Abstract
Background
Insulin-like growth factor-1 receptor (IGF-1R) is a well-studied oncogenic factor that promotes cell proliferation and energy metabolism and is overexpressed in numerous cancers including hepatocellular carcinoma (HCC). Aerobic glycolysis is a hallmark of cancer, and drugs targeting its regulators, including IGF-1R, are being developed. However, the mechanisms of IGF-1R inhibition and the physiological significance of the IGF-1R inhibitors in cancer cells are unclear.
Materials and methods
Cell proliferation was evaluated by cell counting Kit-8 and colony formation assay. Western blot and real-time PCR were accordingly used to detect the relevant proteins, miRNA and gene expression. Luciferase reporter assays were used to illustrate the interaction between miR-342-3p and IGF-1R. The effect of miR-342-3p on glycolysis was determined by glucose uptake, ATP concentration, lactate generation, extracellular acidification rate and oxygen consumption rate assays. In vivo, subcutaneous tumor formation assay and PET were performed in nude mice.
Results
In this study, we demonstrate that by directly targeting the 3′-UTR (3′-untranslated regions) of IGF-1R, microRNA-342-3p (miR-342-3p) suppresses IGF-1R-mediated PI3K/AKT/GLUT1 signaling pathway both in vitro and in vivo. Through suppression of IGF-1R, miR-342-3p dampens glycolysis by decreasing glucose uptake, lactate generation, ATP production, and extracellular acidification rate (ECAR), and increasing oxygen consumption rate (OCR) in hepatoma cells. Importantly, glycolysis regulated by miR-342-3p is critical for its regulating HCC growth both in vitro and in vivo.
Conclusion
Our findings provide clues regarding the role of miR-342-3p as a tumor suppressor in liver cancer mainly through the inhibition of IGF-1R. Targeting IGF-1R by miR-342-3p could be a potential therapeutic strategy in liver cancer.
Introduction
Hepatocellular carcinoma (HCC) is among the most common and lethal cancers worldwide, briefly, standing for the third leading cause of cancer death and the fifth prevalent human cancer.Citation1 Despite much progress made to improve the therapeutic options,Citation2,Citation3 the long-term prognosis of HCC patients still remains poor, with a 5-year survival rate of approximately 30%.Citation4–Citation7 Therefore, further understanding of the underlying molecular mechanisms of HCC tumorigenesis is crucial and urgent.
The Warburg effect (aerobic glycolysis) is a typical feature of cancer cells, exhibiting abnormal metabolism characterized by high glycolysis even in the presence of ample oxygen.Citation8,Citation9 Now, the Warburg effect has been widely recognized as a hallmark of cancer which can facilitate tumor growth with enhanced glucose uptake and lactate production. Therefore, the understanding of the control of this process is crucial to identify potential targets for HCC therapy.
miRNAs are a group of endogenous, small noncoding RNAs interacting preferentially with the 3′-untranslated regions (3′-UTRs) of target mRNAs, negatively regulating gene expression post-transcriptionally.Citation10,Citation11 It has been demonstrated that miRNAs play vital roles in various cancers, and some miRNAs may be crucial for diagnosis, therapy and prognosis in human cancer.Citation12,Citation13 In previous work, researchers have identified miR-342-3p as an important cancer-related miRNA, including colon cancer, cervical cancer, gallbladder cancer and osteosarcoma.Citation14–Citation19 Given that miR-342-3p plays an important suppressive role in cancer, we aimed to determine whether miR-342-3p regulates the Warburg effect in HCC.
In the current study, we demonstrate that miR-342-3p suppresses aerobic glycolysis and cells proliferation through targeting IGF-1R-mediated PI3K/AKT/GLUT1 signaling pathway in liver cancer. Thus, activation of miRNA-342-3p may be a novel, promising way to treat HCC.
Materials and methods
Cell lines, RNA oligonucleotides, reagents
Human liver cancer cell lines HepG2 and MHCC97H, and human embryonic kidney cell line HEK293T were purchased from the American Type Culture Collection (Manassas, VA, USA). Wild-type and mutated miR-342-3p supposed targets on IGF-1R 3′-UTR were cloned into pmir-GLO dual-luciferase miRNA target expression vector (Promega Corporation, Fitchburg, WI, USA). IGF-1R siRNA was purchased from GenePharma (Shanghai, China). Anti-IGF-1R, anti-pIGF-1R (Tyr1135) and anti-GLUT1 antibodies were purchased from Cell Signalling Technology (Danvers, MA, USA). Anti-AKT, anti-pAKT and anti-β-actin antibodies were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). miR-342-3p mimics and miR-342-3p inhibitor were purchased from GenePharma.
Cell growth and colony formation assays
Cell growth was evaluated by a cell counting kit-8 (CCK-8) following the manufacturer’s protocols (Dojindo, Tokyo, Japan). For colony formation assay, transfected cells were seeded in 35 mm plates at a density of 3,000 cells per well. Two weeks later, colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet for 30 min. The number of colonies with diameters of more than 1.5 mm was counted.
miRNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA containing miRNA was extracted from cultured cells using a miRcute miRNA isolation kit (Tiangen, Beijing, China). Target miRNA was reverse transcribed to cDNA with the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen). The expression level of miRNA was identified by a miScript SYBR Green PCR Kit (Qiagen NV, Venlo, the Netherlands) and performed on the CFX96 system (BioRad Laboratories Inc., Hercules, CA, USA). miR-342-3p expression level was assessed by qRT-PCR with the following primers: 5′-GGGTCTCACACAGAAATCGC-3′ (forward), and 5′-CAGTGCGTGTCGTGGAGT-3′ (reverse). The control primers (U6) were 5′-CGCGCTTCGGCAGCACATATACT-3′ (forward) and 5′-ACGCTTCACGAATTTGCGTGTC-3′ (reverse). IGF-1R mRNA expression was determined by qRT-PCR with the following primers: 5′-GAGGAGATGGAGCCTGGCTTC-3′ (forward), and 5′-GTGTCTGTCGGGCAGTGGCAG-3′ (reverse). The control primers for β-actin were 5′-ATCACCATTGGCAATGAGCG-3′ (forward) and 5′-TTGA AGGTAGTTTCGTGGAT-3′ (reverse). Normalized to the corresponding control, the relative quantification value of the target was computed by the comparative cycle threshold methods.
Luciferase reporter assay
Cells seeded in 24-well plates at 1×105 cells per well were cotransfected with luciferase reporters, either wild-type or mutant IGF-1R 3′-UTR, in combination with miR-342-3p mimics or negative control for miRNA mimics using Lipofectamine 2000. Forty-eight hours later, cells were harvested and analyzed for luciferase and β-galactosidase activities according to the manufacture’s instruction (Promega).
Glucose uptake, lactate, and ATP assays
Glucose Uptake Colorimetric Assay kit, Lactate Assay Kit II, and ATP Colorimetric Assay kit were made use of evaluating glucose uptake, lactate production and ATP generation respectively, according to the manufacturer’s instructions (BioVision, Milpitas, CA, USA). For glucose uptake colorimetric assay, cells were seeded at 1×104 per well in a 96-well plate. The cells were starved for glucose by preincubating with 100 μL Krebs-Ringer-Phosphate-HEPES buffer containing 2% BSA for 40 min. Ten microliters of 10 mM 2-DG was added, then the cells were incubated for 20 min. For lactate and ATP assays, 5×105 cells were homogenized in 50 μL corresponding assay buffer provided by the kits. Samples were centrifuged at 4°C, and the soluble fraction was assayed.
Extracellular acidification and oxygen consumption rate assays
The extracellular acidification rate (ECAR) and cellular oxygen consumption rate (OCR) were determined using the Seahorse XFe 96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). Experiments were performed according to the manufacturer’s instructions. ECAR and OCR were measured using Seahorse XF Glycolysis Stress Test Kit and Seahorse XF Cell Mito Stress Test Kit, respectively. Briefly, cells were seeded in a Seahorse XF 96 cell culture microplate at a density of 1×104 per well. For ECAR, after baseline measurements, glucose, the oxidative phosphorylation inhibitor oligomycin, and the glycolytic inhibitor 2-DG were sequentially injected into each well at indicated time points; and for OCR, oligomycin, the reversible inhibitor of oxidative phosphorylation FCCP (p-trifluoromethoxy carbonyl cyanide phenylhydrazone), and the mitochondrial complex I inhibitor rotenone plus the mitochondrial complex III inhibitor antimycin A (Rote/AA) were sequentially injected. Data were computed by Seahorse XF-96 Wave software. ECAR is shown in mpH/min and OCR in pmol/min.
In vivo small-animal PET imaging and lactate assays
The animal study was approved and monitored by the Ethics Committees of the Third Affiliated Hospital of Hebei Medical University (Shijiazhuang, China). All experiments were performed in accordance with the guidelines approved by Institutes of Health for the Care and Use of Laboratory Animals at the Third Affiliated Hospital of Hebei Medical University. For in vivo tumor assay, a total of 1×107 HepG2 cells with stable expression of pCDH or pCDH-miR-342 subcutaneously inoculated into the hind flank of nude mice (6-week-old male nude mice), respectively. All mice were kept for about 60 days until imaged by small-animal positron emission tomography (PET) imaging. An animal PET scanner (Philips Corp., Amsterdam, Netherlands) was used for performing the PET of tumor-bearing mice. After they were intraperitoneally anesthetized by pentobarbital (100 mg/kg; Sinopharm Chemical Reagent, Beijing, China), mice were injected intravenously with 3.7 MBq (100 μCi) of 18F radio-labeled fluorodeoxyglucose (18F-FDG). Five-minute emission scans were implemented to obtain attenuation correction data in the prone position at 60 min after injection. The radioactivity of tumors was measured using a NaI (Tl) well counter (China Atom Corp., Beijing, China). The mice were sacrificed about 60 days after tumor cell inoculation, and tumor tissues were excised for lactate production and protein levels study. Tissues were homogenized in the lactate assay buffer. Insoluble materials were removed after centrifuge. The soluble fraction was assayed directly according to the Lactate Assay Kit II manufacturer’s instructions (BioVision).
Statistical analysis
All the experiments were performed in triplicate and repeated 3 times in vitro. Statistical significance in cell proliferation, glucose uptake, lactate, ATP, ECAR and OCR, as well as luciferase reporter assays was determined by two-tailed Student’s t-test. The statistical analyses were computed by the SPSS 17.0 statistical software package. P-values of less than 0.05 were considered statistically significant.
Results
Aerobic glycolysis is critical for regulating hepatoma cell proliferation by miR-342-3p
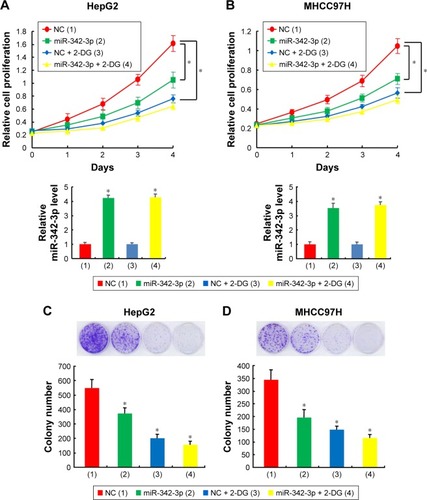
Since aerobic glycolysis plays a crucial role in cancer progression and miR-342-3p acts as a suppressor gene, we investigated whether aerobic glycolysis plays a role in miR-342-3p mediated suppression of hepatoma cell proliferation. Indeed, 2-deoxy-d-glucose (2-DG), the glycolytic inhibitor, weakened the ability of miR-342-3p mimics to inhibit proliferation in HepG2 and MHCC97H hepatoma cells (). Colony formation assays proved similar trends to that of growth curves mentioned above (). These data collectively show that aerobic glycolysis could take part in the regulation of cell proliferation by miR-342-3p.
Figure 1 Aerobic glycolysis is involved in regulating hepatoma cell proliferation by miR-342-3p.
Abbreviation: NC, negative control.
Identification of IGF-1R as a direct target of miR-342-3p
To explore the relationship between miR-342-3p and aerobic glycolysis, we searched for the potential target genes of miR-342-3p using publicly available databases (TargetScan and miRanda). Multiple genes were predicted as the potential miR-342-3p targets, from which we picked out those reported to associate with glycolysis (). Next, we performed Western blot analysis to confirm the potential targets in human kidney embryonic HEK293T cells. As previously reported,Citation27 overexpression of miR-342-3p mimics inhibited the E2F1 expression (). Moreover, miR-342-3p repressed the expression of IGF-1R, a key glycolysis, but not ENO1 (alpha-enolase), another enzyme involved in glycolysis. Therefore, we chose IGF-1R for further study.
IGF-1R is well known to activate intracellular AKT signaling pathway, which subsequently upregulate the expression of GLUT1 on plasma membrane and then enhance glucose metabolism in cancer cells.Citation20,Citation21 Therefore, we tested if miR-342-3p influenced the process mentioned above. As expected, overexpression of miR-342-3p mimics suppressed the levels of IGF-1R expression, the phosphorylation form of AKT and the GLUT1 expression in HepG2 and MHCC97H cells (). On the contrary, anti-miR-342-3p facilitated IGF-1R expression, and increased that of phosphorylation form of AKT and the expression level of GLUT1 (). To determine how miR-342-3p influenced the expression of IGF-1R, we detected the expression of IGF-1R mRNA after transfection of miR-342-3p mimics or miR-342-3p inhibitor into HepG2 and MHCC97H cells. The levels of IGF-1R mRNA were down-regulated upon miR-342-3p overexpression, whereas the levels of IGF-1R mRNA were up-regulated upon miR-342-3p inhibition ().
Figure 2 IGF-1R is a direct target of miR-342-3p.
Abbreviations: NC, negative control; 3′-UTR, 3′-untranslated region.
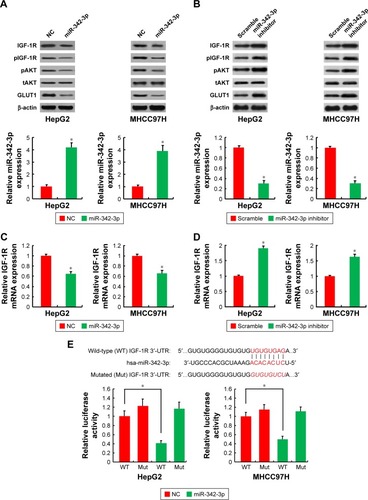
To further identify whether the negative regulation of miR-342-3p on IGF-1R expression were mediated through binding of IGF-1R directly, we transfected HepG2 and MHCC97H cells with wild-type IGF-1R 3′-UTR or mutated IGF-1R 3′-UTR luciferase reporter and miR-342-3p. miR-342-3p reduced the wild-type IGF-1R 3′-UTR reporter activity, but not the luciferase activity of the reporter in which the binding sites for miR-342-3p were mutated (). In short, these results reveal that miR-342-3p inhibits IGF-1R expression by targeting its 3′-UTR directly in hepatoma cells.
miR-342-3p suppresses cell proliferation and glycolysis mainly through inhibition of IGF-1R expression in hepatoma cells
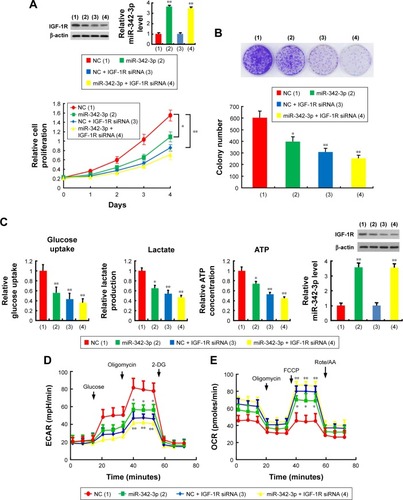
miR-342-3p has been shown to inhibit hepatoma cell proliferation.Citation22 Thus, we tested if these functions mediated by miR-342-3p were dependent on its target IGF-1R. Cell proliferation and colony formation assays determined that IGF-1R knockdown significantly reduced cell proliferation and colony formation ability in hepatoma cells. More importantly, IGF-1R knockdown abolished the ability of miR-342-3p to regulate hepatoma cell proliferation, revealing that miR-342-3p suppresses liver cancer cell proliferation through suppression of IGF-1R expression (). To further validate this, we performed IGF-1R rescue experiment. Transfection of miR-342-3p mimics decreased the proliferation of HepG2 cells. These effects were reversed by IGF-1R reexpression in the miR-342-3p-transfected cell lines (). Next, we showed that miR-342-3p mimics decreased glucose uptake, lactate production and ATP generation; however, these effects were eliminated by IGF-1R knockdown. Again, miR-342-3p mimics decreased glucose uptake, lactate production and ATP generation, which could be reversed by IGF-1R reexpression in the miR-342-3p-transfected cells (). miR-342-3p mimics also exhibited decreased ECAR, which reflects overall glycolytic flux, and increased OCR, an indicator of mitochondrial respiration. When IGF-1R was knockdown, miR-342-3p mimics had no effects on the glycolytic phenotype (), indicating that miR-342-3p represses the glycolytic phenotype via IGF-1R. Taken together, these results suggest that miR-342-3p restrain glycolysis via inhibition of IGF-1R expression in hepatoma cells.
Figure 3 miR-342-3p suppresses hepatoma cell proliferation and glycolysis mainly through inhibition of IGF-1R expression.
Abbreviations: NC, negative control; CCK-8, cell counting kit-8; ECAR, extracellular acidification rate; OCR, oxygen consumption rate; FCCP, p-trifluoromethoxy carbonyl cyanide phenylhydrrazone; Rote/AA, rotenone plus the mitochondrial complex III inhibitor antimycin A.
miR-342-3p inhibits tumor growth and glucose uptake of HCC in nude mice
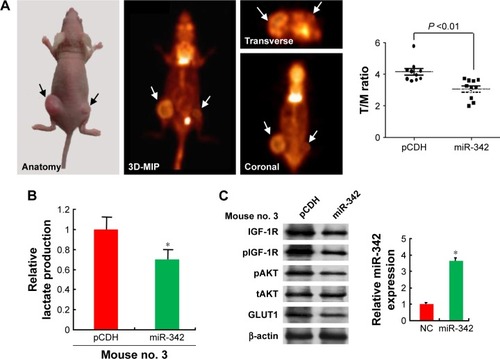
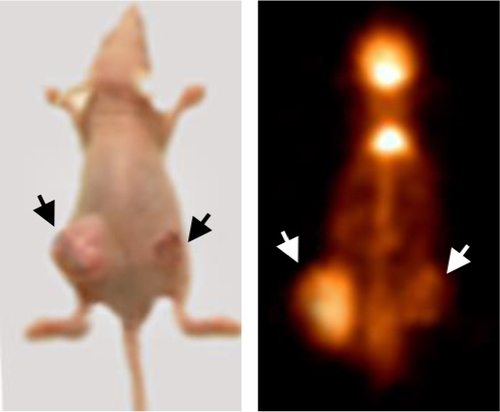
To examine the effects of the miR-342-3p on glycolysis in vivo, we used 18F-FDG microPET scans to measure glucose uptake in tumor xenografts in nude mice. The tumors with miR-342-3p overexpression revealed decreased glucose uptake ( and ). As expected, the tumors with miR-342-3p grew more slowly than the empty control vector. Lactate production analysis of the tumor masses further confirmed that miR-342-3p repressed the lactate production (). In addition, the expression of pAKT, GLUT1, pIGF-1R, IGF-1R, was downregulated by miR-342-3p overexpression, suggesting that the miR-342-3p inhibits tumor growth and glucose uptake in HCC ().
Figure 4 miR-342-3p/IGF1R axis regulates glycolysis and tumor growth in vivo.
Abbreviations: FDG microPET, fluorodeoxyglucose micropositron emission tomography; T/M, tumor to muscle; NC, negative control.
Discussion
We have demonstrated for the first time, to the best of our knowledge, that miR-342-3p regulates Warburg effect in suppression of liver tumor growth both in vitro and in vivo. First, glycolytic inhibitor 2-DG weakens the effect of miR-342-3p to regulate hepatoma cell proliferation. Mechanistically, through targeting IGF-1R 3′-UTR directly, miR-342-3p decreases IGF-1R expression, and then decreases the level of pAKT and GLUT1. miR-342-3p inhibits glucose uptake, lactate production, ATP production and induces a switch from glycolysis to mitochondrial respiration through inhibition of IGF-1R mediated the Warburg effect. In vivo micro-PET imaging demonstrates that miR-342-3p significantly inhibits the glucose uptake of liver tumor and lactate production. These observations collectively suggest that the effect of miR-342-3p on the Warburg effect is crucial for regulating HCC tumorigenesis.
Cancer cells frequently exhibit high level of aerobic glycolysis (the Warburg effect), regarded as a hallmark of cancer.Citation8,Citation9 This metabolic reprogramming gives cancer cells a growth benefit by providing energy for cancer cell progression. Targeting glucose metabolism may offer a selective molecular mechanism by which to kill cancer cells.Citation23 In solid tumors, miR-342-3p has been identified as a tumor suppressor that inhibits tumorigenesis and cancer progression. For example, miR-342-3p suppresses cervical cancer proliferation, migration and invasion by inhibiting FOXM1.Citation24 miR-342-3p targets AGR2 and RAP2B in lung cancer to restrain cancer cell proliferation, migration and invasion.Citation25,Citation26 Tai et al reported that miR-342-3p regulates MYC transcriptional activity through direct repression of E2F1 in human lung cancer.Citation27 In liver cancer, Zhao et al showed that miR-342-3p affects cell proliferation via regulating NF-kB pathway.Citation22 However, whether miR-342-3p regulates the Warburg effect is unknown. We proved that miR-342-3p not only inhibits the Warburg effect but also suppresses hepatoma cell proliferation through inhibition of IGF-1R-mediated Warburg effect.
IGF-1R is a transmembrane protein of the insulin receptor family containing two extracellular α subunits with the ligand-binding site and two transmembrane β subunits with intracellular tyrosine kinase activity. It has been demonstrated that IGF-1R was upregulated in numerous cancers, including HCC, renal cell carcinoma, gastric cancer and so on.Citation28–Citation30 Deletion of the IGF-1R gene results in a 50% decrease in the size of mouse embryos.Citation31 IGF-1R interacts with its ligand IGF-1 and plays crucial role in tumorigenesis, proliferation and metastasis through multiple signaling pathways downstream including PI3K/AKT and MAPK/ERK signal pathways.Citation32,Citation33 It has been reported that activation of IGF-1R stimulates PI3K/AKT pathway, regulating cell proliferation, migration, invasion.Citation34–Citation37 Interference with the function of IGF-1R appears to be potential therapeutic strategy for cancer. In fact, development of drugs targeting IGF-1R is being investigated clinical trials as anticancer treatment.Citation38
AKT is a serine/threonine kinase, which is also called protein kinase B. The activity of AKT relays on the phos-phorylation at its hydrophobic motif (Ser473) and activation loop (Thr308), which can be activated by variety of growth factors such as IGF.Citation39 Phosphorylated AKT (pAKT) is the active form of AKT influencing a diverse range of intracellular functions.Citation40 Zhang et al reported that AKT kinase was upregulated and significantly associated with tumor aggressiveness and poor prognosis in patients with HCC.Citation41 In addition, previous investigations indicated that increased expression of pAKT was associated with poor prognosis in patients with HCC, colorectal cancer, breast cancer, renal cell carcinoma, etc.Citation42–Citation45 Inhibition of AKT signal pathway is a target for cancer therapy. Some researchers reported that the PI3K/AKT pathway was involved in the control of GLUT1 trafficking and activity.Citation21,Citation46,Citation47 Inactivation of PI3K/AKT signaling pathway results in GLUT1 expression inhibition, thus affecting glucose metabolism, and finally suppressing the proliferation of colorectal cancer cells.Citation48
In short, our data establish molecular mechanism of miR-342-3p in regulating IGF-1R-mediated Warburg effect. miR-342-3p activation may be effective for HCC therapy with IGF-1R overexpression.
Conclusion
Our investigation demonstrated that miR-342-3p inhibits hepatoma cell proliferation by targeting IGF-1R directly, dampening glycolysis mediated by PI3K/AKT/GLUT1 axis. These findings outline the importance of the miR-342-3p in the Warburg effect and HCC tumorigenesis. Therefore, upregulation of miR-342-3p may be a prospective way to treat IGF-1R overexpression liver cancer patients.
Acknowledgments
This work was supported by National Natural Science Foundation (81472589, 81672602, 81372834, 81471466, 81572597, 81641023, and 81502264), Logistics Scientific Research project (BWS16J010), Beijing Nova Program (Z171100001117024), and Beijing Capital Special Development Application Program (Z141107002514159).
Supplementary materials
Figure S1 Screening for the target of miR-342-3 in HEK293T cells.
Abbreviation: NC, negative control.
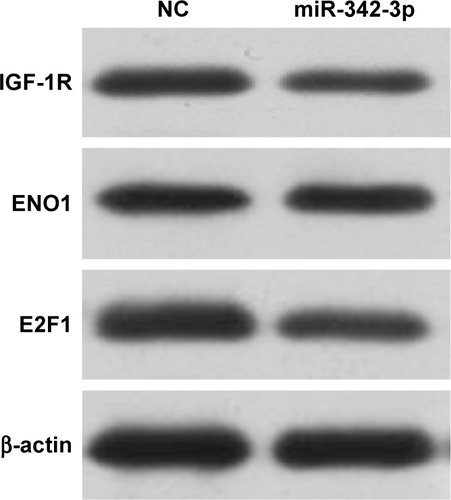
Figure S2 miR-342-3p suppresses proliferation, glucose uptake and the production of lactate and ATP through inhibition of IGF-1R expression in HCC cells.
Abbreviations: NC, negative control; CCK-8, cell counting kit-8.
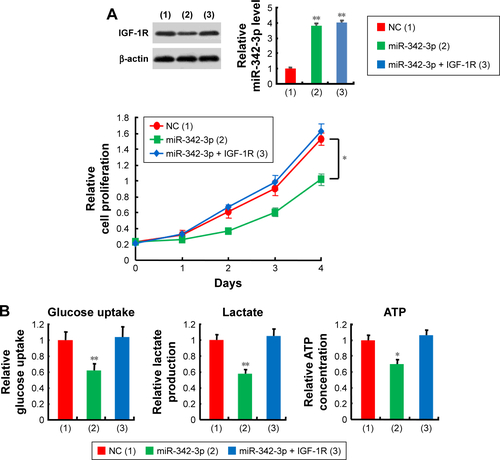
Figure S3 miR-342-3p/IGF-1R axis regulates glycolysis and tumor growth in vivo.
Abbreviation: FDG microPET, fluorodeoxyglucose micropositron emission tomography.
Disclosure
The authors report no conflicts of interest in this work.
References
- FaraziPADePinhoRAHepatocellular carcinoma pathogenesis: from genes to environmentNat Rev Cancer20066967468716929323
- CabreraRNelsonDRReview article: the management of hepatocellular carcinomaAliment Pharmacol Ther201031446147619925500
- FornerAReigMBruixJAlpha-fetoprotein for hepatocellular carcinoma diagnosis: the demise of a brilliant starGastroenterology20091371262919482098
- BruixJShermanMManagement of hepatocellular carcinomaHepatology20054251208123616250051
- LlovetJMBurroughsABruixJHepatocellular carcinomaLancet200336293991907191714667750
- MarreroJAWellingTModern diagnosis and management of hepatocellular carcinomaClin Liver Dis200913223324719442916
- JemalABrayFCenterMMFerlayJWardEFormanDGlobal cancer statisticsCA Cancer J Clin2011612699021296855
- HanahanDWeinbergRAHallmarks of cancer: the next generationCell2011144564667421376230
- Vander HeidenMGCantleyLCThompsonCBUnderstanding the Warburg effect: the metabolic requirements of cell proliferationScience200932459301029103319460998
- BartelDPMicroRNAs: genomics, biogenesis, mechanism, and functionCell2004116228129714744438
- CalinGACroceCMMicroRNA signatures in human cancersNat Rev Cancer200661185786617060945
- GaurAJewellDALiangYCharacterization of microRNA expression levels and their biological correlates in human cancer cell linesCancer Res20076762456246817363563
- SrivastavaSKAhmadAZubairHMicroRNAs in gynecological cancers: small molecules with big implicationsCancer Lett201740712313828549791
- WangQLiPCLiALPlasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of gliomaJ Exp Clin Cancer Res2012319723174013
- TaoKYangJGuoZHuYShengHGaoHYuHPrognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancerAm J Transl Res20146439140125075256
- JungCKJungSHYimSHPredictive microRNAs for lymph node metastasis in endoscopically resectable submucosal colorectal cancerOncotarget2016722329023291527096956
- LeeKHLeeJKChoiDWPostoperative prognosis prediction of pancreatic cancer with seven microRNAsPancreas201544576476825906450
- ZhangSLiuLLvZLiQGongWWuHMicroRNA-342-3p inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting astrocyte-elevated gene-1 (AEG-1)Oncol Res20172591505151528276315
- WangSHMaFTangZHLong non-coding RNA H19 regulates FOXM1 expression by competitively binding endogenous miR-342-3p in gallbladder cancerJ Exp Clin Cancer Res201635116027716361
- PollakMNInsulin and insulin-like growth factor signalling in neoplasiaNat Rev Cancer200881291592819029956
- PhadngamSCastiglioniAFerraresiAMoraniFFolloCIsidoroCPTEN dephosphorylates AKT to prevent the expression of GLUT1 on plasmamembrane and to limit glucose consumption in cancer cellsOncotarget2016751849998502027829222
- ZhaoLZhangYBmiR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-kB pathwayBiochem Biophys Res Commun2015457337037725580008
- HamanakaRBChandelNSTargeting glucose metabolism for cancer therapyJ Exp Med2012209221121522330683
- LiXRChuHJLvTWangLKongSFDaiSZmiR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancerFEBS Lett2014588173298330725066298
- XueXFFeiXYHouWJZhangYJLiuLHuRKmiR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancerCancer Lett201841217017829107102
- XieXLiuHTWangMSmiR-342-3p targets RAP2B to suppress proliferation and invasion of non-small cell lung cancer cellsTumour Biol20153675031503825663460
- TaiMCKajinoTNakatochiMmiR-342-3p regulates MYC transcriptional activity via direct repression of E2F1 in human lung cancerCarcinogenesis201536121464147326483346
- ECLiJShaoDThe insulin-like growth factor-I receptor inhibitor picropodophyllin-induced selective apoptosis of hepatocellular carcinoma cell through a caspase-dependent mitochondrial pathwayOncol Res201321210311024406046
- SichaniMMYazdiFSMoghaddamNAPrognostic value of insulin-like growth factor-I receptor expression in renal cell carcinomaSaudi J Kidney Dis Transpl2010211697420061696
- GrykoMKiślukJCepowiczDExpression of insulin-like growth factor receptor type 1 correlate with lymphatic metastases in human gastric cancerPol J Pathol201465213514025119174
- EfstratiadisAGenetics of mouse growthInt J Dev Biol19984279559769853827
- CaoZLiuL-ZDixonDAZhengJZChandranBJiangB-HInsulin-like growth factor-I induces cyclooxygenase-2 expression via PI3K, MAPK and PKC signaling pathways in human ovarian cancer cellsCell Signal20071971542155317341442
- GuoTFengYLiuQYangXJiangTChenYZhangQMicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1RTumour Biol20143511112691127525117070
- PollakMNInsulin-like growth factors and neoplasiaNovartis Found Symp20042628498 discussion: 98–107, 265–26815562824
- WernerHLeRoithDThe role of the insulin-like growth factor system in human cancerAdv Cancer Res1996681832238712068
- PollakMThe insulin and insulin-like growth factor receptor family in neoplasia: an updateNat Rev Cancer201212315916922337149
- KingHAleksicTHaluskaPMacaulayVMCan we unlock the potential of IGF-1R inhibition in cancer therapy?Cancer Treat Rev20144091096110525123819
- HartogHWesselingJBoezenHMvan der GraafWTThe insulin-like growth factor 1 receptor in cancer: old focus, new futureEur J Cancer200743131895190417624760
- WangLGuoWMaJAberrant SIRT6 expression contributes to melanoma growth: role of the autophagy paradox and IGF-AKT signalingAutophagy Epub20171231
- VivancoISawyersCLThe phosphatidylinositol 3-kinase AKT pathway in human cancerNat Rev Cancer20022748950112094235
- ZhangYGuoXYangMYuLLiZLinNIdentification of AKT kinases as unfavorable prognostic factors for hepatocellular carcinoma by a combination of expression profile, interaction network analysis and clinical validationMol BioSyst201410221522224247267
- ChenYLChenPMMingYZLinPYChuCPChuPYPhosphorylated AKT expression in tumor-adjacent normal tissue is associated with poor prognosis in patients with hepatocellular carcinomaOncol Lett20171467461746629344189
- MalinowskyKNitscheUJanssenKPActivation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancerBr J Cancer201411082081208924619078
- TokunagaEKimuraYOkiEAkt is frequently activated in HER2/neu-positive breast cancers and associated with poor prognosis among hormone-treated patientsInt J Cancer2006118228428916049961
- HagerMHaufeHKemmerlingRHitzlWMikuzGMoserPLKolbitschCIncreased activated Akt expression in renal cell carcinomas and prognosisJ Cell Mol Med2009138B2181218818774962
- WiemanHLWoffordJARathmellJCCytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and traffickingMol Biol Cell20071841437144617301289
- MelstromLGSalabatMRDingXZMilamBMStrouchMPellingJCBentremDJApigenin inhibits the GLUT-1 glucose transporter and the phosphoinositide 3-kinase/Akt pathway in human pancreatic cancer cellsPancreas200837442643118953257
- WuXLWangLKYangDDEffects of Glut1 gene silencing on proliferation, differentiation, and apoptosis of colorectal cancer cells by targeting the TGF-β/PI3K-AKT-mTOR signaling pathwayJ Cell Biochem201811922356236728884839
