Abstract
Objective
This study aimed to determine the expression, clinical significance, and possible biologic function of microRNA-324-3p in nasopharyngeal carcinoma (NPC) tissues.
Methods
In total, 54 NPC and 35 control tissues were collected. The correlation between miR-324-3p expression and the clinicopathologic characteristics was analyzed. A dual-luciferase reporter gene assay was employed to examine the predicted target gene of miR-324-3p. The miR-324-3p expression level in 5–8F cells was determined with quantitative reverse transcription-polymerase chain reaction following the transfection of miR-324-3p mimics and inhibitors. Cell proliferation and the percentage of apoptosis were measured with MTT and flow cytometry. Cell invasion ability was assessed by Transwell invasion assay.
Results
Our results showed that miR-324-3p was downregulated in the NPC tissues. The expression level of miR-324-3p in poorly differentiated NPC was significantly reduced in comparison with that in well/moderately differentiated NPC. The expression level in clinical stages III/IV was lower than that in clinical stages I/II. Moreover, the expression level of miR-324-3p was significantly lower in NPC patients with lymph node metastasis than that in NPC patients without lymph node metastasis. NPC patients with higher levels of miR-324-3p expression also demonstrated a longer survival time. Predictions from bioinformatics indicated the Hedgehog pathway transcription gene GLI3 as the target gene of miR-324-3p, and the dual- luciferase reporter assay showed that miR-324-3p is directly combined with the 3′-untranslated region of GLI3. The overexpression of miR-324-3p suppressed cell proliferation and invasion, and it enhanced apoptosis in 5–8F cells.
Conclusion
miR-324-3p can act as a tumor suppressor in NPC cells by the negative regula- tion of GLI3 gene.
Introduction
Nasopharyngeal carcinoma (NPC) cases were observed to mainly occur in the southern provinces of China and Southeast Asia, while they rarely occur in Europe, the USA, and northern China.Citation1 It is believed that the occurrence of NPC is associated with an Epstein–Barr virus infection, diet, geography, and heredity. Furthermore, previous studies have shown that NPC is controlled by multiple genes, pathways, and pathologic processes.Citation2,Citation3 After decades of research, significant progress has been made in the etiology of NPC. However, the specific mechanisms of this disease remain unclear.
miRNA refers to a small noncoding RNA encoded by an endogenous gene with a length of about 19–25 bp. These miRNAs are able to recognize specific target mRNAs at posttranscriptional levels by promoting mRNA degradation and/or inhibiting the translation process, thereby negatively regulating the expressions of target genes.Citation3 In addition, miRNAs also participate in a series of processes, such as development, hematopoiesis, fat metabolism, organogenesis, cell proliferation, differentiation, and apoptosis, which are quintessential for organisms.Citation2 Hence, miRNA expression disorders or dysfunctions may lead to the occurrence of a variety of diseases, including tumors. Many studies have shown that miRNAs also play a vital role as oncogenes or tumor suppressor genes in tumor cell proliferation, apoptosis, differentiation regulation, tumor development, and biologic functions.Citation2,Citation4 For example, the miR-34 family is directly regulated by p53 and is involved in p53-dependent tumor suppressor p21-dependent pathways,Citation5,Citation6 and its expression is often reduced in the pancreas, colon, and neuroblastomas.Citation7–Citation9 The expression of let-7 is restricted by the regulation of K-Ras and is closely related to the prognosis of lung cancer.Citation10,Citation11 In addition, miR-182 is highly expressed in melanomas, and its function is to inhibit the expression of FOXO3 and MIF, which promote melanoma metastasis.Citation12
At present, the literature on the role of miR-324-3p in the function and mechanism of NPC tumor is undefined, and its role in NPC tumor invasion and metastasis has not been reported. In this study, the expression of miR-324-3p in NPC tissues and control nasopharyngeal mucosa (including normal and inflammatory mucosa) was detected, and the relationship between miR-324-3p and clinical parameters, such as the clinical stage, metastasis, and prognosis of NPC, were evaluated. Further in vitro studies were conducted to reveal the role of miR-324-3p in NPC and to discover new diagnostic and prognostic molecular markers.
Materials and methods
Specimen collection
All clinical biopsy tissues, including 54 cases of NPC (36 males and 18 females) and 34 cases of normal and chronic inflammatory nasopharyngeal mucosal biopsy tissues (21 males and 13 females), were collected from July 2006 to October 2015 from the Guizhou Provincial People’s Hospital. The biopsy tissues were immediately stored in liquid nitrogen. All biopsy specimens were confirmed by pathology. NPC and poorly differentiated squamous cell carcinoma was determined in 49 cases, whereas a high/moderate differentiation of squamous cell carcinoma was determined in 5 cases. According to the TNM classification of malignant tumors, three cases were clinical stage I, 17 were clinical stage II, 23 were clinical stage III, and 11 cases were clinical stage IV. Lymph node metastasis was found in 42 cases and no lymph node metastasis was determined in 12 cases.
Ethics statement
The study was approved by the Ethics Committee of the Guizhou Provincial People’s Hospital. Written informed consent was obtained from each patient prior to surgery. All experimental procedures were carried out in accordance with the approved guidelines and the Declaration of Helsinki.
Cell culture
The 5–8F NPC cell line was purchased from the cell bank at the Chinese Academy of Sciences (Beijing, China). Cells were cultured in RPMI-1640 (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and incubated at 37°C in a humidified 5% CO2 atmosphere.
Cell proliferation and apoptosis
MTT assay was used to measure cell proliferation rate at 0, 24, 48, 72, and 96 h following the manufacturer’s instructions (Sigma-Aldrich Co., St Louis, MO, USA). Cell apoptosis was determined using the Annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (Thermo Fisher Scientific) and analyzed using flow cytometer (BD Biosciences, San Jose, CA, USA).
Luciferase assay
The single purification wild-type or mutant 3′-untranslated region (3′-UTR) seeding region sequence of GLI3 was transfected into pGL3 reporter (Promega, Madison, WI, USA). Then, the established reporter and the 50 nM miRNA inhibitor or mimic were cotransfected into 5–8F cells. A commercial dual-luciferase assay kit (Promega) was used to detect luciferase activity following transfection for 24 h.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Mature miRNA-324-3p expression in cells was detected by using TaqMan miRNA assays (Thermo Fisher Scientific) in an ABI 7500 real-time PCR system. The RT and PCR primers for miRNA-324-3p were purchased from Thermo Fisher Scientific. cDNA transcription process was performed using commercial kits (Takara, Tokyo, Japan). The condition of TaqMan determination was as follows: 95°C for 10 min, 40 cycles for 15 sec at 95°C, and for 60 sec at 60°C. U6 was chosen as the internal control. The relative miRNA levels were calculated using 2−ΔΔCt method.
Plasmid construction and cell transfection
The miRNA-324-3p mimic, miR-324-3p inhibitor, and negative control (Thermo Fisher Scientific) were transfected into 5–8F using Lipofectamine 2000 (Thermo Fisher Scientific) to enhance or inhibit miRNA-324-3p expression in cells. A qRT-PCR was utilized to confirm the expression of miRNA-324-3p.
Western blot analysis
Protein samples were prepared with a lysis buffer by adding protease and phosphatase inhibitors (Thermo Fisher Scientific). These proteins were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes under a semi-dry system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Next, the membranes were blocked with primary antibodies overnight at 4°C. Following two continuous washing, the membranes were then incubated with secondary antibodies. All antibodies including E-cadherin (1:500), vimentin (1:500), and β-actin (1:5,000) were purchased from Santa Cruz Company (Santa Cruz, CA, USA). Protein expression was visualized with a gel image system (Bio-Rad Laboratories Inc.).
Wound healing assay
To assess the ability of the cell migration, 5–8F cells (1×106) were seeded in six-well plates (Corning Life Sciences, Corning, NY, USA) supplemented with RPMI-1640 which contained 10% FBS. A wound was then created in the center of the cell monolayer using a sterile plastic pipette tip (Corning Life Sciences). The cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 72 h to heal the wound. An inverted microscope (Nikon Eclipse Ti; Nikon, Tokyo, Japan) was used to capture images of the cells at 0 and 72 h after wounding.
Transwell invasion assay
5–8F cells (1×106) cultured in 200 μL serum-free RPMI- 1640 medium were seeded onto the upper chambers of Matrigel®-coated Transwell® filters (pore size, 8 μm; Corning Life Sciences). Five hundred microliters of RPMI-1640 containing 10% FBS was added to the lower chambers as a chemoattractant. Following 24 h incubation at 37°C in a humidified 5% CO2 atmosphere, cells that had successfully invaded through the inserts were fixed in 4% paraformaldehyde for 30 min and stained with methylrosanilinium chloride (Sigma-Aldrich Co.). Five preselected microscopic fields were utilized to count the number of invaded cells (magnification, ×200). All experiments were repeated in triplicate.
Statistical analysis
SPSS 13.0 (SPSS Inc, Chicago, IL, USA) was used to statistically analyze the data obtained in this study. Numerical data are represented as mean ± SD. Statistical analysis was carried out using the Student’s t-test to evaluate the statistical differences between normal and gene knock-down cells. The χ2 test was used to compare the percentage distribution in different groups. Fisher’s test was performed in cases when the number of cells obtained was <5. Each experiment was repeated at least twice or was performed in triplicate. Kaplan–Meier survival curve was used to analyze the relationship between miR-324-3p expression and prognosis. P-value <0.05 was considered statistically significant.
Results
Expression of miR-324-3p in NPC and its relationship with clinical features and prognosis
Our study extracted total RNA from 54 cases of NPC and 34 cases of nasopharyngeal mucosa. The relative expression of miR-324-3p was detected using the qRT-PCR method. The results showed that miR-324-3p expression was downregulated in the NPC tissues compared with the control nasopharyngeal epithelium (). The average expression of miR-324-3p in NPC (2.35±0.33) ×10−3 was significantly lower compared to that in nasopharyngeal mucosa (7.87±1.02) ×10−3. The average expression of miR-324-3p was about 3.3 times that of NPC (P<0.05). The difference obtained was statistically significant (P=0.000).
Figure 1 Correlation between miR-324-3p expression and clinical features of NPC.
Abbreviations: Cum, cumulative; NPC, nasopharyngeal carcinoma; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
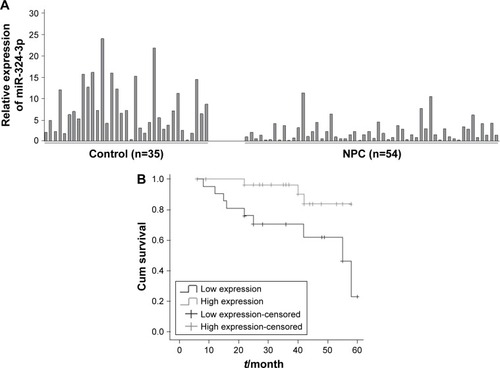
As shown in , the expression of miR-324-3p was significantly lower in poorly differentiated squamous cell carcinoma than in high/moderately differentiated squamous cell carcinoma (P=0.032). The expression of miR-324-3p was significantly lower in clinical stage III/IV NPC than in clinical stage I/II NPC (P=0.007). The expression of miR-324-3p was significantly lower in metastatic NPC than that in NPC without lymph node metastasis (P=0.019), which indicated that the miR-324-3p expression may be closely related to the clinical progression, metastasis, and pathologic differentiation of NPC. In addition, the expression level of miR-324-3p was unrelated to the age and/or sex of the patients with NPC (P>0.05).
Table 1 The relationship between miR-324-3p expression and the clinicopathologic characteristics in NPC patients
Among the 54 patients with NPC, 51 patients were followed up in the study (3 patients with NPC lost contact, leading to a loss rate of 0.06%). The follow-up time was 4 months, and the longest follow-up time was 62 months. Of the patients who were followed up, 23 patients showed a relatively low expression of miR-324-3p and 9 patients died, giving a mortality rate of 39.1%. In total, 28 patients demonstrated a relatively high level of miR-324-3p and 8 cases died, giving a mortality rate of 10.7%. The Kaplan–Meier survival analysis (log-rank test) found that the average survival time was significantly higher in the miR-324 high- expression group than in the miR-324 low-expression group (P=0.016; ). The average survival time was 58.5 and 36.3 months, respectively. Thus, our results suggest that a higher expression level of miR-324 among NPC patients can lead to a longer survival time and may deliver a better prognosis for patients afflicted with NPC.
Effects of miR-324-3p-3p expression on the migration and invasion of NPC cells
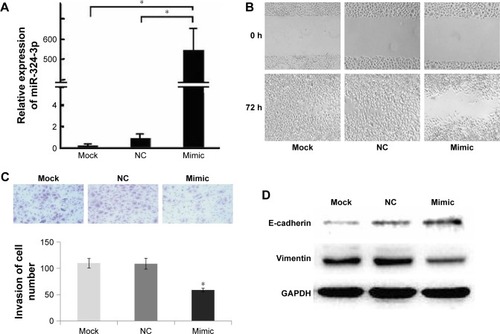
To further elucidate the effect of miR-324-3p on the migration and invasion of NPC, we first used an miR-324-3p mimic to upregulate its expression in NPC cells. The results of qPCR showed that the expression of miR-324-3p in NPC 5–8F cells was significantly upregulated after 48 min of mimic transfection compared to the control (). The scratch healing experiments showed that the scratch healing ability of 5–8F cells was significantly decreased compared to the blank control group and the negative control group (). Furthermore, we confirmed the effect of miR-324-3p on NPC cell invasion ability. The data showed that the cells passing through the polycarbonate membrane were significantly lower than those in the blank control group and the negative control group after the overexpression of miR-324-3p (). To elucidate the molecular mechanism of the regulation of NPC cell invasion and metastasis by miR-324-3p, we examined the expression of E-cadherin and vimentin. The results from the Western blot analysis showed that the expression of E-cadherin was increased in NPC 5–8F cells, while the expression of vimentin was downregulated (; P<0.05). The results highlight that miR-324-3p can promote the invasion and metastasis of NPC cells by inducing changes in epithelial–mesenchymal transition (EMT).
Figure 2 Effects of miR-324-3p-3p expression on migration and invasion of nasopharyngeal carcinoma cells.
Abbreviations: NC, negative control; qPCR, quantitative polymerase chain reaction.
Effect of miR-324-3p on the cell proliferation and apoptosis of NPC cells
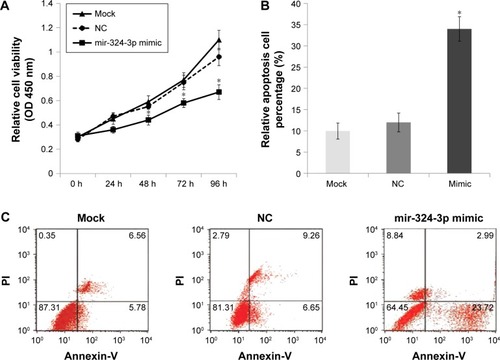
To investigate further the role of miR-324-3p in cell proliferation and the apoptosis process, 5–8F NPC cells were transfected with an miR-324-3p mimic and cultured for a period of 96 h. The cell proliferation rate was determined at 0, 24, 48, 72, and 96 h. The MTT results demonstrated a significantly decreased cell proliferation rate in cells with an overexpression of miR-324-3p (). In addition, enhanced cell apoptosis percentage was observed in the miR-324-3p transfected group in comparison with the control group ().
Figure 3 Effect of miR-324-3p on cell viability and apoptosis.
Abbreviation: NC, negative control.
GLI3 as the target gene of miR-324-3p regulated NPC cell invasion
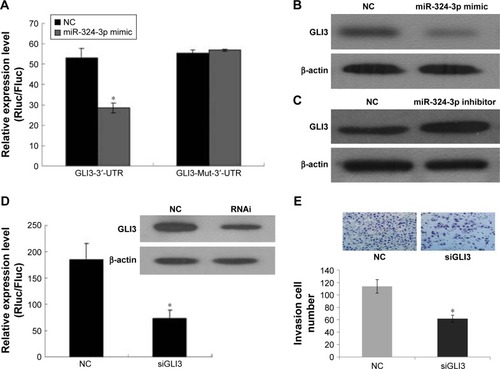
The target genes of hsa-miR-324-3p were predicted using the TargetScan, PicTar, and MicroRNA.org databases. The intersection of three software predictions was selected as possible target genes, where miR-324-3p is a conserved species and the seven bases (GAGAUGA) in its “seed region” are completely complementary to the 3′-UTR (TCATCTC) of GLI3. In addition, the free energy between miR-324-3p and GLI3 is relatively low. Thus, from the perspective of bioinformatics, it can be predicted that GLI3 may act as the target gene for miR-324-3p. To confirm this prediction, our experiment constructed the GLI3 3′-UTR expression vector and its mutant vector. Luciferase results confirmed that miR-324-3p could regulate the expression of the GLI3 gene. The expression of luciferase activity in the miR-324-3p/psiCHECK-2-GLI3-3′-UTR group was significantly inhibited, while the activity of miR-324-3p/psiCHECK-2-GLI3-Mut-3′-UTR and NC/psiCHECK-2-GLI3-Mut-3′-UTR did not exhibit significant changes (). Further, the expression of GLI3 protein was significantly lower in the miR-324-3p inhibitor group than that in the control group (). In contrast, the expression of GLI3 protein was significantly higher in the miR-324-3p mimic group than that in the control group (). Our results showed that miR-324-3p negatively correlated with GLI3 expression in 5–8F cells. We found in our previous study that an miR-324-3p mimic could decrease the migration and invasion ability of NPC cells (). After knocking down the GLI3 genes with siRNA (), the Transwell assay results showed that the number of transmembrane cells was significantly lower in 5–8F cells than that in the NC group (). In short, the results indicate that miR-324-3p may inhibit the invasion of 5–8F cells through a negative regulatory GLI3 expression.
Figure 4 GLI3 as the target gene of miR-324-3p-regulated NPC cell invasion.
Abbreviations: NC, negative control; NPC, nasopharyngeal carcinoma; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; UTR, untranslated region.
Discussion
In this study, the expression of miR-324-3p in NPC tissue samples was detected and the clinical significance of miR-324-3p was analyzed. Results showed that the expression of miR-324-3p in NPC was significantly lower than in the control nasopharyngeal epithelium. The expression of miR-324-3p was also significantly lower in poorly differentiated NPC than that in the control nasopharyngeal epithelium. Moreover, the expression of miR-324-3p was significantly lower in NPC with lymph node metastasis than that in the non-lymph node metastasis group with NPC. Overall, the results showed that miR-324-3p expression was negatively correlated with the clinical progress and metastasis of NPC, while it was positively correlated with the pathologic degree. The survival time of NPC patients with higher miR-324-3p expression levels was significantly longer compared to that in patients with lower miR-324-3p levels. The results implied that variations in the miR-324-3p level in NPC and control nasopharyngeal epithelial tissues could serve as cancer gene regulators that are involved in the occurrence and development of NPC. NPC is a highly malignant tumor that causes death through distant metastasis. Therefore, it is vital to identify effective molecular indicators that can guide the diagnosis and treatment of NPC, as well as reduce distant metastasis from the nasopharynx. The detection of miR-234-3p may become a new part of NPC diagnosis. Moreover, this study may give rise to novel treatments for NPC with new emerging targets that can promote better prognosis for NPC patients.
A new type of gene regulator, miRNA, is closely related to tumor occurrence and development. It is mainly involved in the regulation of the posttranscriptional levels of genes. The miRNAs are complementary to the 3′-UTR of their target mRNA molecules and they regulate the degradation of their target mRNA or inhibit its translation.Citation13 Recent studies have shown that miRNAs play a pivotal role in the pathogenesis of various types of tumors, including NPC. In addition, miRNAs can regulate their downstream target genes, which affects the biologic behavior of tumor cells, such as proliferation, apoptosis, differentiation, movement, invasion, metastasis, and angiogenesis. Therefore, it is important to investigate the role of miRNA in tumorigenesis to enhance our knowledge on tumor diagnosis, treatment, and prognosis.Citation14,Citation15
Many studies have focused on the correlation between the serologic expression of miR-324-3p and the pathologic parameters of different diseases. Cui et alCitation16 reported that miR-324-3p in combination with five other miRNAs (miR-148a, miR-143, miR-628-3p, miR-140-5p, and miR-362-3p) in the serum can help distinguish the diagnosis of enterovirus 71 and Coxsackie virus 16, which is caused by hand, foot, and mouth infection. In addition, renal fibrosis studies have shown that miR-324-3p can be targeted to reduce the PREP protein, which promotes renal disease progression in renal fibrosis.Citation17 In the tumor study, Hu et alCitation18 found the expression of miR-324-3p and three other miRNAs (miR-16, miR-25, and miR-222) to be significantly decreased in the serum of breast cancer patients. Greenberg et alCitation19 found the obvious absence of miR-324-3p in the serum of patients with malignant melanoma, which can be an indicator of disease progression. In hepatocellular carcinoma,Citation20 miR-324-3p can serve as an early marker of liver cancer. A recent study has shown that miR-324-3p can play a regulatory role in the radiotherapy resistance of NPC through regulating its target gene, SMAD7.Citation21 However, functional research studies on miR-324-3p in NPC have not yet emerged.
In this study, we found that upregulated miR-324-3p could significantly affect the cell proliferation and apoptosis process. Moreover, the migration and invasion of NPC cells were also inhibited by regulating the EMT process. At present, tumor cell EMT changes are believed to be related to cancer invasion and metastasis.Citation22 The EMT process includes changes in the morphology and gene expressions of epithelial cells to interstitial cells, which is often associated with the downregulation of the epithelial cell marker E-cadherin and the upregulation of the interstitial cell marker vimentin.
Thus, an important question that arises from our study is how miR-324-3p obtains its regulatory function by binding to its target gene. At present, the main method of identifying the miRNA target gene is to predict the target gene using bioinformatics prediction software, such as TargetScan, PicTar, and MicroRNA.org. In our study, the bioinformatics software predicted that the target gene of miRNA-324-5p is the GLI3 gene. However, bioinformatics often predicts a large false positive for miRNA target genes, whereas miRNAs are complementary to the 3′-UTR of target mRNA by inhibiting mRNA translation or directly degrading mRNA,Citation23,Citation24 which can lead to inhibition of protein synthesis.Citation25 To avoid the false-positive results, we investigated the binding effect of miR-324-3p and GLI3 with the dual-luciferase reporter assay. The dual-luciferase assay showed that miR-324-3p was able to directly modulate the 3′-UTR of GLI3. However, the dual-luciferase reporter gene system was unable to determine whether the GLI3 protein expression was inhibited; hence, miR-324-3p mimics and miR-324-3p inhibitors were transfected into 5–8F cells. The cells were detected by the Western blot method, and the expression levels of miR-324-3p and GLI3 in 5–8F cells were confirmed to be negatively correlated. Furthermore, the results further indicated that GLI3 was the target gene of miR-324-3p. In addition, the results from the Transwell invasion showed that the invasion ability of 5–8F cells was significantly decreased after the overexpression of miR-324-3p or interference with endogenous GLI3.
GLI3 (Homo sapiens GLI family zinc finger 3) is located on chromosome 7p13 and is a member of the GLI family. GLI3 is a downstream gene of the Hedgehog-GLI (HH-GLI) signaling pathway, which was discovered by Ruppert et al in 1988.Citation26 Some studies have found that the Hedgehog (Hh) signaling pathway is directly related to the occurrence and development of tumors, and the transcription factor GLI3 can be used as a marker gene for Hh signaling pathway activation.Citation27 In the Hh signaling pathway, the transmembrane protein PTCH forms a complex with Smo and inhibits its activity. When the ligand Hh binds to the PTCH receptor, the PTCH-Smo complex is dissociated, which results in the activation of Smo. The downstream gene of GLI family regulates gene transcription, thereby activating the Hh signaling pathway. In short, the results suggested that miR-324-3p may affect the expression of GLI3 and result in inhibition of the invasion ability of 5–8F cells.
Conclusion
This study suggests that the expression of miR-324-3p may relate to the carcinogenesis, progression, and prognosis of NPC. As well, miR-324-3p can inhibit the invasion of NPC cells through the negative regulation of GLI3 gene, which sheds light on the role of miR-324-3p and the Hh pathway in NPC. In the future, our study could be utilized to provide a clinical benefit to NPC, as miR-324-3p can be used as a potential therapeutic target, a diagnostic biomarker, and/or a prognostic predictor.
Acknowledgments
We would like to thank all participants of this study.
Disclosure
The authors report no conflicts of interest in this work.
References
- WeiWIKwongDLCurrent management strategy of nasopharyngeal carcinomaClin Exp Otorhinolaryngol20103111220379395
- De SantaFIosueIDel RioAFaziFmicroRNA biogenesis pathway as a therapeutic target for human disease and cancerCurr Pharm Des201319474576423016856
- HeLHannonGJMicroRNAs: small RNAs with a big role in gene regulationNat Rev Genet20045752253115211354
- ManikandanJAarthiJJKumarSDPushparajPNOncomirs: the potential role of non-coding microRNAs in understanding cancerBioinformation20082833033418685719
- HeLHeXLimLPA microRNA component of the p53 tumour suppressor networkNature200744771481130113417554337
- WeiCLWuQVegaVBA global map of p53 transcription-factor binding sites in the human genomeCell2006124120721916413492
- ChangTCWentzelEAKentOATransactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosisMol Cell200726574575217540599
- CorneyDCFlesken-NikitinAGodwinAKWangWNikitinAYMicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growthCancer Res200767188433843817823410
- WelchCChenYStallingsRLMicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cellsOncogene200726345017502217297439
- JohnsonSMGrosshansHShingaraJRAS is regulated by the let-7 microRNA familyCell2005120563564715766527
- TakamizawaJKonishiHYanagisawaKReduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survivalCancer Res200464113753375615172979
- SeguraMFHannifordDMenendezSAberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factorProc Natl Acad Sci U S A200910661814181919188590
- TanGTangXTangFThe role of microRNAs in nasopharyngeal carcinomaTumour Biol2015361697925427638
- WangFLuJPengXIntegrated analysis of microRNA regulatory network in nasopharyngeal carcinoma with deep sequencingJ Exp Clin Cancer Res2016351726795575
- ZhengXHLuLXCuiCChenMYLiXZJiaWHEpstein-Barr virus mir-bart1-5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinomaOncotarget2016744972498026701721
- CuiLQiYLiHSerum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth diseasePLoS One2011611e2707122087245
- MacconiDTomasoniSRomagnaniPMicroRNA-324-3p promotes renal fibrosis and is a target of ACE inhibitionJ Am Soc Nephrol20122391496150522822076
- HuZDongJWangLESerum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controlsCarcinogenesis201233482883422298638
- GreenbergEBesserMJBen-AmiEA comparative analysis of total serum miRNA profiles identifies novel signature that is highly indicative of metastatic melanoma: a pilot studyBiomarkers201318650250823902289
- WenYHanJChenJPlasma miRNAs as early biomarkers for detecting hepatocellular carcinomaInt J Cancer201513771679169025845839
- XuJAiQCaoHLiuQMiR-185-3p and miR-324-3p predict radiosensitivity of nasopharyngeal carcinoma and modulate cancer cell growth and apoptosis by targeting SMAD7Med Sci Monit2015212828283626390174
- RibattiDEpithelial-mesenchymal transition in morphogenesis, cancer progression and angiogenesisExp Cell Res201735311528257786
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- WuLFanJBelascoJGMicroRNAs direct rapid deadenylation of mRNAProc Natl Acad Sci U S A2006103114034403916495412
- LlaveCXieZKasschauKDCarringtonJCCleavage of Scarecrow- like mRNA targets directed by a class of Arabidopsis miRNAScience200229755892053205612242443
- RuppertJMKinzlerKWWongAJThe GLI-Kruppel family of human genesMol Cell Biol198888310431132850480
- CohenMMJrThe hedgehog signaling networkAm J Med Genet A2003123A152814556242
