
- ZhangYXueCZhuXZhuXXianHHuangZSuppression of microRNA-125a-5p upregulates the TAZ-EGFR signaling pathway and promotes retinoblastoma proliferationCell Signal201628885086027094723
- CalinGADumitruCDShimizuMFrequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemiaProc Natl Acad Sci U S A20029924155241552912434020
- WangWLiJZhuWMicroRNA-21 and the clinical outcomes of various carcinomas: a systematic review and meta-analysisBMC Cancer20141481925376700
- DaiSLZhouJPanCPrognostic value of microRNA-145 in patients with various cancers: a meta-analysisCancer Biomark201515450751325792469
- QinCHuangRYWangZXPotential role of miR-100 in cancer diagnosis, prognosis, and therapyTumour Biol20153631403140925740059
- ShiWAlajezNMBastianuttoCSignificance of Plk1 regulation by miR-100 in human nasopharyngeal cancerInt J Cancer201012692036204819739117
- HensonBJBhattacharjeeSO’DeeDMFeingoldEGollinSMDecreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancyGenes Chromosomes Cancer200948756958219396866
- FeberAXiLPennathurAMicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinomaAnn Thorac Surg20119151523153021420070
- HuangLLinJXYuYHZhangMYWangHYZhengMDownregu-lation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervixPLoS One201273e3376222438992
- LiuJLuKHLiuZLSunMDeWWangZXMicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1BMC Cancer20121251923151088
- LuoJChenBJiXXZhouSWZhengDOverexpression of miR-100 inhibits cancer growth, migration, and chemosensitivity in human NSCLC cells through fibroblast growth factor receptor 3Tumour Biol Epub2015828
- TorresATorresKPesciADeregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinomaBMC Cancer20121236922920721
- PengDXLuoMQiuLWHeYLWangXFPrognostic implications of microRNA-100 and its functional roles in human epithelial ovarian cancerOncol Rep20122741238124422246341
- AzizmohammadiSAzizmohammadiSSafariAThe role and expression of miR-100 and miR-203 profile as prognostic markers in epithelial ovarian cancerAm J Transl Res2016852403241027347348
- WangSXueSDaiYReduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancerDiagn Pathol2012715923173870
- CaoYHZhangHHXuHFDuanYJLiQHuangBPrognostic role of microRNA-100 in patients with bladder cancerGenet Mol Res2015144159481595426662386
- WangGChenLMengJChenMZhuangLZhangLOverexpres-sion of microRNA-100 predicts an unfavorable prognosis in renal cell carcinomaInt Urol Nephrol201345237337923378187
- SunJChenZTanXMicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinomaMed Oncol201330141123292834
- ZhouSYangBZhaoYXuSZhangHLiZPrognostic value of microRNA-100 in esophageal squamous cell carcinomaJ Surg Res2014192251552025218280
- ChenPZhaoXMaLDownregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinomaMol Cell Biochem20133831–2495823842624
- ChenPXiQWangQWeiPDownregulation of microRNA-100 correlates with tumor progression and poor prognosis in colorectal cancerMed Oncol2014311023525216869
- ZhangSYuanWTangWXuCMaJExpression of microRNA-100 and its relation with prognosis of colorectal cancerZhonghua Zhong Liu Za Zhi2015378603608 Chinese26714601
- DhayatSAAbdeenBKohlerGSenningerNHaierJMardinWAMicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage IIClin Epigenetics2015713226705427
- MotawiTKRizkSMIbrahimTMIbrahimIACirculating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive bio-markers for bladder cancer diagnosisCell Biochem Funct201634314214826916216
- WellsGASheaBO’connellDThe Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analysesOttawa, ONOttawa Hospital Research Institute2000
- ShaoYGengYGuWHuangJNingZPeiHPrognostic significance of microRNA-375 downregulation in solid tumors: a meta-analysisDis Markers2014201462618525404787
- TierneyJFStewartLAGhersiDBurdettSSydesMRPractical methods for incorporating summary time-to-event data into meta-analysisTrials200781617555582
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- EggerMSmithGDSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- RaneJKErbHHNappoGInhibition of the glucocorticoid receptor results in an enhanced miR-99a/100-mediated radiation response in stem-like cells from human prostate cancersOncotarget Epub2016621
- LeiteKRTomiyamaAReisSTMicroRNA-100 expression is independently related to biochemical recurrence of prostate cancerJ Urol201118531118112221255804
- de OliveiraJCScrideliCABrassescoMSDifferential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological featuresLeuk Res201236329329822099053
- LiXJLuoXQHanBWDuanFTWeiPPChenYQMicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathwaysBr J Cancer201310982189219824030073
- BaiJGuoAHongZKuaiWUpregulation of microRNA-100 predicts poor prognosis in patients with pediatric acute myeloid leukemiaOnco Targets Ther2012521321923055746
- WangTLvMShenSCell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancerPLoS One201278e4326822937028
- ZhangBZhaoRHeYMicroRNA 100 sensitizes luminal A breast cancer cells to paclitaxel treatment in part by targeting mTOROncotarget2016755702571426744318
- XuCZengQXuWmiRNA-100 inhibits human bladder urothelial carcinogenesis by directly targeting mTORMol Cancer Ther201312220721923270926
- BiYJingYCaoYOverexpression of miR-100 inhibits growth of osteosarcoma through FGFR3Tumour Bio201536118405841126018508
- CattoJWMiahSOwenHCDistinct microRNA alterations characterize high- and low-grade bladder cancerCancer Res200969218472848119843843
- YangGGongYWangQWangYZhangXThe role of miR-100-mediated Notch pathway in apoptosis of gastric tumor cellsCell Signal20152761087110125703026
- KlemmFBleckmannASiamLβ-catenin-independent WNT signaling in basal-like breast cancer and brain metastasisCarcinogenesis201132343444221173432
- JiangQHeMGuanSMicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/beta-catenin signaling pathwayTumour Biol20163745001501126537584
- BhushanLKandpalRPEphB6 receptor modulates micro RNA profile of breast carcinoma cellsPLoS One201167e2248421811619
- FuHLWu dePWangXFAltered miRNA expression is associated with differentiation, invasion, and metastasis of esophageal squamous cell carcinoma (ESCC) in patients from Huaian, ChinaCell Biochem Biophys201367265766823516093
- XiaoFBaiYChenZDownregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100Eur J Cancer20145081541155424559685
- ChenJZhengBWangCPrognostic role of microRNA-100 in various carcinomas: evidence from six studiesTumour Biol20143543067307124258109
- ChenDZhouYDuHCheGRefute the conclusion made by Jie et al. in “prognostic role of microRNA-100 in various carcinomas: evidence from six studies”Tumour Biol20143587393739624903380
- KomatsuSIchikawaDTakeshitaHPrognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinomaExpert Opin Biol Ther201212Suppl 1S53S5922519435