Abstract
Breast cancer, the most common cancer among women, is a clinically and biologically heterogeneous disease. Numerous prognostic tools have been proposed, including gene signatures. Unlike proliferation-related prognostic gene signatures, many immune-related gene signatures have emerged as principal biology-driven predictors of breast cancer. Diverse statistical methods and data sets were used for building these immune-related prognostic models, making it difficult to compare or use them in clinically meaningful ways. This study evaluated successfully published immune-related prognostic gene signatures through systematic validations of publicly available data sets. Eight prognostic models that were built upon immune-related gene signatures were evaluated. The performances of these models were compared and ranked in ten publicly available data sets, comprising a total of 2,449 breast cancer cases. Predictive accuracies were measured as concordance indices (C-indices). All tests of statistical significance were two-sided. Immune-related gene models performed better in estrogen receptor-negative (ER−) and lymph node-positive (LN+) breast cancer subtypes. The three top-ranked ER− breast cancer models achieved overall C-indices of 0.62–0.63. Two models predicted better than chance for ER+ breast cancer, with C-indices of 0.53 and 0.59, respectively. For LN+ breast cancer, four models showed predictive advantage, with C-indices between 0.56 and 0.61. Predicted prognostic values were positively correlated with ER status when evaluated using univariate analyses in most of the models under investigation. Multivariate analyses indicated that prognostic values of the three models were independent of known clinical prognostic factors. Collectively, these analyses provided a comprehensive evaluation of immune-related prognostic gene signatures. By synthesizing C-indices in multiple independent data sets, immune-related gene signatures were ranked for ER+, ER−, LN+, and LN− breast cancer subtypes. Taken together, these data showed that immune-related gene signatures have good prognostic values in breast cancer, especially for ER− and LN+ tumors.
Introduction
Breast cancer is the most common cancer among women. As a clinically and biologically heterogeneous disease, accurate prognostic predictions are very important for breast cancer treatment planning. In the past, the pathological Tumor-Node-Metastasis Classification of Malignant Tumors staging system of the American Joint Committee on Cancer was the predominant prognostic prediction tool. In recent decades, molecular breast cancer subtypes have been identified based on pathological markers including estrogen receptor (ER), progesterone receptor, human epidermal growth factor 2 (HER2), and Ki67. Measured expression levels of these biomarkers are routinely used in clinical practices to stratify patients for prognostic predictions and to select treatments.Citation1–Citation4 In particular, four distinct subtypes of breast cancers have been defined: luminal A-like, luminal B-like, HER2-positive, and triple negative. These subtypes were first adopted by the St Gallen International Expert Consensus panel in 2013 as a basis for selecting adjuvant systemic therapy regimens (ie, endocrine therapy, chemotherapy, or anti-HER2 therapy).Citation2 Nevertheless, diagnosis and treatment decision-making for breast cancer still relies on classical histopathological and immunohistochemical techniques. More quantitative diagnostic approaches and individualized treatment plans remain unmet needs. To meet these needs, multigene prognostic models have emerged, including 21-gene recurrence score,Citation5–Citation7 PAM50,Citation8–Citation13 Mammaprint®,Citation14–Citation16 and Breast Cancer Index™.Citation17–Citation20
Historically, prognostic multigene predictors strictly consisted of proliferation-associated genes.Citation21,Citation22 The tests were highly effective at classifying recurrence risks for ER-positive (ER+) and lymph node–negative (LN−) breast cancer subtypes; prognoses for these subtypes were well predicted by Ki67 indices or levels of DNA synthesis or cell cycle regulatory genes. Although most ER-negative (ER−) or LN-positive (LN+) breast cancers are highly proliferative, multigene predictors do not have discriminative power for prognoses within either subtype. Therefore, alternative prognostic signatures must be proposed for these subtypes that are not limited to proliferation-related genes.Citation23,Citation24 Recent preclinical studies have underscored inflammation and the stromal immune landscape as important drivers of breast cancer. There has been growing interest in establishing prognostic multigene signatures focused on immune-related genes, such as the STAT-1 gene, interferons, and immune response genes.Citation25–Citation32
Currently, no immune-related signature has been widely accepted in clinical practice; it is unknown whether one test is the most robust. In this study, all published immune-related prognostic gene models were systematically reviewed to evaluate performances across independent data sets. The aims of this study were two-fold: first, to investigate the prognostic effects of immune-related gene signatures and, second, to identify the most robust signature for the prognostication of nonmetastatic breast cancer.
Methods
The design of this study had two major sections. First, published literature describing prognostic immune-related gene signatures and publicly available genomic data sets were systematically reviewed. Second, a multistudy evaluation of immune-related prognostic models that met specific inclusion criteria was performed. The literature search strategies as well as inclusion criteria of prognostic models and data sets are detailed in the following subsections.
Literature search strategies
Systematic literature searches of articles published between July 2006 and March 2017 were performed independently by two authors (KC and JZ). In both cases, Internet-based searches of the PubMed and Web of Science platforms were performed without language or regional restrictions, using combinations of the following keywords: “breast cancer,” “breast neoplasm,” “breast carcinoma,” “immune-related gene,” “immune response gene,” “prognostic model,” and “immune-related predictor.” Reference lists of retrieved articles were manually screened to identify relevant articles. If a single study generated multiple publications, then either the highest quality publication or the most recent publication was chosen for subsequent analyses.
Inclusion criteria of prognostic models
Published prognostic models of immune-related gene signatures were evaluated for inclusion in the current study. Eligible studies included the following: 1) a claim of providing prognostic value for nonmetastatic breast cancer cases; 2) a model that was based on mRNA expression levels, as quantified by microarray technologies; 3) a model that was developed using a training data set and then validated with a test data set; 4) a training data set of at least 30 subjects; 5) defined outcome measurements of either overall survival, disease-free survival (DFS), or recurrence-free survival (RFS); 6) a model that was fully specified or could be rederived from the specified data set; and 7) a model for which ≥80% of genes could also be identified in the validation data set.
Inclusion criteria of data sets and samples
Publicly available microarray data sets were selected as validation data sets for this study. Eligible data sets included the following: 1) gene expression data gathered from human nonmetastatic breast cancer sources; 2) continuous time-to-event survival data; 3) at least 40 samples with 15 deaths; 4) measurements of gene expression profiles using U133A GeneChip technologies (Affymetrix/Thermo Fisher Scientific, Santa Clara, CA, USA); and 5) clinical characteristics of the enrolled subjects, including ER levels and LN status.
Evaluation of published prognostic models on independent data
Immune-related gene prognostic models that met the inclusion criteria were evaluated on the included data sets. CEL files containing pixel intensity calculations of the validation data sets were downloaded from public databases (such as the Gene Expression Omnibus). Gene expression levels were normalized based upon background corrections. For each prognostic model, the risk was calculated for each sample in the validation data set. The risk score calculation was defined as follows: score = Σwixi/Σ|wi|, where xi was the expression level of the involved gene in the model, and wi was either +1 or −1, depending on the correlation between the gene and the prognosis of the paired subject described in the original publication.Citation23,Citation24,Citation33
Concordance indices (C-indices) were calculated as previously described.Citation34 For each model tested on each validation data set, the C-index was defined as the probability that a patient was predicted to have a lower risk score when compared to another patient who survived longer than the first patient. A C-index of 0.5 indicated that the model did not predict risk better than random guess. A score of 1 indicated perfect prediction of risk. C-indices generated across all of the validation data sets were synthesized into a summary value using random effects modeling. All computations were completed using R (version 3.3.2).
Stratification by ER status and LN status
The discriminative power of each immune-related prognostic model for predicting breast cancer prognosis was evaluated in patients depending on ER and LN status. Patients were stratified into four subgroups (ER+ versus ER− and LN+ versus LN−). C-indices were computed in each of these four subgroups for every data set and then synthesized into a final assessment.
Statistical analyses
All outcomes were reported with 95% CIs. Statistical heterogeneities between data sets were assessed using χ2 tests, with significance levels set to P<0.05. Statistical tests were two-sided, and P-values <0.05 were considered statistically significant. Random effects modeling was used if there was significant heterogeneity; otherwise, fixed effects modeling was reported.
Results
A systematic review of prognostic immune-related gene signatures for breast cancer was performed. After excluding redundant literature, a total of 393 papers were investigated. From these papers, eight models emerged as candidates that passed the study inclusion criteria and were selected for further analyses (). summarizes the features of the included models. The performance of each model was evaluated in each of ten data sets that were selected based on above-stated inclusion criteria ().
Table 1 Features of eight published models that prognosticate breast cancer outcomes
Table 2 Features of ten publicly available microarray data sets used to validate published risk scores
Heterogeneity analyses
Heterogeneities across data sets were assessed using each immune-related gene model. Based on the results, C-indices of summary values were synthesized using either random effects or fixed effects models. Both methodologies produced similar summary values, resulting in similar ranking patterns for the immune-related gene models. Given that significant heterogeneity was identified in several models (), the random effects modeling methodology was chosen to synthesize C-indices across the prognostic models.
Table 3 Heterogeneity analyses for eight publicly available models
Evaluation of prognostic models on independent data sets
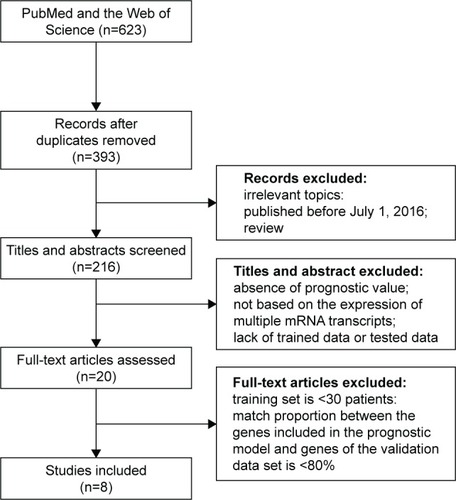
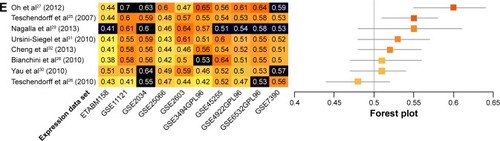
Prognostic accuracy of each model was evaluated in every data set, with the exception of the training data set used by the original authors. Evaluations were quantified by C-indices (), and the models were ranked by summary values, from high to low. The top-ranked model was authored by Oh et al,Citation27 with a summary C-index of 0.61. Two other models had better than chance predictive values, with summary C-indices of 0.54 and 0.53, respectively.Citation25,Citation32
Figure 2 Performance assessment of published immune-related risk scores.
Abbreviations: C-indices, concordance indices; ER, estrogen receptor; LN, lymph node.
Stratification by known prognostic factors
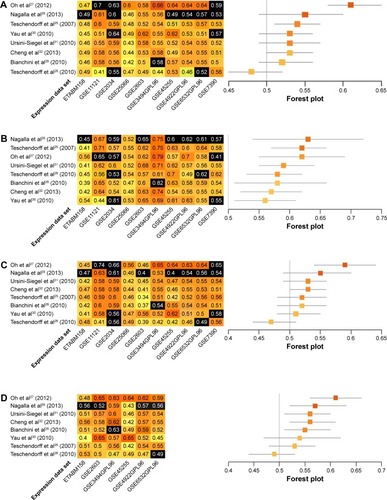
Further analyses were performed within subgroups that were stratified by ER status (ER+ versus ER−), or LN status (LN+ versus LN−). Only a subset of data sets, those in which these factors were recorded, could be assessed. Most immune-related gene models performed better in ER− data sets. The top-ranked three models for ER− breast cancer were authored by Nagalla et al,Citation29 Teschendorff et al,Citation25 and Oh et al.Citation27 These models achieved summary C-indices between 0.62 and 0.63. The remaining five modelsCitation26,Citation28,Citation30–Citation32 also provided predictions that were better than chance, with summary C-indices varying from 0.56 to 0.59 (). Interestingly, summary C-indices in data from ER+ subjects were lower than in data from ER− subjects. The two top-ranking models for ER+ subjects were those authored by Oh et alCitation27 and Cheng et alCitation32 with summary C-indices of 0.59 and 0.53, respectively. The other six models showed no predictive advantage above chance in ER+ subjects ().
Data were also stratified by the status of invasion to the LNs. The prognostic models performed better in LN+ data sets. The four top-ranking models for LN+ were designed by Oh et al,Citation27 Nagalla et al,Citation29 Ursini-Siegel et al,Citation31 and Cheng et al.Citation32 Summary C-indices in these models ranged from 0.56 to 0.61. In the remaining four models,Citation25,Citation26,Citation28,Citation30 no predictive advantages above chance were observed (). Synthesized C-indices for LN− subjects were lower than those calculated for LN+ subjects. Only two models, originally published by Oh et alCitation27 and Teschendorff et al,Citation25 predicted outcomes better than chance ().
Univariate and multivariate analyses
Univariate analyses were used to investigate correlations between risk scores and the presence of ERs or LN invasion. Risk scores from the prognostic models designed by Nagalla et al,Citation29 Teschendorff et al,Citation25 Cheng et al,Citation32 Bianchini et al,Citation28 Yau et al,Citation30 and Ursini-Siegel et alCitation31 significantly correlated with ER status (range of odds ratios: 1.01–1.06; P<0.05). Risk scores from the models of Oh et alCitation27 and Yau et alCitation30 significantly correlated with LN status (odds ratios: 1.02 and 1.22, respectively; P<0.05). In multivariate analyses that controlled for the status of ER, LN, HER2, and age, DFS was associated with risk scores in the models of Oh et alCitation27 (P=0.0443), Cheng et alCitation32 (P=0.0391), and Bianchini et alCitation28 (P=0.0343). These findings demonstrated that modeling immune-related gene signatures may provide unique information to prognosticate breast cancer cases.
Discussion
Gene expression profile studies on breast cancer have uncovered new insights into the genetic pathways that contribute to tumorigenesis. This research has inspired the development of gene expression signatures to predict patient outcomes. It is generally understood that proliferation-associated genes account for the majority of predictive power of many previously reported prognostic genetic tools, such as the 21-gene Oncotype DX test (Genomic Health, Inc., Redwood City, CA, USA).Citation5,Citation7 However, these prognostic models are only applicable for low proliferation tumors, as found in ER+ and LN− breast cancer subtypes. Therefore, developing new prognostic signatures, independent of proliferation phenotypes, is a challenging unmet need.Citation23,Citation24
In recent years, research revealed that immune-related gene signatures were associated with both clinical outcomes and pathological complete response rates in breast cancer.Citation25,Citation35–Citation37 A whole-transcriptome analysis of the NCCTG-N9831 adjuvant trastuzumab trial in early-stage HER2+ breast cancer revealed that tumors enriched for immune-functioning genes had increased RFS rates. Conversely, trastuzumab did not increase RFS in patients whose tumors lacked elevated immune-functioning gene expression scores. These data suggested that the increased expression of a subset of immune-functioning genes predicted benefit from adjuvant trastuzumab.Citation38 Additionally, the Finland Herceptin trial reported that higher levels of tumor-infiltrating lymphocytes at diagnosis had a significant inverse association with distant recurrence rates in primary ER−/HER2− breast cancers.Citation39
Previously, investigators pooled genomic data to identify predictors of clinical outcomes in breast cancer.Citation25–Citation32 The current study is the largest pooled analysis of immune-related prognostic gene models for breast cancer; all published immune-related prognostic gene models were evaluated. For ER− breast cancer, the top-ranking risk score was achieved by the model of Nagalla et al.Citation29 This model comprised three distinct immunity-related metagenes indicative of tumor–immune cell interactions. The first metagene, the B/P cluster, consisted primarily of immunoglobulin-encoding genes that were expressed in B-cell and plasma cell populations. The second metagene was the T/NK cluster, which contained components of the T-cell receptor–cluster of differentiation 3 complex and granzymes. This was largely restricted to T cells and natural killer cells. The third metagene was the M/D cluster, which included the major histocompatibility complex class II and myeloid-specific markers. These were expressed at the highest levels in monocytes and dendritic cells. This prognostic model covered the most comprehensive array of immune-related genes in the cohort studied here. This may explain why the model of Nagalla et al had the highest prognostic value.
In the current study, seven of ten validation data sets were training data sets used to develop the model of Nagalla et al.Citation29 This reduced the statistical power for the validation of this model. Therefore, the model of Teschendorff et al,Citation25 the second best prognostic model for ER− breast cancer, may have the most clinical value. This model employed a methodology called profile analysis using clustering and kurtosis (PACK). PACK was a semi-supervised algorithm, consisting of two main steps. First, a feature selection criterion was performed. Second, a supervised step correlated the selected features with specific phenotypes.Citation40 Despite being a conservative procedure that removed true positives, PACK efficiently removed false positives, providing more reliable identifications of prognostic markers. Another advantage of PACK was that it tested prognostic genes by applying two statistical tools: singular value decompositionCitation41 and the shrunken centroids classifier.Citation42 Two of the potential prognostic genes identified by the model of Teschendorff et alCitation25 were associated with breast cancer clinical outcomes. Specifically, C1QA, a gene involved in the classical complement pathway, harbored a single nucleotide polymorphism that correlated with distant breast cancer metastases.Citation43 Two other studies also found prognostic value for OPN in metastatic breast cancer.Citation44,Citation45
In the current study, the prognostic model by Oh et alCitation27 ranked first in ER+ breast cancer and third in ER− breast cancer. This was the only model that integrated proliferation- and immunity-related genes into a single prognostic model for nonmetastatic breast cancer patients. Additionally, this model used a large data set, which reduced size-based sampling problems. Prognostic genes were selected based upon both statistical significance and biological relevance. Together, these factors explain the success of the model of Oh et al in prognosticating breast cancer outcomes.
The current study also revealed that immune-related gene models performed best in ER− and LN+ breast cancer subtypes. This finding directly opposed current proliferation-related gene models. ER− and LN+ breast cancers tend to present as high-grade diseases that frequently harbor p53 mutations, leading to worsen prognoses. These molecular and pathological tendencies may explain why differences in clinical outcome within the ER− subgroup cannot be explained by the differential activation of cell cycle pathways. Thus, proliferation-related gene models do not have prognostic power in this breast cancer subtype. Unlike proliferation-related functional gene signatures, immune-related gene expression profiles have more individualized properties. Immune-related models have achieved statistical significance in both high and low proliferation breast neoplasms.Citation29 Therefore, immune-related gene signature–based tools may be advantageous for the prognostication of ER− and LN+ breast cancers. Future use of these tools may complement more established proliferation-based gene expression panels.
It is of high clinical relevance to identify patients who will experience poor outcomes prior to the initiation of therapy. Patients with lower immune-related gene expression scores may require increased follow-up frequency and systemic management after traditional adjuvant therapy (eg, metronomic chemotherapy for triple-negative breast cancer or ovary functional suppression for ER+ breast cancer). Alternatively, these patients might be candidates for future clinical trials evaluating therapeutic approaches to enhance immune activity within breast tumors, such as by targeting checkpoint proteins.Citation46
The coverage of immune-related gene signatures included many critical and well-studied immunomodulatory genes, such as TGFβ, CD28, and CTLA4. Members of the immunomodulatory Siglec gene family were not included in these models. Most of the Siglec family members contain immunoreceptor tyrosine-based inhibitory motifs in regions exposed to cytosolic compartments. Siglec proteins are inhibitory receptors for both innate and adaptive immune cells and may attenuate reductions in tumor cell immunity. CD169/Siglec-1 is bound to cell surface–associated (MUC1) receptors that were expressed on breast cancer cells.Citation47 High densities of CD169+ macrophages in regional LNs surrounding colorectal tumors, endometrial carcinomas, malignant melanomas, and breast neoplasms were associated with improved clinical prognoses. These findings may be due to increases in cytotoxic immune cell infiltrates in these tumors.Citation48–Citation50 Therefore, expression levels of Siglec immunomodulatory gene family members could affect prognostic breast cancer outcomes. Functions of Siglec gene products have been well studied in oncogenic model systems. Further investigations should concentrate on clinical oncology. Future immune-related prognostic gene models for breast cancer should consider assessing Siglec gene expression levels.
The study reported here had several limitations. First, the selected models included only intracellular immune-related genes. Therefore, it remains unknown whether interstitial genes in the microenvironment, such as CCL-18,Citation51 can prognosticate breast cancer patient outcomes. Second, useful gene expression profiles that could be included in this study were limited by incomplete clinical information. This caveat was an unavoidable inadequacy found in all publicly available data sets. Third, evaluations of these gene models were limited to the breast cancer subtypes discussed above. Few authors have investigated the impact of immune-related prognostic gene models on conventional breast cancer subtypes. Few studies have published validation data sets containing clinical information that specifies patient molecular subtypes. Rather, previous studies published information specifying ER status and regional LN involvement. This allowed, in the current study, for the stratification of summary C-indices by these characteristics. The model by Nagalla et alCitation29 was the only tool in which subgroup analyses were performed in subtypes. In those studies, basal-like breast tumors were positively associated with T/NK, M/D, and B/P metagenes. Similarly, luminal B tumors trended toward association with the T/NK metagene and were significantly associated with the M/D metagene. None of the metagenes were significantly associated with the luminal A, HER2-enriched, or claudin-low tumor subtypes.
Despite these limitations, the current analysis revealed the top-ranked immune-related prognostic gene models for ER+, ER−, LN+, and LN− breast cancer subtypes. Moreover, these data showed that immune-related gene signatures had good prognostic value in breast cancer, especially for ER− and LN+ tumors. Therefore, immune-related gene models may be clinically relevant and complementary to current proliferation-related gene models. The use of immune-related gene models may fulfill an unmet need in breast cancer prognostication.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant numbers: 81402201, 81372817, and 11671409), the National Natural Science Foundation of Guangdong Province (grant number: 2014A030310070), and a grant ([2013]163) from the Key Laboratory of Malignant Tumor Molecular Mechanisms and Translational Medicine of the Guangzhou Bureau of Science and Information Technology.
Supplementary material
Full forms of genes provided in
STAT1: Signal transducer and activator of transcription 1
IFN: Interferons
IR: Immune response gene
C1QA: Complement C1q subcomponent subunit A
LY9: T-lymphocyte surface antigen Ly-9
TNFRSF17: Tumor necrosis factor receptor superfamily member 17
SPP1: Bone sialoprotein I
XCL2: Chemokine (C motif) ligand 2
HLA-F: HLA class I histocompatibility antigen, alpha chain F
IL2: Interleukin 2
IL12: Interleukin 12
IFNG: Interferon gamma
IL4: Interleukin 4
IL13: Interleukin 13
TGFB: Transforming growth factor beta
UBE2C: Ubiquitin-conjugating enzyme E2 C
PRC1: Protein regulator of cytokinesis 1
CCNB2: G2/mitotic-specific cyclin-B2
CDC20: The cell-division cycle protein 20
CD3D: T-cell surface glycoprotein CD3 delta chain
CD52: Cluster of differentiation 52
CCL19: Chemokine (C-C motif) ligand 19
TRBV20-1: T-cell receptor beta variable 20-1
LYN: LYN Proto-oncogene, Src family tyrosine kinase
STK17A: Serine/threonine kinase 17a
CSF1R: Colony-stimulating factor 1 receptor
PIM2: Pim-2 proto-oncogene, serine/threonine kinase
LCK: LCK proto-oncogene, Src family tyrosine kinase
STK17B: Serine/threonine kinase 17b
BTK: Bruton tyrosine kinase
HCK: HCK proto-oncogene, Src family tyrosine kinase
PRKCB1: Protein kinase C beta
PRKCQ: Protein kinase C theta
FYN: FYN proto-oncogene, Src family tyrosine kinase
ITK: IL2 Inducible T-cell kinase
ZAP70: Zeta chain of T-cell receptor associated protein kinase 70
MAP4K1: Mitogen-activated protein kinase 1
STK10: Serine/threonine kinase 10
B/P metagenes: B/P cluster consisted of genes (mostly immunoglobulin-encoding genes)
M/D metagenes: Genes of M/D cluster (including major histocompatibility complex class II and myeloid-specific markers)
T/NK metagenes: Expression of genes in T/NK cluster (such as components of the T cell receptor–cluster of differentiation 3 complex and granzymes)
CXCL13: C-X-C motif chemokine ligand 13
HAPLN1: Hyaluronan and proteoglycan link protein 1
HRBL: Arf-GAP domain and FG repeats-containing protein 2
MATN1: Matrilin 1, cartilage matrix protein
PRTN3: Proteinase 3
SSX3: SSX family member 3
RPS28: Ribosomal protein S28
EXOC7: Exocyst complex component 7
ABO: Alpha 1-3-N-acetylgalactosaminyltransferase and alpha 1-3-galactosyltransferase
CLIC5: Chloride intracellular channel 5
RFXDC2: Regulatory factor X domain containing 2
ZNF3: Zinc finger protein 3
PRRG3: Proline rich and Gla domain 3
CD247: CD247 molecule
CD3E: CD3e molecule
CD4: CD4 molecule
CD28: CD28 molecule
CTLA4: Cytotoxic T-lymphocyte associated protein 4
CXCL11: C-X-C motif chemokine ligand 11
CXCL13: C-X-C motif chemokine ligand 13
CXCL9: C-X-C motif chemokine ligand 9
CXCR3: C-X-C motif chemokine receptor 3
HLA–DQA1: Major histocompatibility complex, Class II, DQ alpha 1
IL1B: Interleukin 1 beta
ITK: IL2 Inducible T-cell kinase
LAG3: Lymphocyte activating 3
LCK: LCK Proto-oncogene, Src family tyrosine kinase
NFATC2: Nuclear factor of activated T-cells 2
PPIA: Peptidylprolyl isomerase A
PTPRC: Protein tyrosine phosphatase, receptor type C
SLAMF6: SLAM family member 6
SLAMF7: SLAM family member 7
SLAMF9: SLAM family member 9
TCRA: T-cell receptor alpha locus
RELT: RELT, TNF receptor
CD27: CD27 molecule
VTCN1: V-set domain containing T-cell activation inhibitor 1
ATRN: Attractin
CD69: CD69 molecule
ICOS: Inducible T-cell costimulator
IL7R: Interleukin 7 receptor
B2M: Beta-2-microglobulin
CD8B: CD8b molecule
GZMB: Granzyme B
IL12B: Interleukin 12B
LILRB3: Leukocyte immunoglobulin like receptor B3
TIA1: TIA1 Cytotoxic granule associated RNA binding protein
CD72: CD72 molecule
IGHG1: Immunoglobulin heavy constant gamma 1 (G1m marker)
IGHM: Immunoglobulin heavy constant Mu
IGHG2: Immunoglobulin heavy constant gamma 2 (G2m marker)
IGJ: A J chain is a protein component of the antibodies IgM and IgA.
C1S: Complement C1s subcomponent
C3: Complement C3
PTPRC: Protein tyrosine phosphatase, receptor type C
CD53: CD53 molecule
LCP2: Lymphocyte cytosolic protein 2
LAPTM5: Lysosomal protein transmembrane 5
DOCK2: Dedicator of cytokinesis 2
IL10RA: Interleukin 10 receptor subunit alpha
CYBB: Cytochrome B-245 beta chain
CD48: CD48 molecule
ITGB2: Integrin subunit beta 2
EVI2B: Ecotropic viral integration site 2B
Disclosure
The authors report no conflicts of interest in this work.
References
- PerouCMSorlieTEisenMBMolecular portraits of human breast tumoursNature2000406679774775210963602
- GoldhirschAWinerEPCoatesASPersonalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013Ann Oncol20132492206222323917950
- CoatesASWinerEPGoldhirschATailoring therapies–improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015Ann Oncol20152681533154625939896
- SotiriouCNeoSYMcShaneLMBreast cancer classification and prognosis based on gene expression profiles from a population-based studyProc Natl Acad Sci U S A200310018103931039812917485
- PaikSTangGShakSGene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancerJ Clin Oncol200624233726373416720680
- AlbainKSBarlowWEShakSPrognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trialLancet Oncol2010111556520005174
- PaikSShakSTangGA multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancerN Engl J Med2004351272817282615591335
- ParkerJSMullinsMCheangMCSupervised risk predictor of breast cancer based on intrinsic subtypesJ Clin Oncol20092781160116719204204
- PratACheangMCMartinMPrognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancerJ Clin Oncol201331220320923233704
- MartinMPratARodriguez-LescureAPAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancerBreast Cancer Res Treat2013138245746623423445
- FilipitsMNielsenTORudasMThe PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancerClin Cancer Res20142051298130524520097
- ScottEMamawalaMEpsteinJIIntermediate and longer-term outcomes from a prospective active-surveillance program for favorable-risk prostate cancer Tosoian JJ, Mamawala M, Epstein JI, Landis P, Wolf S, Trock BJ, Carter HB. J Clin Oncol. 20;33(30):3379–3385. [Epub 2015 Aug 31]. doi:10.1200/JCO.2015.62.5764Urol Oncol2017353121122
- SestakIDowsettMZabagloLFactors predicting late recurrence for estrogen receptor-positive breast cancerJ Natl Cancer Inst2013105191504151124029245
- AaldersKCKuijerAStraverMECharacterisation of multifocal breast cancer using the 70-gene signature in clinical low-risk patients enrolled in the EORTC 10041/BIG 03-04 MINDACT trialEur J Cancer2017799810528477490
- CardosoFvan’t VeerLJBogaertsJ70-Gene signature as an aid to treatment decisions in early-stage breast cancerN Engl J Med2016375871772927557300
- MakamaMDrukkerCARutgersEJAn association study of established breast cancer reproductive and lifestyle risk factors with tumour subtype defined by the prognostic 70-gene expression signature (MammaPrint®)Eur J Cancer20177551328214658
- SestakIZhangYSchroederBECross-stratification and differential risk by breast cancer index and recurrence score in women with hormone receptor-positive lymph node-negative early-stage breast cancerClin Cancer Res201622205043504827252417
- SgroiDCChapmanJABadovinac-CrnjevicTAssessment of the prognostic and predictive utility of the Breast Cancer Index (BCI): an NCIC CTG MA.14 studyBreast Cancer Res2016181126728744
- SgroiDCSestakICuzickJPrediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: a prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study populationLancet Oncol201314111067107624035531
- MathieuMCMazouniCKestyNCBreast Cancer Index predicts pathological complete response and eligibility for breast conserving surgery in breast cancer patients treated with neoadjuvant chemotherapyAnn Oncol20122382046205222112967
- PusztaiLGene expression profiling of breast cancerBreast Cancer Res200911Suppl 3S1120030862
- CheangMCChiaSKVoducDKi67 index, HER2 status, and prognosis of patients with luminal B breast cancerJ Natl Cancer Inst20091011073675019436038
- WirapatiPSotiriouCKunkelSMeta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signaturesBreast Cancer Res2008104R6518662380
- DesmedtCHaibe-KainsBWirapatiPBiological processes associated with breast cancer clinical outcome depend on the molecular subtypesClin Cancer Res200814165158516518698033
- TeschendorffAEMiremadiAPinderSEEllisIOCaldasCAn immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancerGenome Biol200788R15717683518
- TeschendorffAEGomezSArenasAImproved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modulesBMC Cancer20101060421050467
- OhEChoiYLParkTLeeSNamSJShinYKA prognostic model for lymph node-negative breast cancer patients based on the integration of proliferation and immunityBreast Cancer Res Treat2012132249950921667120
- BianchiniGIwamotoTQiYPrognostic and therapeutic implications of distinct kinase expression patterns in different subtypes of breast cancerCancer Res201070218852886220959472
- NagallaSChouJWWillinghamMCInteractions between immunity, proliferation and molecular subtype in breast cancer prognosisGenome Biol2013144R3423618380
- YauCEssermanLMooreDHWaldmanFSninskyJBenzCCA multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancerBreast Cancer Res2010125R8520946665
- Ursini-SiegelJCorySZuoDReceptor tyrosine kinase signaling favors a protumorigenic state in breast cancer cells by inhibiting the adaptive immune responseCancer Res201070207776778720924104
- ChengWYOu YangTHAnastassiouDDevelopment of a prognostic model for breast cancer survival in an open challenge environmentSci Transl Med20135181181ra150
- IgnatiadisMSinghalSKDesmedtCGene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysisJ Clin Oncol201230161996200422508827
- UnoHCaiTXTianLWeiLJEvaluating prediction rules for t-year survivors with censored regression modelsJ Am Stat Assoc2007102478527537
- AlexeGDalginGSScanfeldDHigh expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence ratesCancer Res20076722106691067618006808
- SchmidtMBohmDvon TorneCThe humoral immune system has a key prognostic impact in node-negative breast cancerCancer Res200868135405541318593943
- BianchiniGQiYAlvarezRHMolecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancersJ Clin Oncol201028284316432320805453
- PerezEAThompsonEABallmanKVGenomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group n9831 Adjuvant Trastuzumab TrialJ Clin Oncol201533770170825605861
- LoiSMichielsSSalgadoRTumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trialAnn Oncol20142581544155024608200
- LiLChaudhuriAChantJTangZPADGE: analysis of heterogeneous patterns of differential gene expressionPhysiol Genomics200732115415917925482
- AlterOBrownPOBotsteinDGeneralized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organismsProc Natl Acad Sci U S A200310063351335612631705
- TibshiraniRHastieTNarasimhanBChuGDiagnosis of multiple cancer types by shrunken centroids of gene expressionProc Natl Acad Sci U S A200299106567657212011421
- RacilaERacilaDMRitchieJMTaylorCDahleCWeinerGJThe pattern of clinical breast cancer metastasis correlates with a single nucleotide polymorphism in the C1qA component of complementImmunogenetics20065811816465510
- AllanALGeorgeRVantyghemSARole of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancerAm J Pathol2006169123324616816376
- de Silva RudlandSMartinLRoshanlallCAssociation of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancerClin Cancer Res20061241192120016489073
- AliHRGlontSEBlowsFMPD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytesAnn Oncol20152671488149325897014
- NathDHartnellAHapperfieldLMacrophage-tumour cell interactions: identification of MUC1 on breast cancer cells as a potential counter-receptor for the macrophage-restricted receptor, sialoadhesinImmunology199998221321910610356
- OhnishiKKomoharaYSaitoYCD169-positive macrophages in regional lymph nodes are associated with a favorable prognosis in patients with colorectal carcinomaCancer Sci201310491237124423734742
- OhnishiKYamaguchiMErdenebaatarCPrognostic significance of CD169-positive lymph node sinus macrophages in patients with endometrial carcinomaCancer Sci2016107684685226991548
- ShiotaTMiyasatoYOhnishiKThe clinical significance of CD169-positive lymph node macrophage in patients with breast cancerPLoS One20161111e016668027861544
- ChenJYaoYGongCCCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3Cancer Cell201119454155521481794