Abstract
Background and objective
In an attempt to discover a new biomarker for early diagnosis and prognosis of esophageal squamous cell carcinoma (ESCC), the regulation mechanism of large intergenic non-coding RNA–regulator of reprogramming (lincRNA-ROR) as a microRNA (miRNA) sponge was studied.
Patients and methods
ROR expression in 91 pairs of ESCC tissue samples and matched adjacent tissues was quantified with real-time fluorescent quantitative polymerase chain reaction (qRT-PCR). The ROR–miRNA–mRNA regulatory network was built with 161 esophageal cancer (EC) tissues and 11 adjacent tumor tissues from The Cancer Genome Atlas (TCGA) database. A total of 96 cases of ESCC from TCGA database were collected for analysis on survival rates. The regulatory relationship between ROR, miR-145 and FSCN1 was verified in ESCC cells via qRT-PCR, dual luciferase reporter (DLR) assay, RNA immunoprecipitation (RIP) and Western blotting. The transwell method was used to detect cell migration and invasion.
Results
ROR expression in ESCC tumor tissues was significantly higher than in the adjacent tissues, p<0.001. The survival rate of ESCC patients with high ROR expression levels was lower than that of patients with low ROR expression levels (p<0.001). ROR overexpression could downregulate miR-145 by up to 50% was proven by RIP, DLR assay, and qRT-PCR. Two effective binding sites of ROR to miR-145 were verified by DLR assay. One of the sites has never been cited in the literature. The Western blotting results showed that FSCN1 was a downstream target of ROR/miR-145 (p<0.05). Transwell assays were used to show that overexpression of ROR enhanced migration and invasion behavior of ESCC and miR-145 hindered these effects.
Conclusion
ROR acted as a competitive endogenous RNA (ceRNA) of miR-145 in ESCC. A novel, effective miR-145 binding site of ROR was discovered. The ROR/miR-145/FSCN1 pathway was shown to take part in the metastasis of ESCC. ROR is likely an oncogene biomarker for ESCC early diagnosis and prognosis.
Introduction
Esophageal cancer (EC) is the eighth most common cancer, the second most common cancer in China, and the sixth most common cause of cancer death worldwide.Citation1–Citation3 Previous research indicated that the incidence of EC in China is ~23.9/100,000, with a 5-year survival rate of 20%–30%.Citation4–Citation6 In China, >90% of EC are esophageal squamous cell carcinoma (ESCC).Citation3,Citation7 ESCC has a high propensity for lymphatic spread via their rich network of submucosal lymphatic vessels, with a 5-year survival rate of 15%–34%.Citation4,Citation7,Citation8 Identifying biomarkers that could predict the prognosis of ESCC is needed. This would be instructive for therapeutic decision making as well as screening and early detection.
There are various long non-coding RNAs (lncRNAs) in ESCC that are proven to be dysregulated, such as HOTAIR, SPRY4-IT1, UCA1, and TUG1.Citation9–Citation13 LncRNAs are RNA polymerase II transcripts that are longer than 200 nucleotides and lack an open reading frame. There are >10,000 types of lncRNAs that are thought to play crucial roles in development and differentiation of human disease, particularly tumor development.Citation14–Citation21
Among lncRNAs, the large intergenic non-coding RNAs (lincRNAs) are numerous and their functions are better understood. Several studies have proposed a model that suggests lincRNA function as a competitive endogenous RNA (ceRNA) when modulating concentration and biological functions of microRNAs (miRNA).Citation22–Citation24
The regulator of reprogramming (ROR, linc-ROR) was first identified in induced pluripotent stem cells (iPSCs).Citation25 ROR is ~2.6 kbp in length. It is also known as lincRNA-ST8SIA3, and it is located on chromosome 18 (hg19 chr18: 57, 054, 571–557, 072, 119) containing four exons. ROR is highly expressed in self-renewing human embryonic stem cells, iPSCs, and various cancer cells, including breast cancer, hepatocellular carcinoma, endometrial cancer, and pancreatic cancer.Citation26–Citation30 There is a positive correlation between the expression level of ROR and the undifferentiation degree of the carcinoma tissues. ROR has been proven to have many functions in human cancers, including promoting the epithelial–mesenchymal transition, enhancing the hypoxia resistance of the tumor tissue, and reducing the sensitivity of the tumor to chemotherapy.Citation26–Citation32 ROR is a known negative regulator of the p53 pathway.Citation33,Citation34 ROR competitively inhibits cancer-suppressive miRNAs, such as miR-145, miR-205, and miR-124, in order to upregulate cancer-promoting transcription factors including Oct4, Sox2, and Nanog.Citation23,Citation25,Citation35–Citation37
The function of ROR in ESCC was investigated by building an ROR–miRNA–mRNA regulatory network based on The Cancer Genome Atlas (TCGA) database. Our previous study proved that the miR-145 was downregulated in ESCC and promoted lymph node metastases by regulating FSCN1. This corresponds with the aforementioned regulatory network.Citation38 It is proposed that ROR can accelerate the development of ESCC by sponging miR-145 and upregulating FSCN1. This study implemented an expression detection of ROR in both cell lines and tissues of ESCC, in order to study the relationship between ROR and ESCC incidence risk. The cox survival analysis of 96 cases collected from TCGA database was done to study the effects of ROR on ESCC prognosis. FSCN1 has been reported to be upregulated in many human carcinomas and positively correlated with the clinical aggressiveness of tumors, particularly invasion and metastasis,Citation39–Citation41 indicating a poor prognosis. Transwell migration and invasion assays were performed to determine whether ROR could stimulate ESCC invasion and migration by regulating miR-145/FSCN1.
Patients and methods
Cell lines and culture conditions
EC cell lines, EC109 and EC9706, and immortalized esophageal epithelial cell line, H5E46, were provided by the Key Laboratory of Environmental Medicine Engineering of Southeast University, Ministry of Education, China. The cells were cultured in a humidified incubator at 37°C with 5% CO2 in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA).
Clinical tissue samples
Huaian is considered a high-risk region for EC in China. The 91 pairs of ESCC tissues and the matched tissues adjacent to carcinoma (>5 cm away from the cancer tissue) were all collected from the Department of Thoracic Surgery of Huaian First People’s Hospital of Jiangsu Province, 2010. The patients whose tissue samples were used had provided written informed consent for their use in research. The inclusion criteria of the volunteers were as follows: 1) diagnosed with ESCC by pathological section or endoscope; 2) without radiotherapy or chemotherapy treatment; 3) >20 years of residence in Huaian; and 4) Han nationality. Ethical approval was provided by the institutional review board of the Southeast University, Affiliated Zhongda Hospital (Nanjing, China).
The extraction and purification of total RNA
The total RNA was extracted using the Trizol (Thermo Fisher Scientific) method. The RNA was then purified by DNase and a RNA centrifugal column purification kit (Tiangen, Beijing, China) in order to erase genome pollution. The purified RNA samples were stored at −80°C.
Real-time fluorescent quantitative polymerase chain reaction (qRT-PCR)
Reverse transcription (RT) of ROR was performed with a PrimeScript RT Master Mix Kit (Takara, Shiga, Japan). The RT reaction was performed in a gradient gene amplification instrument (Eppendorf, Hamburg, Germany). The fluorescent quantitative PCR was performed with a SYBR Premix Ex Taq II Kit (Takara), in accordance to the manufacturer’s instructions. The reaction was performed in a StepOnePlus System (Thermo Fisher Scientific). The ROR expression analysis was normalized with β-actin. The sequences of ROR and β-actin primers are listed in .
Table 1 Primers of ROR and ACTB
The RT reaction of miR-145 was implemented with deoxy-ribonucleoside triphosphate, moloney murine leukemia virus RT, 5× moloney murine leukemia virus buffer, RNase inhibitor (Thermo Fisher Scientific), and miRNA-specific Bulge-Loop RT Primer (RIBOBIO, Guangzhou, China). The reaction was performed in a gradient gene amplification instrument (Eppendorf).
MiR-145 was downregulated in ESCC, so prior to the fluorescent quantitative PCR preamplification of miR-145 for 12 cycles was added. The reaction system was a 20 μL mixture of cDNA 1 μL, Bulge-Loop Forward and Reverse Primers each 1.6 μL, 10× PCR buffer 2 μL, 2.5 mM deoxy-ribonucleoside triphosphate 1.6 μL, Taq DNA polymerase 0.2 μL (Takara), and RNase-free water 12 μL. The reaction condition was 94°C for 5 min, 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, 12 cycles, and 72°C for 5 min. The reaction kit and the quantitative PCR instrument were the same as ROR. The analysis of the miR-145 expression was normalized with U6. Quantitative PCR reaction of cell samples was repeated three times.
Establishment of stable ROR overexpressing ESCC cell line
EC109 was used to establish ROR high-level expression cell line, where it was transfected by the lentivirus expressing ROR and green fluorescent protein with puromycin resistance. The multiplicity of infection (MOI) was 20. The cells were cultured in a complete RPMI-1640 medium with puromycin (2 μg/mL) after the green fluorescence was observed with a fluorescence microscope. The stable expression cell line was established at the 8th passage. The transfection efficiency of the ROR was confirmed with qRT-PCR. The ROR expression in the positive group was 16,000 times higher than that in the negative control (NC) group.
ROR–miRNA–mRNA regulatory network
A total of 161 EC tumor samples and 11 adjacent tumor samples were collected from patients by TCGA database. The screening criteria were as follows: p<0.05, false discovery rate (FDR) <0.1, and fold change (FC) ≥2. A random variance model t-test was used to filter the differentially expressed genes in ESCC. The target miRNAs of ROR were predicted by miRanda. The target mRNAs of the miRNAs were predicted by miRanda and Targetscan.
RNA immunoprecipitation (RIP)
The RIP assay was performed in accordance to the manufacturer’s instructions of the EZ-Magna RIP Kit (EMD Millipore, Billerica, MA, USA). The EC109 cells were divided into two groups: the ROR overexpressing group and the NC group. Each sample was immunoprecipitated by mouse IgG and Argonaute2 (AGO2) antibodies (EMD Millipore). The RIP procedure was subjected to qRT-PCR for detection of miR-145. The miR-145 expression was relatively low in ESCC cells. A 12-cycle pre-amplification was performed after RT and before quantitative PCR. The cycle threshold values of the miR-145 in the anti-AGO2 groups were normalized with the input groups.
DL Rassay
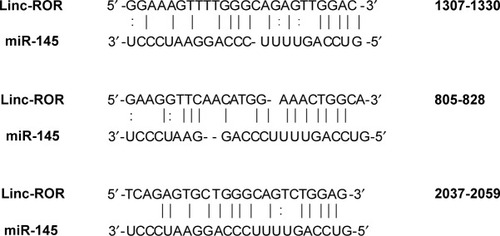
The grouping of cells (EC109) is shown in . The cells were seeded in 48-well plates at a density of 2.5×104 cells per well, 24 h prior to transfection. The cells were cotransfected with a mixture of conditioned vector pmiR-GLO (Genewiz, Suzhou, China) and miRNA mimics. The three binding sites between miR-145 of the ROR are shown in . They were mutated in this experiment.Citation26,Citation29 Site 805–828 has not been reported in previous literature. The miR-145 and the NC mimics were obtained from TranSheepBio (Shanghai, China). The luciferase activity was measured with a DLR assay system (Promega Corporation, Madison, WI, USA) and a LB942 Modular Multimode Microplate Reader (Berthold, Bad Wildbad, Germany) after 48 h. The hLuc activity was normalized with the hRluc activity for comparison.
Table 2 Grouping of DLR assay
Western blotting
The cells were washed with phosphate-buffered saline and lysed in a radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) that contained a protease inhibitor. The protein concentrations were quantified with a BCA Protein Assay Kit (Thermo Fisher Scientific). Proteins were separated with a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to the polyvinylidene fluoride (PVDF) membranes. The membranes were incubated overnight at 4°C with rabbit monoclonal anti-human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000, Cell Signaling Technology) and rabbit polyclonal anti-human Fascin (1:10,000, Abcam, Cambridge, MA, USA). The anti-rabbit IgG and horseradish peroxidase-linked antibody (1:2,000; Cell Signaling Technology, Danvers, MA, USA) were washed and applied to the cells at room temperature for 1 h. The signals were detected with the SuperSignal West Femto Trial Kit (Thermo Fisher Scientific).
Transwell migration and invasion assay
The transwell assay was performed with a 6.5 mm in diameter and an 8-mm pore size chamber (Corning Incorporated, Corning, NY, USA). The invasion assay was performed 24 h after transfection, where 3×105 cells in the RPMI-1640 medium supplemented with 0.1% FBS were seeded onto the Matrigel-coated membrane matrix of the upper chamber. The lower chamber was filled with RPMI-1640 medium, supplemented with 10% FBS. The chamber was incubated for 24 h, and the cells that were invading the bottom of the membrane were fixed with methyl alcohol and stained with 0.1% crystal violet. The number of invaded cells was counted with a FSX100 bioimage navigator (Olympus Corporation, Tokyo, Japan).
The migration assay was performed 24 h after transfection, where 2×105 cells in RPMI-1640 medium were seeded in the upper chamber. The lower chamber was filled with a RPMI-1640 medium supplemented with 10% FBS. The fixation, staining, and counting methods of migrated cells were the same as the invasion assay.
Statistical analysis
The quantitative variables were compared by the Student’s t-test or the analysis of variance (ANOVA). The data were expressed as the mean value ± SD. P<0.05 was considered statistically significant. A cox regression analysis was used to detect the relationship between the risk of ESCC incidence and the ROR expression. The survival analysis was used to study the relation between the ROR expression and the prognosis of ESCC. The statistical analysis and the graph presentation were performed by SPSS 18.0 and GraphPad Prism 5 software.
Results
Highly upregulated ROR in ESCC cells and tissues
The ROR expression levels in ESCC cell lines and clinical sample tissues were determined by the qRT-PCR assay. and show that the expression level of ROR in the two ESCC cell lines was 10.8 times and 29.5 times higher than the normal esophageal epithelial cell line.
Table 3 Relative expression of the ROR in the three cell lines
Figure 2 The ROR expression was higher in ESCC cell lines EC109 and EC9706 than the immortalized esophageal epithelial cell line H5E46 (**p<0.01).
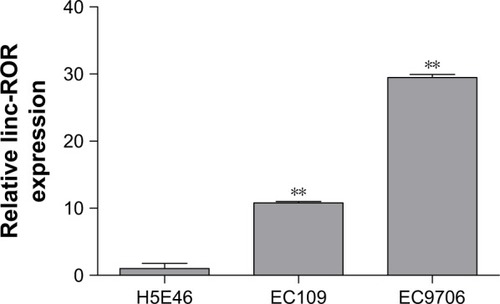
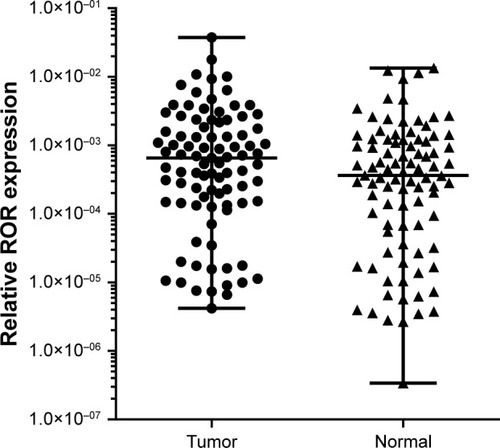
The qRT-PCR results of the 91 pairs of ESCC tumor tissues and the adjacent tissues are shown in . The ROR expression in ESCC tissues was significantly higher than normal esophageal epithelial tissues. The high ROR level could increase the risk of ESCC. The relative ROR expression in tumor tissue and normal tissue is shown in .
Table 4 ROR expression was higher in the ESCC tissues than in the adjacent tissues
High ROR expression level could reduce the survival rate of ESCC patients
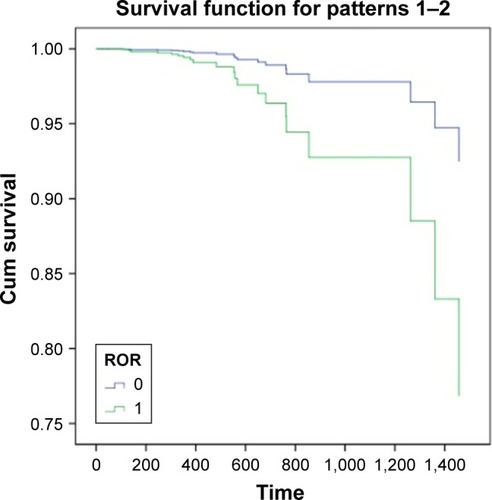
The 96 cases of ESCC provided by TCGA database were collected to perform the survival analysis of ROR expression, using age as the covariate. The SPSS 18.0 results are shown in . This result showed that high ROR level reduced the survival rate of ESCC patients.
An ROR–miRNA–mRNA regulatory network was built with TCGA database
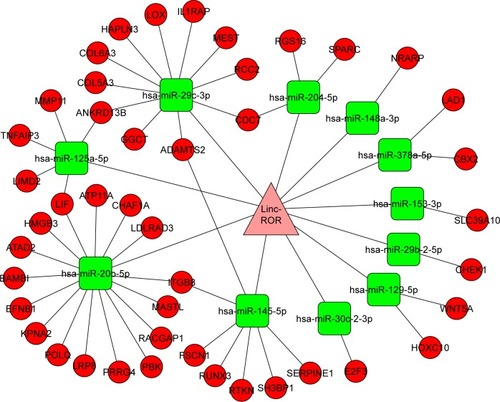
There were 82 miRNAs (p<0.001, FDR <0.1, and FC ≥2) and 1,398 mRNAs (p<0.001, FDR <0.05, and FC ≥2) after the differential screening of the TCGA EC data with the random variance model t-test. Out of the 82 miRNAs, 11 were predicted to bind to ROR by miRanda and were downregulated in the EC. The target miRNAs and the binding sites are shown in . Out of the 1,398 mRNAs, 45 were targets of the 11 miRNAs and were upregulated in EC. The ROR–miRNA–mRNA regulatory network is shown in . Our previous work suggested that miR-145 decreased in ESCC and could regulate FSCN1.Citation38 These findings were in accordance with this network, so the research of the ROR/miR-145/FSCN1 pathway in ESCC commenced.
Table 5 The 11 target miRNAs of ROR and their binding sites
ROR could competitively bind to miR-145 as an miRNA sponge in ESCC
ROR could downregulate miR-145 expression
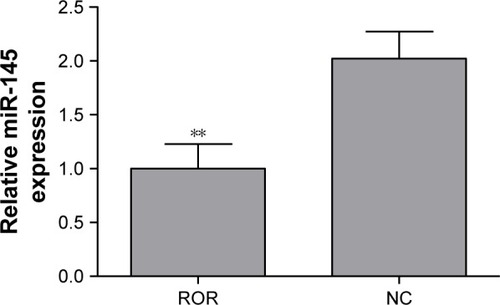
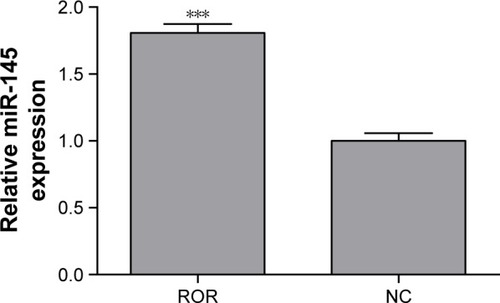
The regulative relations of ROR and miR-145 were studied by implementing qRT-PCR of miR-145 in the ROR overexpressing group and the NC group cells. The results showed that the overexpression of ROR downregulated the expression of miR-145 in EC109 ( and ).
Table 6 The downregulation of miR-145 induced by ROR over-expression
ROR could bind to miR-145
The binding relation of the ROR and the miR-145 was verified by performing an anti-AGO2 RIP assay to explore the effects of ROR overexpression in miR-145. MiR-145 of the ROR overexpressing group immunoprecipitated by AGO2 antibodies was significantly higher than the NC group (). The cycle threshold values were normalized with the input. It can be suggested that ROR could bind to miR-145 and was recruited to the AGO2-related RNA-induced silencing complexes (RISCs).
Two predicted binding sites of ROR to miR-145 were proved effective
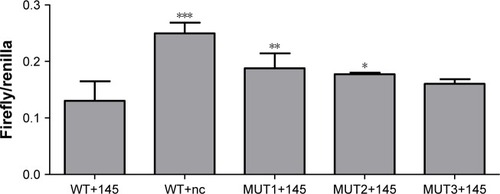
A DLR assay was implemented to verify the predicted binding sites of ROR and miR-145. The vector utilized was pmiR-GLO, and hLuc/hRluc was for the fluorescence signal intensity. and show that group 2 (WT+miR-145) signal was significantly lower than group 1 (WT+nc mimic), group 3 (MUT1+miR-145), and group 4 (MUT2+miR-145). This meant that ROR could bind to miR-145 via at least two binding elements. Site 1307–1330 and the novel 805–828 site were effective miR-145 binding sites that are likely the targets of ESCC molecular therapy.
Table 7 The DLR fluorescent signal intensity
An ROR/miR-145/FSCN1 axis was proved to be working in ESCC
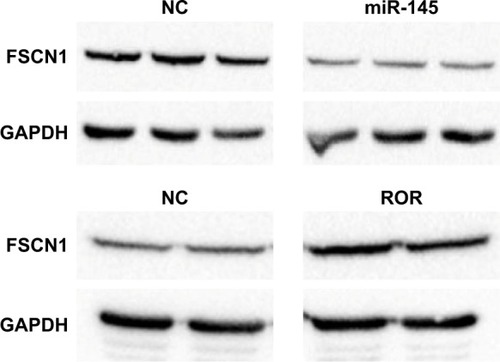
The TargetScan algorithm found FSCN1 to be among the highest scoring predicted mRNA targets of miR-145. Previous literature suggested that miR-145 could suppress the metastasis of ESCC by targeting the FSCN1.Citation38 The regulative relations of ROR, miR-145, and FSCN1 were further explored by Western blotting.
The Western blotting results showed that miR-145 down-regulated FSCN1 (). ROR indirectly upregulated FSCN1. The ROR/miR-145/FSCN1 pathway was proven and played a role in ESCC progress on the protein level.
ROR could promote migration and invasion of ESCC cells
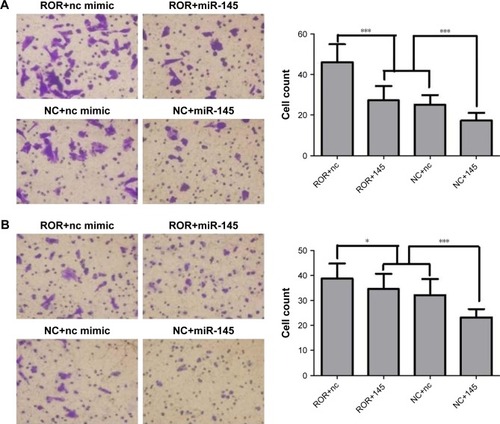
Previous research has shown that FSCN1 promotes tumor metastasis in various cancers.Citation39–Citation41 ROR functions in migration and invasion of ESCC were explored, in order to verify its regulation of miR-145/FSCN1. The EC109 cells transfected with lentivirus (that were overexpressing ROR or only the vector) were chosen as the treatment (ROR) group or the NC group. The regulating relationship between ROR and miR-145/FSCN1 was supplemented with miR-145 mimics or NC mimics as a treatment of the following Transwell migration and invasion assays. The grouping of the EC109 cells is shown in .
Table 8 Grouping type
The cell migration and the invasion assay results are shown in . ROR enhanced the migratory and the invasive capacities of EC109. It seems that miR-145 had the opposite function, where it restrained the effects of ROR to some extent. The mechanisms could be regulating tumor invasion and metastasis-related gene FSCN1 by sponging miR-145.
Figure 10 ROR promoted migration and invasion of ESCC cells (*p<0.05 and ***p<0.001).
Abbreviations: ESCC, esophageal squamous cell carcinoma; NC, negative control; ROR, regulator of reprogramming.
Discussion
ROR is a typical lincRNA that plays important regulatory roles in the pathogenesis and progression of tumors.Citation23,Citation35,Citation36 Previous studies suggested that ROR could promote cell reprogramming and enhance the malignancy of multiple tumors, including breast cancer and pancreatic cancer.Citation23,Citation25,Citation27,Citation30 ROR could even enhance the therapeutic resistance of tumors.Citation28,Citation31,Citation37 But its clinical significance and prognostic value in ESCC remain largely unknown. In this study, ROR expression in both ESCC cells and tissues was tested. The qRT-PCR results showed that ROR was significantly upregulated in ESCC cells and tissues than the normal. The results showed that the ROR expression was positively correlated with the ESCC incidence risk. The cox survival analysis of 96 ESCC patients, recorded in TCGA database, showed that the survival time of patients with a higher ROR expression was significantly shorter than the patients with a lower ROR expression. This suggested that the high ROR expression level also predicted a poor prognosis of ESCC. ROR is likely a biomarker for early screening and poor prognosis of ESCC.
Recent literature has suggested that ROR could act as a molecular sponge in many tumors, where it absorbed and inhibited the activity of miRNAs. MiR-145 seems to suppress the development of various tumors by promoting apoptosis or inhibiting proliferation and invasion.Citation23,Citation25–Citation27,Citation29,Citation33–Citation36 ROR is predicted by miRanda to have multiple binding sites with miR-145. It was speculated that ROR could enhance the malignance of ESCC by competitively binding to miR-145.
Previous literature showed that miR-145 was downregulated in ESCC. The reduced expression of miR-145 showed a trend of lymph node metastasis by regulating FSCN1.Citation38 FSCN1 was higher in ESCC tissues than normal epithelium. Its overexpression was associated with poor prognosis or lymph node metastasis in several human epithelial cancers, including colonic, gastric, breast, skin, urothelial carcinomas, and ESCC.Citation42 In this study, it was suggested that ROR could promote metastasis and invasion of ESCC by regulating miR-145 and FSCN1. An ROR–miRNA–mRNA regulatory network was built in ESCC that contained 11 downregulated miRNAs and 95 upregulated mRNAs based on TCGA database. The results were in agreement with our hypothesis that the ROR/miR-145/FSCN1 pathway existed in ESCC.
The qRT-PCR showed that ROR overexpression down-regulated miR-145. The RIP results confirmed that ROR bound to miR-145 in ESCC cells. The binding relations of ROR and miR-145 were further studied by predicting binding sites with miRanda. The top three sites obtained high scores, but only two of the sites had been previously reported. The DLR assay was used to prove that the novel site 805–828 effectively bound to miR-145, meaning that this site could be a new target for ESCC molecular therapy. Western blotting was used to verify the regulation of miR-145 and ROR on FSCN1 expression. The results proved that miR-145 decreased FSCN1 and the overexpression of ROR upregulated FSCN1. This verified the ROR/miR-145/FSCN1 axis in ESCC. Previous researchCitation38,Citation39 led us to perform transwell migration and invasion assays, which proved that ROR enhanced metastasis and invasion of ESCC. miR-145 counteracted the effects of ROR to some extent.
Conclusion
In this study, it is supposed that ROR is an oncogene of ESCC, which positively correlates with ESCC incidence and results in poor prognosis. ROR can promote metastasis and invasion of ESCC by regulating miR-145/FSCN1. ROR is likely a biomarker that can be used for early screening and will suggest poor prognosis for ESCC patients. ROR can probably be an effective target of miR-145 in molecular therapy for ESCC.
Acknowledgments
This work was supported by National Natural Science Foundation of China grants (81573108, 81573191, and 81172747), New Century Excellent Talents in University from Ministry of Education (NCET-13-0124), Commonweal Technology Application Research Project of Zhejiang Province (2016C33218), and Graduate Research and Innovation Program of Colleges and Universities of Jiangsu Province (KYLX15_0174).
Disclosure
The authors report no conflicts of interest in this work.
References
- SakaiNSSamia-AlyEBarberaMFitzgeraldRCA review of the current understanding and clinical utility of miRNAs in esophageal cancerSemin Cancer Biol201323651252124013023
- KanoMSekiNKikkawaNmiR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinomaInt J Cancer2010127122804281421351259
- TangWRChenZJLinKSuMAuWWDevelopment of esophageal cancer in Chaoshan region, China: association with environmental, genetic and cultural factorsInt J Hyg Environ Health20152181121825455641
- LiWZhengJDengJQIncreased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cellsGastroenterology201414671714.e51726.e524631494
- YuanYHuYZhaoYChenLExpressions of Notch signaling-associated proteins in esophageal squamous cell carcinomaZhonghua Wei Chang Wai Ke Za Zhi201518990991326404689
- JiangMSuMJiXA case-control study on risk factors for esophageal cancer in Huaian District of Jiangsu Province, ChinaTumor2013338701705
- DengHYWangYCNiPZLinYDChenLQLong noncoding RNAs are novel potential prognostic biomarkers for esophageal squamous cell carcinoma: an overviewJ Thorac Dis201688E653E65927621894
- XuXHChenZLZhaoXHMicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinomaBiochem Biophys Res Commun2012421464064522450326
- GeXSMaHJZhengXHHOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathwayCancer Sci2013104121675168224118380
- XieHWWuQQZhuBLong noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosisTumor Biol201435877437754
- LiJYMaXZhangCBOverexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinomaInt J Clin Exp Pathol20147117938794425550835
- WangXHGaoZKLiaoJlncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathwayJ Toxicol Environ Health A2016799–1040741827267823
- XuYTWangJQiuMTUpregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinomaTumor Biol201536316431651
- BatistaPJChangHYLong noncoding RNAs: cellular address codes in development and diseaseCell201315261298130723498938
- KornienkoAEGuenzlPMBarlowDPPaulerFMGene regulation by the act of long non-coding RNA transcriptionBMC Biol2013115923721193
- FaticaABozzoniILong non-coding RNAs: new players in cell differentiation and developmentNat Rev Genet201415172124296535
- JiangMHuangOXieZQA novel long non-coding RNA-ARA: adriamycin resistance associatedBiochem Pharmacol201487225428324184505
- SongHYuXXiaTGuoJXiaoBAssociations between long non-coding RNAs and tumors, and their clinical valuesChin J Cell Biol2012347704712
- PangQQGeJXShaoYFIncreased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significanceTumor Biol201435654415447
- WuYSZhangLWangYLong noncoding RNA HOTAIR involvement in cancerTumor Biol2014351095319538
- LuXXFangYWangZTDownregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6Cell Tissue Res2013354389189624026436
- PopadinKGutierrez-ArcelusMDermitzakisETAntonarakisSEGenetic and epigenetic regulation of human lincRNA gene expressionAm J Hum Genet20139361015102624268656
- WangYXuZYJiangJFEndogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewalDev Cell2013251698023541921
- UlitskyIBartelDPlincRNAs: genomics, evolution, and mechanismsCell20131541264623827673
- LoewerSCabiliMNGuttmanMLarge intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cellsNat Genet201042121113111721057500
- EadesGWolfsonBZhangYSLiQLYaoYZhouQlincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6Mol Cancer Res201513233033825253741
- HouPZhaoYLiZLincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasisCell Death Dis20145e128724922071
- TakahashiKYanIKKogureTHagaHPatelTExtracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemo-sensitivity in human hepatocellular cancerFEBS Open Bio20144458467
- ZhouXGaoQWangJZZhangXLiuKDuanZLinc-RNA-RoR acts as a “sponge” against mediation of the differentiation of endometrial cancer stem cells by microRNA-145Gynecol Oncol2014133233333924589415
- ZhanHXWangYLiCLincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathwayCancer Lett2016374226127126898939
- TakahashiKYanIKHagaHPatelTModulation of hypoxia-signaling pathways by extracellular linc-RoRJ Cell Sci201412771585159424463816
- RezaeiMEmadi-BaygiMHoffmannMJSchulzWANikpourPAltered expression of LINC-ROR in cancer cell lines and tissuesTumor Biol201637217631769
- ZhangAZhouNJHuangJGThe human long non-coding RNA-RoR is a p53 repressor in response to DNA damageCell Res201323334035023208419
- HuangJZhouNWatabeKLong non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1)Cell Death Dis20145e100824457952
- GaoSWangPHuaYQROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancerOncotarget2016721608161826636540
- ChengECLinHFRepressing the repressor: a lincRNA as a microRNA sponge in embryonic stem cell self-renewalDev Cell20132511223597480
- LiCGZhaoZMZhouZPLiuRLinc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axisCancer Chemother Pharmacol20167861199120727785603
- LiuRLiaoJYangMThe cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1PLoS One201273e3398722457808
- XieJJXuLYZhangHHRole of fascin in the proliferation and invasiveness of esophageal carcinoma cellsBiochem Biophys Res Commun2005337135536216185662
- HashimotoYSkacelMAdamsJCRoles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker?Int J Biochem Cell Biol20053791787180416002322
- HashimotoYItoTInoueHPrognostic significance of fascin overexpression in human esophageal squamous cell carcinomaClin Cancer Res20051172597260515814639
- ZhangHXuLXiaoDFascin is a potential biomarker for early-stage oesophageal squamous cell carcinomaJ Clin Pathol200659995896416524962