Abstract
Purpose
The AVAglio trial established the beneficial effect of add-on bevacizumab (BEV) for the treatment of newly diagnosed glioblastomas (nd-GBMs) that led to the approval of BEV for the treatment of these patients in Japan. However, the rationality of using BEV as a first-line treatment for nd-GBMs remains controversial. The purpose of this study was to analyze the outcomes of a case series of nd-GBM patients.
Patients and methods
The outcomes of 69 nd-GBM patients treated after 2006 were retrospectively analyzed. Clinical and genetic analyses were performed, and estimates of progression-free survival (PFS) and overall survival (OS) were calculated using the Kaplan–Meier method. Since add-on BEV therapy was only used for partially resected GBMs (pr-GBMs) after its approval in 2013, the patients were subdivided into 3 treatment groups: Type I, partial removal with temozolomide (TMZ)/BEV and concurrent radiotherapy (CCRT); Type II, partial removal with TMZ and CCRT; and Type III, gross total removal with TMZ and CCRT.
Results
The PFS rate of Type I patients was significantly higher than that of Type II patients (P=0.014), but comparable to that of Type III patients. Differences in OS rates between Type I and Type II patients were less apparent (P=0.075), although the median OS of Type I patients was ~8 months higher than that of Type II patients (17.4 vs 9.8 months, respectively). The clinical deterioration rate during initial treatment was significantly (P=0.024) lower in Type I than in Type II patients (7.7% vs 47.4%, respectively). Differences in OS rates between Type I and Type II patients with a poor performance status (PS) were significant (P=0.017).
Conclusion
Our findings suggest that add-on BEV can prevent early clinical deterioration of pr-GBM patients and contribute to a prolonged survival, especially for those with a poor PS.
Introduction
Glioblastoma (GBM) is a common type of brain tumor that has an especially poor prognosis. Temozolomide (TMZ) treatment and concurrent radiotherapy (CCRT) followed by maintenance with TMZ (the Stupp regimen) have been regarded as the global standard for patients with newly diagnosed GBMs (nd-GBMs).Citation1 The AVAglio trial,Citation2 mainly conducted in European and Asian countries, has established the beneficial effect of adding bevacizumab (BEV; a humanized monoclonal antibody targeting vascular endothelial growth factor) to the Stupp regimen for improving progression-free survival (PFS) rates in GBM patients. However, no significant improvements in overall survival (OS) rates were reported.Citation2 In the Japanese population of the AVAglio trial,Citation3 although the effect of BEV on OS was not statistically significant, the median OS was longer in the BEV-treated group compared to the placebo group. Subsequently, BEV was approved in Japan, in 2013, as a first-line treatment for nd-GBMs. Currently, Japan is the only country where BEV is routinely available for treating nd-GBMs in a clinical setting. BEV for recurrent GBMs had been approved in a limited number of countries.Citation4 However, BEV was not approved in such countries for nd-GBMs because of controversies surrounding its clinical benefits, as demonstrated by another randomized study (the Radiation Therapy Oncology Group 0825 study),Citation5 which failed to establish a favorable outcome of BEV treatment for OS and quality of life. The rationality of using BEV as a first-line treatment for nd-GBMs remains controversial, and there is no global consensus.
At our institute and affiliated hospitals, based on the concept that BEV treatment can contribute to the control of tumor growth through regression of the existing tumor vasculature,Citation6 BEV has been used for the treatment of nd-GBMs when gross total removal could not be achieved. Herein, we report on a case series in which we analyze the outcomes of GBM patients and evaluate the rationality of using BEV as a first-line treatment option for patients with nd-GBMs.
Patients and methods
Patients
Since TMZ was approved in Japan in 2006, 87 patients with nd-GBMs, histologically confirmed by qualified neuropathologists according to the criteria of the World Health Organization (WHO), were registered in our brain tumor database between 2006 and 2015. Patients who had refused adjuvant treatment (n=2), who had undergone immunotherapy of their own choosing (n=2), who had infratentorial tumors (n=3) or Type I neurofibromatosis (n=1), and whose genetic status was unknown due to the lack of available tissue samples (n=2) were excluded from our analysis. Genetic alterations were determined using snap-frozen tumor tissue samples obtained surgically. The present investigation was approved by the ethics committee of Kyushu University and Kyushu Medical Center. All participants had provided written informed consent. Research was conducted in accordance with the 1964 Declaration of Helsinki (as revised in Fortaleza, Brazil, October 2013). According to the results, patients with IDH1 (n=5), BRAF (n=2), and H3F3A (n=1) mutations were also excluded since these are known to be genetic markers of distinct biological subgroups of GBMs.Citation7,Citation8 In total, 69 patients (79.3%) were included in the final analysis to evaluate their outcomes. Since the approval of TMZ in Japan in 2006, patients with nd-GBMs were planned to be treated with maximal safe resection and TMZ with CCRT, followed by maintenance with TMZ. After the approval of BEV in Japan in 2013, the above-mentioned adjuvant treatment regimen was applied only to those patients who had undergone gross total removal (defined as the removal of >90% of the tumor using contrast-enhanced magnetic resonance imaging). Instead, patients who had not undergone gross total removal (ie, partial removal or biopsy) were treated with BEV plus TMZ and CCRT. Consequently, the 69 nd-GBM patients enrolled in this study were divided into 3 treatment groups: Type I, partial removal with TMZ/BEV and CCRT (n=13); Type II, partial removal with TMZ and CCRT (n=19); and Type III, gross total removal with TMZ and CCRT (n=37).
BEV administration
BEV was administered intravenously at a dose of 10 mg/kg body weight every 2 weeks (commencing 28 days after craniotomy or 14 days after stereotactic biopsy), followed by subsequent cycles every 2 weeks as the add-on treatment for nd-GBM patients receiving TMZ and CCRT. Maintenance treatment with BEV of the same dose commenced 4 weeks after the completion of CCRT and was performed in combination with maintenance treatment with TMZ. TMZ maintenance therapy was performed for a maximum of 24 cycles. Each physician was permitted to modify treatment intervals and/or doses, based on the patient’s condition. Use of steroid was limited during the perioperative phase for the purpose of controlling the symptomatic edema.
Genetic analyses
Tissue sampling and DNA preparation were performed according to our previous study.Citation9 The detection of hot spot mutations in the IDH1–2, BRAF, and H3F3A genes and promoter mutations in the TERT gene was performed according to our previous study with modifications as described in the Supplementary materials.Citation10 MGMT methylation status was assessed using a methylation-specific polymerase chain reaction-based method as described previously.Citation11
Statistical analyses
The main outcome measures of this study were postoperative PFS and OS, with censoring at the date of last follow-up for survivors. Disease progression was assessed according to the Response Assessment in Neuro-Oncology criteria.Citation12 OS and PFS rates were calculated using the Kaplan–Meier method, and the between-group differences in survival distributions were compared using the Wilcoxon test. Multivariate Cox proportional hazards regression models were applied to estimate the hazard ratios (HRs) and 95% CIs of putative prognostic factors. All statistical analyses were conducted using JMP Pro 11 Version 11.0.0 (SAS Institute Inc., Cary, NC, USA). Probability (P) values were 2-sided, and a threshold of 0.05 was used to determine statistical significance.
Results
First-line treatments
Since our study has a retrospective design, we analyzed the effect of BEV on partially resected GBM (pr-GBM) patients by comparing Type I patients with Type II patients who were used as a historical control. No significant bias of clinical or molecular factors was observed between Type I and Type II patients (). In both Type I and Type II treatment groups, the median Karnofsky performance status (KPS) score was ~60. This confirms that our case series of pr-GBM patients included a considerably higher proportion of patients with a poor performance status (PS) than previous clinical trials.Citation2,Citation5
Table 1 Clinical and molecular characteristics of newly diagnosed, partially resected glioblastoma patients (n=32)
Treatments after progression of pr-GBMs
In the Type I treatment group, with the exception of 2 patients who died of unrelated diseases without recurrence, disease progression was observed in 9 of the remaining 11 patients. Of these 9 patients, 4 patients continued BEV treatment and 1 patient received reirradiation to the recurrent lesion with concurrent BEV treatment. The remaining 4 patients were treated with best supportive care. In the Type II treatment group, of the 18 patients with disease progression (with the exception of 1 patient who died of an unrelated disease without recurrence), 2 patients continued TMZ treatment, 10 patients were treated with best supportive care, and 3 patients underwent resection of the recurrent lesion, followed by maintenance with TMZ (including 1 patient who was treated with Cyber-Knife Radiosurgery after resection). The remaining 3 patients received BEV treatment (these recurrences occurred after Japanese approval of BEV in 2013). In summary, after disease progression in a total of 27 pr-GBM patients (Type I and Type II treatment groups combined), 14 (51.8%) patients did not receive second-line therapy, and the ratios of these patients were comparable between the Type I and Type II treatment groups (44.4% vs 55.6%, 2-sided Fisher’s exact test, P=0.71).
Survival analyses
To verify the rationality of using BEV as a treatment for pr-GBMs, we evaluated the outcome of Type I patients by comparing them with Type II patients. The PFS rate of Type I patients was significantly higher than that of Type II patients (P=0.014; ), but comparable to that of Type III patients. The median PFS rates across the 3 groups were 10.0, 2.6, and 8.5 months, respectively. According to the multivariate Cox proportional hazards model of PFS, BEV treatment proved to be a significant prognostic factor (P=0.022) among the clinical and molecular markers analyzed (). Moreover, the differences in OS rates between Type I and Type II patients were not found to be statistically significant (P=0.075), although the median OS of Type I patients was ~8 months higher than that of Type II patients (17.4 vs 9.8 months, respectively; ). The multivariate Cox proportional hazards model showed BEV as a marginal significant prognostic factor (P=0.054) among the clinical and molecular markers analyzed ().
Figure 1 Kaplan–Meier estimates of PFS rate in newly diagnosed glioblastoma patients (n=69) stratified according to treatment type.
Abbreviations: BEV, bevacizumab; CCRT, concurrent radiotherapy; GTR, gross-total removal; PFS, progression-free survival; PR, partial removal; TMZ, temozolomide.
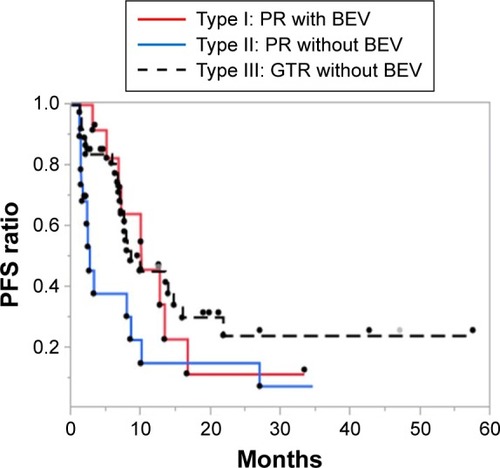
Figure 2 Kaplan–Meier estimates of OS rate in newly diagnosed glioblastoma patients (n=69) stratified according to treatment type.
Abbreviations: BEV, bevacizumab; CCRT, concurrent radiotherapy; GTR, gross-total removal; OS, overall survival; PR, partial removal; TMZ, temozolomide.
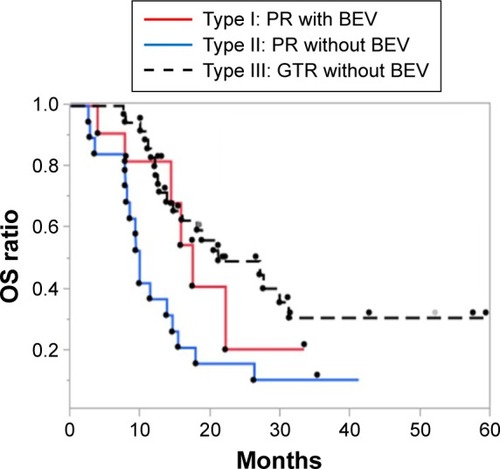
Table 2 Prognostic factors of PFS in newly diagnosed, partially resected glioblastoma patients (n=32)
Table 3 Prognostic factors of OS in newly diagnosed, partially resected glioblastoma patients (n=32)
To elucidate the benefits of BEV for the maintenance of PS in patients with pr-GBMs, we evaluated the differences in KPS scores before and after the initial treatment for Type I and II patients. The rate of clinical deterioration (defined as a ≥20 point reduction in KPS scores, according to the AVAglio studyCitation2) during the initial treatment was significantly lower (2-sided Fisher’s exact test, P=0.024) in Type I patients (n=1; 7.7%) than in Type II patients (n=9; 47.4%). This suggests that add-on BEV contributes to the prevention of early clinical deterioration in Type I patients.
We hypothesized that the benefit of BEV treatment (ie, prevention of early clinical deterioration) may contribute more favorably to the prolongation of survival in patients with a poor PS. To investigate this, we evaluated the survival outcomes of patients (n=36) with a poor PS, defined as having a KPS score of ≤70 that is equivalent to a PS of ≥2 according to the criteria of the WHO, indicating that patients cannot carry out normal daily activities. The OS rate of Type I patients (n=10) was significantly higher than that of Type II patients (n=11; P=0.017; ), but comparable to that of Type III patients (n=15). According to the multivariate Cox proportional hazards model of OS in patients with a poor PS, BEV treatment appeared to be a significant prognostic factor (P=0.026) among the clinical and molecular markers analyzed (). These findings suggest that the contribution of add-on BEV to the prolongation of OS was more apparent in patients with a poor PS.
Figure 3 Kaplan–Meier estimates of OS rate in newly diagnosed glioblastoma patients with a poor performance status (n=36) stratified according to treatment type.
Abbreviations: BEV, bevacizumab; CCRT, concurrent radiotherapy; GTR, gross-total removal; OS, overall survival; PR, partial removal; TMZ, temozolomide.
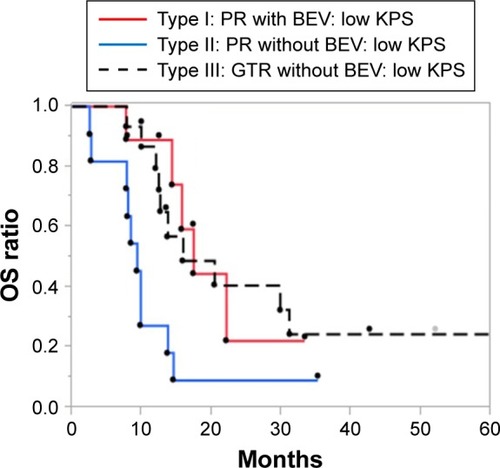
Table 4 Prognostic factors of OS in newly diagnosed, partially resected glioblastoma patients with a poor performance status (n=21)
BEV toxicity
During the course of CCRT, the BEV-related toxicities that led to treatment being discontinued included Grade II neutropenia and thrombocytopenia that were each observed in a single patient. With the exception of myelosuppression, discontinuation of maintenance treatment with BEV due to treatment-related toxicities occurred in 3 Type I patients, including 2 patients with deep vein thrombosis (DVT) and 1 patient who presented with gradually progressive brain atrophy after 1 year of maintenance with BEV.
Discussion
In our retrospective analysis of PFS, add-on BEV to the initial treatment of patients with pr-GBMs was associated with a better outcome and significantly lower HR compared to TMZ treatment alone, as similarly shown in previous Phase III clinical trials.Citation2,Citation5 We also demonstrated that the PS is well maintained in patients treated with BEV compared to patients treated without BEV. Furthermore, in an analysis of OS, we showed a favorable outcome of add-on BEV treatment for patients with pr-GBMs, especially those with a poor PS. Taken together, our findings from this retrospective study suggest that add-on BEV treatment may contribute to the prevention of early clinical deterioration and lead to a prolongation of OS more favorably in pr-GBM patients with a poor PS. Our result might also support the rescue use of BEV for patients whose condition deteriorates.
Previous large clinical trialsCitation2,Citation5 have failed to demonstrate a prolongation of OS in nd-GBM patients treated with add-on BEV in addition to the standard initial treatment. With respect to pr-GBM patients as well, a beneficial effect was not demonstrated in a subanalysis of data from the AVAglio trial,Citation2 which revealed no effect of BEV treatment on the outcomes of patients who underwent partial resections;Citation13 thus, revealing a discrepancy between previous findings and the findings of our present study. The most notable difference between the backgrounds of the patients in our study compared to the patients of the aforementioned clinical trials is their PS. In the AVAglio trial,Citation2 a WHO PS of ≤2 (equivalent to a KPS score of ≥60) was an inclusion criterion and ~50% of the enrolled patients maintained a WHO PS of 0 (equivalent to a KPS score of 90–100). Conversely, KPS scores in ~50% of pr-GBM patients in the present study were ≤70. In real clinical settings, as demonstrated in the present study, general GBM populations include considerably more patients with a poor PS, in contrast to the AVAglio trial.Citation14 The strict selection criteria for participation in clinical trials can lead to such specificity of the enrolled patients.Citation15
The beneficial effect of add-on BEV for patients with a poor PS was suggested even in a subanalysis of the AVAglio trialCitation2 that revealed lower HRs for PFS in patients with a WHO PS of 1–2 compared to patients with a WHO PS of 0. Another exploratory analysis of the AVAglio trialCitation15 revealed that BEV prolonged the OS of patients who did not receive second-line therapy after disease progression. The primary purpose of this exploratory analysis was to evaluate the impact of poststudy crossover. However, the authorsCitation15 took into consideration the fact that a greater proportion of patients with poor prognostic features may be included in this analysis. In the present study, 51.7% of pr-GBM patients did not receive second-line therapy after progression, and the ratio was considerably higher than that of the AVAglio trialCitation15 (24.4%). These findings suggest that pr-GBM patients are less likely to receive second-line therapy after progression. That is, BEV is used as a treatment option for recurrent tumors would not contribute reliably to the favorable outcome of pr-GBM patients. Taken together, it is assumed that BEV would be more beneficial for the treatment of nd-GBM patients with a progressive clinical course (eg, pr-GBM patients with a poor PS) than for the usual patient populations enrolled in clinical trials.
Our study has several limitations, including its non-randomized retrospective design and the small number of enrolled patients from a limited number of institutions, and using a historical control group that may include a considerable proportion of patients who might not have received BEV as salvage treatment. Use of historical controls might be problematic because insights and techniques of treatment for glioma are changing over time. To obtain more credible results and to elucidate further the precise benefits of BEV treatment in real clinical settings, future clinical trials, without limiting criteria (eg, PS), are warranted.
In our case series, DVT was the only apparent adverse event that resulted in the discontinuation of BEV treatment. In the safety data from the AVAglio trial,Citation16 the incidence of arterial thromboembolic events was higher in the BEV-treated group compared to the placebo control group (5.9% vs 1.6%, respectively). A meta-analysisCitation17 revealed a trend toward a significant association between BEV treatment and the risk of developing DVT and pulmonary embolisms. Another complication suspected to be associated with the discontinuation of BEV treatment was brain atrophy. A previous studyCitation18 reported that prolonged BEV treatment was associated with brain atrophy. In a laboratory investigation,Citation19 long-term BEV treatment was found to be associated with a reduction in the dendritic length of hippocampal neurons. Nonetheless, our treatment approach can contribute to relatively long-term survival for patients with pr-GBMs (ie, it should lead to more frequent adverse events derived from prolonged treatment). Although a previous reportCitation20 suggested that discontinuation of BEV treatment could lead to a rebound phenomenon, recent studiesCitation21,Citation22 revealed that treatment intervals of BEV were not associated with a poor outcome. For the purpose of maximizing the clinical benefits to patients, further efforts to address the appropriate timings of BEV discontinuation will be required.
Acknowledgments
The authors wish to thank Dr Hiroshi Muratani (Ando Hospital, Fukuoka, Japan) for his provision of clinical data and Ms Fumie Doi (Kyushu University) for her technical assistance. This work was supported by a Japanese Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) Award (Grant No 26462185, 25293311, 15K15529 and 16K10779).
Supplementary materials
Detection of hot spot mutations in glioblastoma tissues
Mutation detection of the IDH1 (codon 132), IDH2 (codon 172), BRAF (codon 600), and H3F3A (codons 27 and 34) genes was performed by high-resolution melt (HRM) analysis and subsequent Sanger sequencing. Primer sequences for the amplification of genomic DNA were designed using Primer3PlusCitation1 (http://bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/). In silico polymerase chain reaction (PCR) applications (http://genome.ucsc.edu/cgi-bin/hgPcr) were used to verify the theoretical specificity of the forward and reverse primers. Details of the primer sequences and their amplicon sizes are provided in . Whole HRM reactions were prepared using 16.6 ng of DNA, 7.47 pmol/L of each of the forward and reverse primers, and 10 µL of MeltDoctor HRM Master Mix (Applied Biosystems®, Tokyo, Japan) in a total volume of 20 µL, according to the manufacturer’s protocol. An ABI 7,500 Fast Real-Time PCR System (Applied Biosystems) was used for amplification. The cycling conditions were as follows: 1) an initial denaturation step at 95°C for 10 minutes; 2) 40 cycles of 95°C for 15 seconds, followed by 60°C for 1 minute; and 3) a dissociation cycle of 95°C for 15 seconds, followed by 60°C for 1 minute, and 95°C for 15 seconds. Mutations were determined according to our previous study.Citation2 Thereafter, HRM products were purified using ExoSap-IT (Affymetrix Inc., Santa Clara, CA, USA) for mutation genotyping. Cycle sequencing was performed using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). After purification of the products, electrophoresis and analysis were conducted using an ABI PRISM® 310 Genetic Analyzer (Applied Biosystems).
PCR and sequencing of the TERT promoter were performed according to a previous study with slight modifications.Citation3 In our study, we designed the following oligonucleotide primers using Primer3Plus:Citation1 5′-GGCCGATTCGACCTCTCT-3′and 5′-CAGCGCTGCCTGAAACTC-3′. PCR reactions were performed in 10 µL volumes containing ~20 ng of DNA, 0.1 µL of TaKaRa LA Taq® DNA polymerase (TaKaRa Bio Inc., Shiga, Japan), 5 µL of 2× GC Buffer I, 1.6 µL of the deoxyribonucleoside triphosphate mixture (2.5 mM of each dNTP), and 1.67 µL of each of the primers (2 µM). The cycling conditions were as follows: 1) an initial denaturation step at 95°C for 5 minutes; 2) 35 cycles of 94°C for 30 seconds, followed by 62°C for 30 seconds, and 72°C for 30 seconds; and 3) a final elongation step at 72°C for 7 minutes. The PCR products were gel purified and sequenced on an ABI 310® PRISM Genetic Analyzer (Applied Biosystems).
Table S1 Primer sequences
References
- UntergasserANijveenHRaoXBisselingTGeurtsRLeunissenJAPrimer3Plus, an enhanced web interface to primer3Nucleic Acids Res200735Web Server issueW71W7417485472
- HataeRHataNYoshimotoKPrecise detection of IDH1/2 and BRAF hotspot mutations in clinical glioma tissues by a differential calculus analysis of high-resolution melting dataPLoS One2016118e016048927529619
- ChenCHanSMengLLiZZhangXWuATERT promoter mutations lead to high transcriptional activity under hypoxia and temozolomide treatment and predict poor prognosis in gliomasPLoS One201496e10029724937153
Disclosure
The authors report no conflicts of interest in this work.
References
- StuppRMasonWPvan den BentMJRadiotherapy plus concomitant and adjuvant temozolomide for glioblastomaN Engl J Med20053521098799615758009
- ChinotOLWickWMasonWBevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastomaN Engl J Med2014370870972224552318
- TakanoSIshikawaENakaiKBevacizumab in Japanese patients with malignant glioma: from basic research to clinical trialOnco Targets Ther201471551156225228814
- CohenMHShenYLKeeganPPazdurRFDA drug approval summary: bevacizumab (avastin) as treatment of recurrent glioblastoma multiformeOncologist200914111131113819897538
- GilbertMRDignamJJArmstrongTSA randomized trial of bevacizumab for newly diagnosed glioblastomaN Engl J Med2014370869970824552317
- O’ConnorJPCaranoRAClampARQuantifying antivascular effects of monoclonal antibodies to vascular endothelial growth factor: insights from imagingClin Cancer Res200915216674668219861458
- SchindlerGCapperDMeyerJAnalysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytomaActa Neuropathol2011121339740521274720
- SturmDWittHHovestadtVHotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastomaCancer Cell201222442543723079654
- HataNYoshimotoKHataeRDeferred radiotherapy and upfront procarbazine-ACNU-vincristine administration for 1p19q codeleted oligodendroglial tumors is associated with favorable outcome without compromising patient performance, regardless of WHO gradeOnco Targets Ther201697123713127895504
- HataeRHataNYoshimotoKPrecise detection of IDH1/2 and BRAF hotspot mutations in clinical glioma tissues by a differential calculus analysis of high-resolution melting dataPLoS One2016118e016048927529619
- ArakiYMizoguchiMYoshimotoKQuantitative digital assessment of MGMT immunohistochemical expression in glioblastoma tissueBrain Tumor Pathol2011281253121249460
- WenPYMacdonaldDRReardonDAUpdated response assessment criteria for high-grade gliomas: response assessment in neurooncology working groupJ Clin Oncol201028111963197220231676
- SandmannTBourgonRGarciaJPatients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trialJ Clin Oncol201533252735274426124478
- NaritaYShibuiSCommittee of Brain Tumor Registry of Japan Supported by the Japan Neurosurgical Society. Trends and outcomes in the treatment of gliomas based on data during 2001–2004 from the brain tumor registry of JapanNeurol Med Chir (Tokyo)2015554286295
- ChinotOLNishikawaRMasonWUpfront bevacizumab may extend survival for glioblastoma patients who do not receive second-line therapy: an exploratory analysis of AVAglioNeuro Oncol20161891313131827006178
- SaranFChinotOLHenrikssonRBevacizumab, temozolomide, and radiotherapy for newly diagnosed glioblastoma: comprehensive safety results during and after first-line therapyNeuro Oncol2016187991100126809751
- LiXHuangRXuZRisk of adverse vascular events in newly diagnosed glioblastoma multiforme patients treated with bevacizumab: a systematic review and meta-analysisSci Rep201551469826423913
- BagAKKimHGaoYProlonged treatment with bevacizumab is associated with brain atrophy: a pilot study in patients with high-grade gliomasJ Neurooncol2015122358559325711673
- LatzerPSchlegelUTheissCMorphological changes of cortical and hippocampal neurons after treatment with VEGF and bevacizumabCNS Neurosci Ther201622644045026861512
- ZunigaRMTorcuatorRJainRRebound tumour progression after the cessation of bevacizumab therapy in patients with recurrent high-grade gliomaJ Neurooncol201099223724220151176
- AndersonMDHamzaMAHessKRPuduvalliVKImplications of bevacizumab discontinuation in adults with recurrent glioblastomaNeuro Oncol201416682382824596117
- HertensteinAHielscherTMennOImpact of tapering and discontinuation of bevacizumab in patients with progressive glioblastomaJ Neurooncol2016129353353927422128
