Abstract
Esophageal squamous cell carcinoma (ESCC) is among the most common malignancies, with a low 5-year overall survival rate. In previous studies, we and others have found that 9p21.3 was the most frequently deleted region in ESCC. The MTAP gene, which is located close to CDKN2A/B in 9p21.3, encodes methylthioadenosine phosphorylase. This enzyme plays an important role during the process of adenosine transfer. In the present study, we found that MTAP is deleted at the genomic level in 19.1% (64/341) of primary ESCC tumors, and decreased mRNA and protein expression were present in 31.1% (28/90) and 33.3% (6/18) of ESCCs, respectively. Further statistical analysis showed a positive correlation between deletion and decreased mRNA expression of MTAP in the ESCC tissues tested (coefficient: 0.826; P=1.17×10−23). Knockdown of MTAP expression using small interfering RNA-mediated silencing promoted the invasion and migration of ESCC cells. Also, overexpression of MATP using pcDNA3.1-MTAP plasmid decreased the cell invasion and migration. At the molecular level, MTAP knockdown downregulated E-cadherin and p-GSK3β but upregulated Slug expression. Our results indicated that MTAP deletion results in the decreased expression in ESCCs and that it plays a role in promoting the mobility and inducing the epithelial-to-mesenchymal transition of ESCC cells via the GSK3β/Slug/E-cadherin axis. The data suggest that MTAP might function as a tumor suppressor gene in ESCC.
Introduction
Esophageal cancer is among the most common malignancies. Eastern Asia, and Eastern and Southern Africa are the highest-risk areas, and esophageal squamous cell carcinoma (ESCC) is the most prevalent type.Citation1,Citation2 The 5-year overall survival rate for ESCC patients is only 15%–25%.Citation3 Significant progress on the study of gene deletions in human cancers has been made in recent years.Citation4–Citation7 Identification and investigation of genetic deletions might not only help reveal the mechanisms that underlie the tumorigenesis and development of ESCC but also provide potential biomarkers for the detection and therapy of the disease.
In previous studies, we and others have used large-scale genomic techniques to show that 9p21.3 was the most frequently deleted region in ESCC. These techniques included array-based comparative genomic hybridization (array-CGH), single-nucleotide polymorphism (SNP) arrays, and whole-exome sequencing.Citation8–Citation11 Moreover, our multiregional intratumor heterogeneity study showed that 9p21.3 homozygous deletion was an early eventCitation10 that was also found especially in the precancerous lesions of the esophageal squamous epithelia.Citation12
CDKN2A and CDKN2B have been reported in various studies to be the most commonly deleted genes within 9p21.3.Citation11 The MTAP gene, which is located close to CDKN2A/B in 9p21.3, encodes methylthioadenosine phosphorylase. This enzyme plays an important role during the process of adenosine transfer. MTAP is frequently deleted in human cancers.Citation13–Citation15 Kim et al found homozygous deletions in MTAP in xenografts that were established from the thoracic duct lymph of ESCC patients.Citation16
In the present study, we analyzed the deletion and expression of MTAP in the primary ESCC tumors and cell lines and assessed the relationship between the deletion and expression of MTAP. Furthermore, we investigated the impact of decreased MTAP expression on the malignant phenotypes of esophageal squamous carcinoma cells.
Materials and methods
Cell culture and tissue specimens
The human ESCC cell lines KYSE30, KYSE150, KYSE180, KYSE410, KYSE450, and KYSE510 were generously provided by Dr Y Shimada (Kyoto University, Kyoto, Japan). The ESCC cell line EC109 was purchased from the cell bank of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, San Diego, CA, USA), penicillin (100 U/mL), and streptomycin (100 mg/mL).
ESCC tissues and adjacent morphologically normal operative margins were procured from surgical resection specimens collected by the Department of Pathology in the Cancer Hospital, Chinese Academy of Medical Sciences, Beijing, China. All samples used in this study were residual specimens collected after sampling for pathological diagnosis. None of the patients received treatment prior to surgery, and all patients signed the informed consent forms of the Cancer Hospital, CAMS/PUMC for sample collection and molecular analysis. This study was approved by the Ethics Committee/Institutional Review Board of the Cancer Institute/Hospital, PUMC/CAMS (No NCC2015G-06).
Copy number alteration analysis
The MTAP copy number alterations were analyzed using our in-house array-CGH data (GSE46452Citation12), and the data derived from other array-CGH and SNP array platforms are available in Gene Expression Omnibus (GEO; GSE54993 and GSE54994,Citation9 GSE47630,Citation17 GSE17958Citation18) and The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/docs/publications/esca_2016Citation19) databases. A mean log2 ratio of MTAP <;−0.75 was classified as a deletion.
mRNA expression analysis
The MTAP mRNA expression of primary ESCC tumors and cell lines was analyzed using the reads per kilobase per million (RPKM) mapped reads value from the TCGA data (https://tcga-data.nci.nih.gov/docs/publications/esca_2016Citation19) and GEO data (GSE23964), respectively. The mean RPKM value of the esophageal epithelial cells is 5.198±0.408, which is available in NCBI website (https://www.ncbi.nlm.nih.gov/gene/4507). The ratio of the RPKM value in each tumor versus that in esophageal epithelial cells was transformed to the log2 ratio value. Mean log2 ratios of MTAP <;−1.5 were classified as decreased expression.
Small interfering RNA (siRNA), plasmid construction, and transfection
Two duplex MTAP siRNAs, siRNA-1 (5′-TCACTACCATACCTCAGAT-3′) and siRNA-2 (5′-GGTCTTAAAGACCCTGAAA-3′), and a nonsilencing siRNA (5′-TTCTCCGAACGUGUCACGTTT-3′) were designed and chemically synthesized (GeneChem, Shanghai) for transient transfection.
The CDS region of MTAP was amplified by reverse-transcript PCR using the upstream primer 5′-AAAGGATCCATGGCCTCTGGCACC-3′, and downstream primer 5′-CCCGAATTCTTAATGTCTTGGTAATAAAACAGA-3′, and then cloned into pcDNA3.1.
The cells were transfected with siRNAs or plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The cells were harvested 48 hours after transfection. The transfection efficiency was determined by Western blot analysis.
Invasion and migration assays
For the migration assay, 8×104 parental, nonsilencing siRNA- and MTAP siRNA-treated cells were seeded on fibronectin-coated polycarbonate membranes inserted in Transwell (Costar, Cambridge, MA, USA). RPMI-1640 that contained 20% FBS was added to the lower chamber. After incubation for 18 hours at 37°C in a CO2 incubator, the insert was washed with the phosphate-buffered saline, and the cells on the top surface of the insert were removed by wiping with a cotton swab. For the invasion assay, the procedure was similar to the migration assay, except that the transwell membrane was coated with 300 ng/μL matrigel (BD Biosciences, San Jose, CA, USA). The cells that had migrated to the bottom surface of the insert were fixed with methanol, stained with 0.5% crystal violet, and subjected to microscopic inspection.
Western blot analysis
Immunoblotting was conducted with primary antibodies against MTAP (Abcam, ab126770; 1:1,000), E-cadherin (Cell Signaling Technology, #3195; 1:1,000), p-GSK3β (Cell Signaling Technology, #9323; 1:1,000), or GSK3β (Cell Signaling Technology, #12456; 1:1,000). β-actin (Sigma, A19781; 1:5,000) was used as a loading control. The signals were visualized using the super-enhanced chemiluminescence detection reagent (Applygen Technologies, Inc., Beijing, China).
Statistical analysis
Statistical analysis was performed with the SPSS software program (version 17.0). Mann–Whitney test or Kruskal–Wallis test was performed for the evaluation of the association between MTAP deletion and clinicopathological parameters. The correlation between MTAP deletion and mRNA downregulation was analyzed using Spearman’s relative analysis. P-values <;0.05 were considered to be statistically significant.
Results
MTAP deletions in ESCC and the association with clinicopathological parameters
We performed in-house array-CGH on 59 primary ESCC tumorsCitation12 and further analyzed additional two array-CGH data and two SNP array data measuring ESCCs,Citation9,Citation17–Citation19 for a total of 341 ESCC cases. Overall, MTAP was deleted in 19.1% of the primary ESCC tumors (; ).
Figure 1 MTAP deletion in ESCC analyzed by array-based comparative genomic hybridization.
Abbreviation: ESCC, esophageal squamous cell carcinoma.
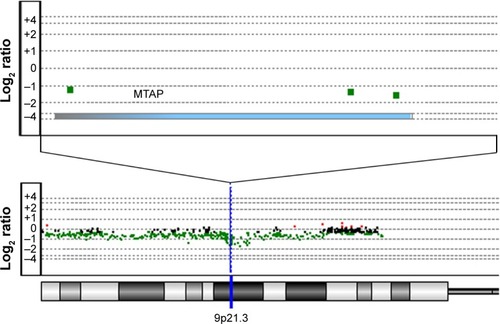
Table 1 MTAP deletions in ESCC
We then analyzed the relationship between MTAP deletion and the clinicopathological parameters (). MTAP deletion was significantly correlated with age (P=0.001) but not with gender, pathological T staging (pT), lymph node metastasis (LNM), and grade.
Table 2 Relationship between MTAP deletion and clinicopathological parameters of ESCC patients
Decreased expression of MTAP mRNA and protein in ESCC
To determine whether the genomic deletion of MTAP results in the downregulation of its expression, we analyzed the relationship between the copy number and mRNA expression of MTAP in 90 ESCC cases using the online dataCitation19 in which both copy number alterations and mRNA expression were detected in each case. The mRNA expression was decreased in 31.1% (28/90) of ESCCs. Overall, the MTAP mRNA expression levels were associated with the copy number levels. Reduced MTAP mRNA expression was present in 90.9% (20/22) of the cases with deletions compared with 5.9% (4/68) of the cases without deletions (). Furthermore, the MTAP mRNA levels in MTAP-deleted cases were much lower than those in the cases without the deletions (; mean level: 0.63±0.11 vs 5.34±0.41, P<;0.0001). Further statistical analysis showed a positive correlation between deletion and decreased mRNA expression of MTAP in the ESCC tissues tested (coefficient: 0.826; P=1.17×10−23; ). We also analyzed the mRNA levels in 16 cell lines using the GEO data GSE23964 and confirmed that the mean mRNA levels in the ESCC cell lines (6.58±0.69) were lower than those in the esophageal epithelial cell lines (7.64±0.10). Moreover, the mRNA levels in four ESCC cell lines were markedly decreased between 1- and 3.8-fold compared with the levels in the epithelial cell lines ().
Figure 2 Decreased expression of MTAP mRNA and protein in ESCC.
Abbreviations: ESCC, esophageal squamous cell carcinoma; N, esophageal epithelial cells from the paired surgical margins; T, tumors; RPKM, reads per kilobase per million mapped reads.
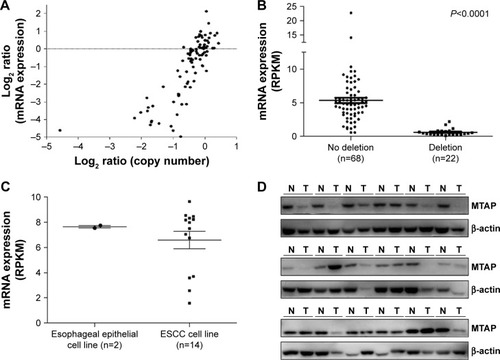
Table 3 Correlation of deletion and mRNA expression of MTAP in ESCC
We further examined the expression of MTAP protein in 18 ESCC cases, and the Western blot results showed that the MTAP protein was downregulated in 33.3% (6/18) of the primary ESCC tumors compared with those in operative margin tissues ().
MTAP knockdown increased invasion and migration of ESCC cells
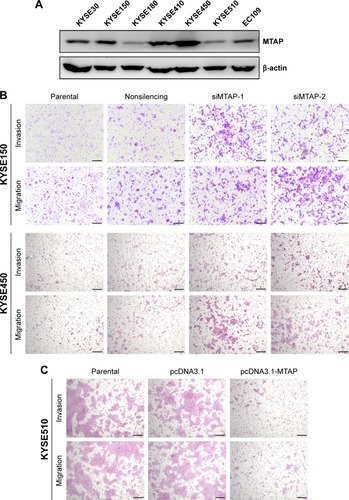
We next investigated whether the decreases in expression affected the malignant phenotype of the ESCC cells. The MTAP protein expression of ESCC cell lines was detected by Western blotting. The levels of MTAP protein are relatively higher in KYSE30, KYSE150, KYSE410, and KYSE450 than those in KYSE180, KYSE510, and EC109 (). We then knocked down MTAP in KYSE150 and KYSE450 using siRNAs, and overexpressed MTAP in KYSE510 using the constructed plasmids pcDNA3.1-MTAP. The transwell assays showed that knockdown of MTAP enhanced the invasion and migration of KYSE150 and KYSE450 cells (), and that overexpression of MTAP decreased the invasion and migration in KYSE510 cells ().
Figure 3 MTAP knockdown increased the motility of ESCC cells.
Abbreviations: ESCC, esophageal squamous cell carcinoma; nonsilencing, nontargeting siRNA control; siMTAP, MTAP-specific siRNA; siRNA, small interfering RNA.
MTAP knockdown regulated the expression of epithelial-to-mesenchymal transition (EMT)-related molecules in ESCC cells
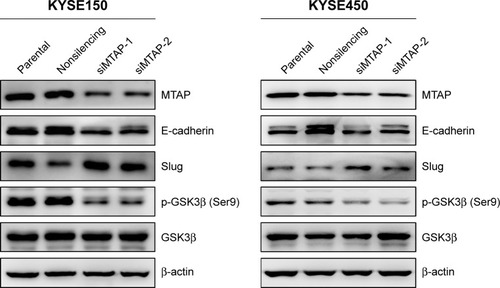
Based on the morphological changes affected by MTAP knockdown, we measured the expression of E-cadherin and Slug, as well as the phosphorylation of GSK3β, which are associated with cell motility and EMT. After MTAP knockdown, both E-cadherin and p-GSK3β were downregulated, whereas Slug was upregulated in the KYSE150 cells ().
Figure 4 MTAP knockdown regulated the expression of proteins related with cell motility in ESCC.
Abbreviations: siMTAP, MTAP-specific siRNA; ESCC, esophageal squamous cell carcinoma.
Discussion
Genomic deletion is one of the major processes that causes tumorigenesis and the development of human cancers. Studies have shown that the most frequent homozygous deletion region in ESCC is 9p21.3,Citation11 which is in an early event during the clonal evolutionary process of ESCC.Citation10 The common deletion peaks are at CDKN2A/B genes,Citation11 inactivation of which are associated with tumorigenesis and cancer development.Citation20–Citation23 The MTAP gene, which is located ~30 kb distal to CDKN2A, is usually co-deleted with CDKN2A in several human cancers.Citation24–Citation27 By analyzing the copy number alterations using high-throughput array-based genomic data from several studies, we found that MTAP deletion occurred in 19.1% of ESCCs. No significant correlation was observed between MTAP deletion and gender, pT, LNM, and grade, except for age. However, a slight higher frequency of MTAP deletion was present in LNM-positive patients than that in LNM-negative ones.
The correlation between MTAP deletion and loss of expression has been found in multiple types of cancers, including gastrointestinal stromal tumors,Citation28 laryngeal squamous cell carcinoma,Citation29 and glioblastoma multiforme.Citation30 In the present study, we established a significantly positive correlation between copy number and mRNA level of MTAP in ESCC. We also found reduced MTAP protein expression in ESCC, which is similar to the observations in lung cancer, liver cancer, lymphoma, and so on.Citation27,Citation31,Citation32
It has been observed that deletion and loss of MTAP expression are associated with poor outcomes for several human cancers,Citation27,Citation33,Citation34 and MTAP inactivation contributes to cell proliferation and invasion of cancer cells.Citation13,Citation14,Citation33–Citation35 However, the role of MTAP in ESCC is currently unknown. In this study, our data indicate that the loss of MTAP expression enhanced the invasion and migration of ESCC cells, and overexpression of MTAP decreased the cell invasion and migration, which suggested that MTAP expression might play a role in the inhibition of cell motility.
EMT is involved in the metastatic process of malignant tumors, and EMT activation promotes the invasion and metastasis of cancer cells.Citation36,Citation37 Our data showed that MTAP knockdown in ESCC cells led to a downregulation of E-cadherin expression, which has been well established as a hallmark of the EMT process in human cancers.Citation38,Citation39 We further found an upregulation of the oncogenic transcriptional repressor Slug in MTAP-knockdown ESCC cells, which indicated that upregulated Slug represses E-cadherin expression through the Slug/E-cadherin axis similar to the process in non-small-cell lung cancers.Citation40,Citation41 It has been reported that Slug expression is stabilized by the inactivation of GSK3β in epithelial cancers.Citation41 In this study, we also detected the decreased phosphorylation of GSK3β at Ser9 after knockdown of MTAP. Collectively, our findings suggest that MTAP expression inhibits cell motility and EMT through GSK3β/Slug/E-cadherin axis. Further investigation should be performed to determine whether deletion of MTAP plays a role in the tumorigenesis or progressions of ESCC.
In summary, our data show that frequent deletion and decreased expression of MTAP occur in primary ESCC tumors and that decreased expression of MTAP enhances the motility and EMT of ESCC cells through the GSK3β/Slug/E-cadherin axis. Together, our current findings suggest that MTAP might act as a tumor suppressor gene in ESCC.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81520108023), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-001 and 2016-I2M-3-007), Fundamental Research Funds for the Central Public-interest Scientific Institution (2016ZX310178), and the Beijing Nova Program (Z171100001117017).
Disclosure
The authors report no conflicts of interest in this work.
References
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- FerlayJSoerjomataramIDikshitRCancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012Int J Cancer20151365E359E38625220842
- PennathurAGibsonMKJobeBALuketichJDOesophageal carcinomaLancet2013381986440041223374478
- MatsuyamaHIkemotoKEguchiSCopy number aberrations using multicolour fluorescence in situ hybridization (FISH) for prognostication in non-muscle-invasive bladder cancer (NIMBC)BJU Int2014113466266723890221
- HidaTHamasakiMMatsumotoSImmunohistochemical detection of MTAP and BAP1 protein loss for mesothelioma diagnosis: comparison with 9p21 FISH and BAP1 immunohistochemistryLung Cancer20171049810528213009
- PereaJGarciaJLPerezJNOMO-1 gene is deleted in early-onset colorectal cancerOncotarget2017815244292443628416736
- CamposCZLosi GuembarovskiRde OliveiraCECGlutathione S-transferases deletions may act as prognosis and therapeutic markers in breast cancerClin Exp Med Epub2017428
- LinDCHaoJJNagataYGenomic and molecular characterization of esophageal squamous cell carcinomaNat Genet201446546747324686850
- SongYLiLOuYIdentification of genomic alterations in oesophageal squamous cell cancerNature20145097498919524670651
- HaoJJLinDCDinhHQSpatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinomaNat Genet201648121500150727749841
- LinDCWangMRKoefflerHPGenomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patientsGastroenterology Epub2017727
- ShiZZShangLJiangYYConsistent and differential genetic aberrations between esophageal dysplasia and squamous cell carcinoma detected by array comparative genomic hybridizationClin Cancer Res201319215867587824009147
- KryukovGVWilsonFHRuthJRMTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cellsScience201635162781214121826912360
- MavrakisKJMcDonaldER3rdSchlabachMRDisordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5Science201635162781208121326912361
- ZhaoMZhaoZConcordance of copy number loss and down-regulation of tumor suppressor genes: a pan-cancer studyBMC Genomics201617Suppl 753227556634
- KimDHMutoMKuwaharaYArray-based comparative genomic hybridization of circulating esophageal tumor cellsOncol Rep20061651053105917016592
- SawadaGNiidaAUchiRGenomic landscape of esophageal squamous cell carcinoma in a Japanese populationGastroenterology201615051171118226873401
- BassAJWatanabeHMermelCHSOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomasNat Genet200941111238124219801978
- Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer AgencyIntegrated genomic characterization of oesophageal carcinomaNature2017541763616917528052061
- OrtizBWhiteJRWuWHChanTADeletion of Ptprd and Cdkn2a cooperate to accelerate tumorigenesisOncotarget20145166976698225138050
- McNealASLiuKNakhateVCDKN2B loss promotes progression from benign melanocytic nevus to melanomaCancer Discov20155101072108526183406
- YangHKircherDAKimKHActivated MEK cooperates with Cdkn2a and Pten loss to promote the development and maintenance of melanomaOncogene201736273842385128263969
- TuQHaoJZhouXCDKN2B deletion is essential for pancreatic cancer development instead of unmeaningful co-deletion due to juxtaposition to CDKN2AOncogene Epub2017911
- SchmidMMalickiDNoboriTHomozygous deletions of methylthioadenosine phosphorylase (MTAP) are more frequent than p16INK4A (CDKN2) homozygous deletions in primary non-small cell lung cancers (NSCLC)Oncogene19981720266926759840931
- HustinxSRHrubanRHLeoniLMHomozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapyCancer Biol Ther200541838615662124
- KrasinskasAMBartlettDLCieplyKDacicSCDKN2A and MTAP deletions in peritoneal mesotheliomas are correlated with loss of p16 protein expression and poor survivalMod Pathol201023453153820081810
- SuCYChangYCChanYCMTAP is an independent prognosis marker and the concordant loss of MTAP and p16 expression predicts short survival in non-small cell lung cancer patientsEur J Surg Oncol20144091143115024969958
- HuangHYLiSHYuSCHomozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumorsClin Cancer Res200915226963697219887491
- CondeLVilasecaIAlosLMethylthioadenosine phosphorylase inactivation depends on gene deletion in laryngeal squamous cell carcinomaHistopathology20126161082108823020581
- CrespoITaoHNietoABAmplified and homozygously deleted genes in glioblastoma: impact on gene expression levelsPLoS One201279e4608823029397
- CzechBDettmerKVallettaDExpression and function of methylthioadenosine phosphorylase in chronic liver diseasePLoS One2013812e8070324324622
- MarceSBalagueOColomoLLack of methylthioadenosine phosphorylase expression in mantle cell lymphoma is associated with shorter survival: implications for a potential targeted therapyClin Cancer Res200612123754376116778103
- KimJKimMAMinSYJeeCDLeeHEKimWHDownregulation of methylthioadenosin phosphorylase by homozygous deletion in gastric carcinomaGenes Chromosomes Cancer201150642143321412930
- LiCFFangFMKungHJDownregulated MTAP expression in myxofibrosarcoma: a characterization of inactivating mechanisms, tumor suppressive function, and therapeutic relevanceOncotarget2014522114281144125426549
- MarjonKCameronMJQuangPMTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axisCell Rep201615357458727068473
- JollyMKWareKEGiljaSSomarelliJALevineHEMT and MET: necessary or permissive for metastasis?Mol Oncol201711775576928548345
- SantamariaPGMoreno-BuenoGPortilloFCanoAEMT: present and future in clinical oncologyMol Oncol201711771873828590039
- ThieryJPEpithelial-mesenchymal transitions in tumour progressionNat Rev Cancer20022644245412189386
- BolosVPeinadoHPerez-MorenoMAFragaMFEstellerMCanoAThe transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressorsJ Cell Sci2003116Pt 349951112508111
- ShihJYTsaiMFChangTHTranscription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinomaClin Cancer Res200511228070807816299238
- KaoSHWangWLChenCYGSK3beta controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of SlugOncogene201433243172318223851495
