Abstract
Introduction
Acute myeloid leukemia (AML) is a common malignancy of the hematopoietic system. In bone marrow samples of AML patients, PDIA3 expression was higher than that in the samples of healthy controls. We aimed at exploring the effect of PDIA3 siRNA on proliferation, apoptosis, migration, and invasion of AML HL-60 and HEL cells.
Materials and methods
RT-PCR was performed to identify PDIA3 expression. Cell proliferation was assessed by MTT. Flow cytometry analysis and transwell were used to detect cell apoptosis, migration and invasion. Gene set enrich-ment analysis (GSEA) was employed to explore the PDIA 3-associated pathways in AML. Western blotting was used for protein expression detection.
Results
PDIA3 siRNA significantly inhibited the proliferation of AML cells at 24 and 48 h. PDIA3 siRNA notably enhanced the percentage of apoptotic cells. The migration and invasion abilities of HL-60 and HEL cells in the PDIA3 siRNA group were significantly suppressed compared with those in the control and siNC groups. GSEA of the Cancer Genome Atlas dataset showed that Kyoto Encyclopedia of Genes and Genomes oxidative phosphorylation and amino sugar and nucleotide sugar metabolism pathways could be correlated with PDIA3 expression; this was further confirmed in AML cells by Western blotting. MAPK signaling was also blocked by PDIA3 siRNA.
Conclusion
PDIA3 siRNA effectively enhanced apoptosis, and suppressed proliferation, invasion, and migration of AML cells by regulating oxidative phosphorylation and amino sugar and nucleotide sugar metabolism pathways, and MAPK signaling, which can provide novel therapeutic targets for AML.
Introduction
Acute myeloid leukemia (AML) comprises a heterogeneous group of hematological malignancies characterized by expansion of clonal myeloid blasts in the bone marrow, blood, and other tissues. About 21,380 new AML cases and 10,590 deaths from this disease were reported in 2017 by the American Cancer Society.Citation1 Despite the advancements in modern chemotherapy, the prognosis of patients with AML has remained poor.Citation2,Citation3 Identifying the mechanisms involved in AML metastasis may lead to innovative treatment methods and improved patient outcomes.
PDIA3, known as ERp57 or glucose-regulated protein, has a molecular weight of 58 kDa and is a thiol-oxidoreductase chaperone belonging to the protein disulfide isomerase (PDI) family.Citation4 PDIA3 expression has been observed in various types of human cancers, including ovarian, mammary, uterine, pulmonary, and gastric cancers, and hepatocellular carcinoma.Citation4 It is reported that PDIA3 expression is significantly increased in hepatocellular carcinoma, and high PDIA3 expression is associated with cancer cell proliferation and poor prognosis.Citation5 PDIA3 is also highly expressed in a newly established serous ovarian cancer cell line, YDOV-139.Citation6 PDIA3 expression is increased in primary ductal breast cancer with lymph node metastasis.Citation7 However, no studies have yet demonstrated the exact role of PDIA3 in AML, and the pathways that signal PDIA3 to exert its function have not been elucidated. Consequently, a possible involvement of PDIA3 subfamily in the development and progression of AML, a malignancy originating from the hematopoietic system, was proposed.
We previously found that PDIA3 expression was remarkably increased in AML patients. Therefore, in this study, we focused on the effect of PDIA3 siRNA on the migration and invasion in HL-60 and HEL cells, which markedly expressed PDIA3. Moreover, the underlying mechanisms have been investigated to provide a basis for the treatment of AML.
Materials and methods
Patients and tissue samples
Bone marrow cells were collected from 20 patients newly diagnosed with AML admitted to Ningbo First Hospital of Zhejiang University. Twenty healthy bone marrow donors served as normal controls. Preoperative clinical and pathological follow-up data were completed by all the patients. Written informed consent was obtained from the study subjects before the use of these clinical samples, and the study protocol was approved by the ethics committee of Ningbo First Hospital of Zhejiang University.
Cell culture
6TCM, HL-60, K-562, THP-1, HEL, and A3 cells were obtained from the Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, People’s Republic of China) and cultured in RPMI medium (St Louis, MO, USA) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2.
siRNA transfection
For siRNA transfection, HL-60 and HEL cells, which were selected for transfection due to high PDIA3 expression confirmed by Western blot, were seeded into 12-well tissue culture plates at a density of 6 × 104 cells/well. The PDIA3 siRNA or control siRNA (GenePharma, Shanghai, People’s Republic of China) were then transfected into cells by using Lipofectamine™ 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. After 48 h, the transfected cells were collected and processed for subsequent experiments.
Cell proliferation
Cell proliferation was evaluated by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Cells were seeded into 96-well plates at a density of 2,000 cells per well and cultured for indicated times, followed by incubation at 5% CO2 and 37°C following the manufacturer’s instructions. Absorbance was measured at 490 nm using a microplate reader absorbance test plate (Molecular Devices, San Jose, CA, USA). Cells were detected from three wells per group.
Annexin V-propidium iodide apoptosis detection
The cell apoptosis rate was detected by flow cytometry using an Annexin V-FITC apoptosis detection kit (Nanjing Keygen Biotechnology Co. Ltd., Nanjing, People’s Republic of China). Following 48-h transfection, the cells were collected, washed with ice-cold PBS, and then resuspended in 500 μL binding buffer at a concentration of 1 × 106 cells/mL. Subsequently, 5 μL Annexin V-FITC was added and incubated for 10 min at room temperature. After addition of 5 μL propidium iodide, the cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
Cell invasion and migration assay
Invasion and migration activity of HL-60 and HEL cells were determined using a 24-well transwell chamber coated with or without Matrigel (Becton Dickinson) on the upper surface of the membrane with 8 μm pore size (Sigma-Aldrich). Briefly, the transfected cells (1 × 104 cells/well) were added in a volume of 100 μL serum-free medium to the upper transwell chamber. The lower chamber was filled with medium containing 10% FBS. After 24-h incubation, the cells that had invaded or migrated to the lower surface of the filter were counted visually under a microscope (OLYMPUS).
Western blot analysis
Protein concentrations were determined using a BCA protein assay kit (Thermo Fisher Scientific). An equal amount of proteins per sample was loaded and separated by 10% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by electrotransfer onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). Following blocking with 5% skimmed-milk powder in Tris-buffered saline containing 0.05% Tween-20 for 1 h, the membranes were incubated with antibodies specific for MAT1, caspase-9, RhoC, CDK1, phosphorylation (p-) P38, P38, p-JNK, JNK, p-extracellular regulated protein kinase (ERK), ERK, PCNA, and GADPH overnight at 4°C. Subsequently, the membranes were washed and incubated with 1:1,000 dilutions of horseradish peroxidase-conjugated secondary antibodies for 1 h. The bands were visualized using enhanced chemiluminescence detection kit (Santa Cruz Biotechnology, Dallas, TX, USA).
Bioinformatics analysis
The gene set enrichment analysis (GSEA) of pathways and genes was performed based on the Cancer Genome Atlas (TCGA) dataset using the GSEA version 2.0 from the Broad Institute at MIT. In this analysis, the gene sets of fewer than 10 genes were excluded. The t-statistic mean of the genes was computed in each Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Using a permutation test 1,000 times, P ≤ 0.01 was chosen as the significance level cutoff for the most significant pathways related to PDIA3 expression.
Statistical analysis
All results are presented as the mean ± SD of three independent experiments. Data for multiple comparisons were subjected to one-way analysis of variance with SPSS version 13.0 followed by Dunnett’s test. P < 0.05 was considered statistically significant.
Results
High PDIA3 expression in the bone marrow of AML patients as well as in AML cell lines
We first identified PDIA3 expression in the bone marrow of AML patients and controls by RT-PCR. As displayed in , compared with that of normal control, the bone marrow of AML patients showed a remarkable increase in mRNA expression of PDIA3 (P < 0.05), suggesting that overexpression of PDIA3 may be involved in the initiation and/or progression of AML.
Figure 1 PDIA3 expression in 20 bone marrow tissues of acute myeloid leukemia (AML) and normal tissues.
Abbreviation: GADPH, glyceraldehyde 3-phosphate dehydrogenase.
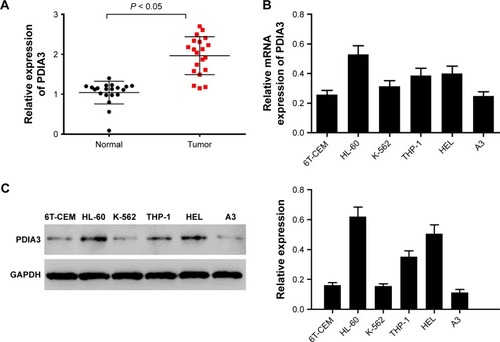
PDIA3 expression was also examined in AML cell lines by RT-PCR and Western blot analysis. As shown in , results from RT-PCR and Western blot showed that mRNA and protein levels of PDIA3 in HL-60 and HEL cells were higher than those in other cell lines. As a result, HL-60 and HEL cell lines were used to conduct further investigations.
Effect of PDIA3 siRNA on apoptosis of HL-60 and HEL cells
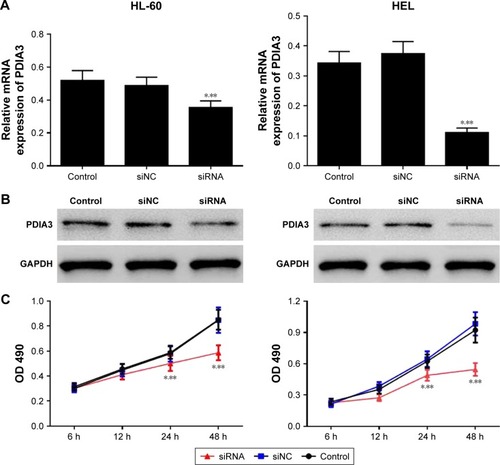
PDIA3 mRNA was interfered in HL-60 and HEL cell lines as previously described. The interference efficiency was then identified by RT-PCR and Western blot analysis. RT-PCR and Western blot analysis showed that protein levels declined dramatically in PDIA3 siRNA group in both the cell lines compared with the control and mock groups (). Cell proliferation was then determined by MTT assay. As shown in , the viability of both HL-60 and HEL cells in the siRNA group was significantly decreased at 24 and 48 h compared with the control and siNC groups (P < 0.01).
Figure 2 PDIA3 siRNA inhibits cell proliferation.
Abbreviation: GADPH, glyceraldehyde 3-phosphate dehydrogenase.
PDIA3 siRNA induced apoptosis and inhibited invasion and migration of HL-60 and HEL cells
After 48 h of transfection, cell apoptosis was analyzed using flow cytometer. As depicted in , compared with the control and siNC groups, the PDIA3 siRNA group displayed a significant increase in the percentage of apoptotic HL-60 and HEL cells in all the phases (P < 0.01).
Figure 3 PDIA3 siRNA induces cell apoptosis and inhibits cell migration and invasion.
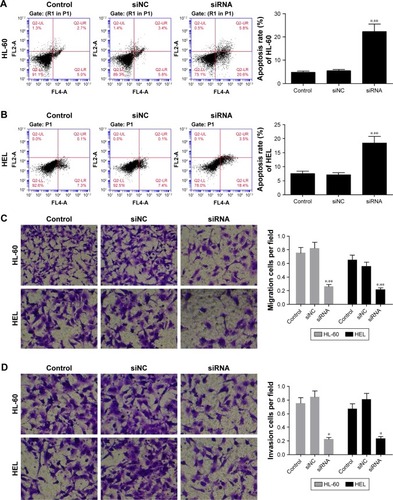
The transwell invasion test results are presented in . Transfection of PDIA3 siRNA resulted in a significantly weakened invasive ability of HL-60 and HEL cells as compared with that of the control and siNC groups (P < 0.01). Consistently, as shown in , the migration ability of HL-60 and HEL cells in the PDIA3 siRNA group was dramatically declined than that of control and mock cells (P < 0.01). Our data suggested that interference of PDIA3 reduces the migration and invasion ability of HL-60 and HEL cells.
PDIA3 siRNA regulated the expression of MAT1, PCNA, caspase-9, RhoC, and CDK1
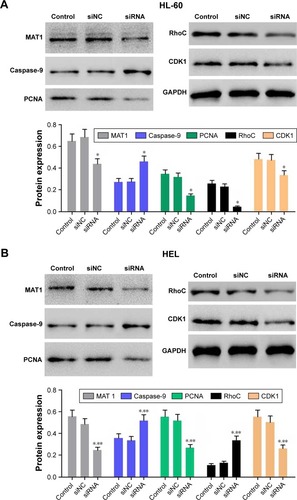
To elucidate the potential mechanism underlying PDIA3-induced cell invasion and migration, the expression levels of PCNA, MAT1, caspase-9, RhoC, and CDK1, which are closely associated with cell proliferation, invasion, and migration, were measured by Western blotting. reveals a significant decrease in protein levels of PCNA, CDK1, RhoC, and MAT1, but an evident increase in caspase-9 expression in the PDIA3 siRNA group compared with those in the control and siNC groups (P < 0.01). The results indicated that PDIA3 siRNA may exert an antitumor effect via regulation of MAT1, PCNA, caspase-9, RhoC, and CDK1 expression.
Figure 4 Effect of PDIA3 siRNA on expressions of PCNA, CDK1, MAT1, RhoC, and caspase-9.
Abbreviation: GADPH, glyceraldehyde 3-phosphate dehydrogenase.
PDIA3-associated pathways in AML
To further explore the role of PDIA3 in AML, we performed GSEA in AML samples with higher PDIA3 expression versus those with lower PDIA3 expression based on TCGA dataset. Our data implied that higher PDIA3 expression in AML samples was positively correlated with KEGG oxidative phosphorylation and amino sugar and nucleotide sugar metabolism pathways ().
Figure 5 Mechanisms of how PDIA3 exerts its function in AML.
Abbreviations: GADPH, glyceraldehyde 3-phosphate dehydrogenase; NES, normalized enrichment score.
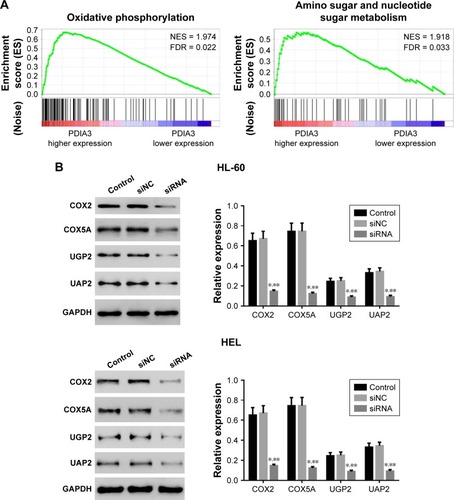
To further validate the GSEA results, we then detected the protein expression of oxidative phosphorylation (COX2 and COX5A) and amino sugar and nucleotide sugar metabolism pathway-related proteins (UGP2 and UAP1) in PDIA3-silenced AML cells. The levels of detected protein were significantly decreased in both HEL and HL-60 cells () after the downregulation of PDIA3.
Effect of PDIA3 siRNA on MAPK signaling
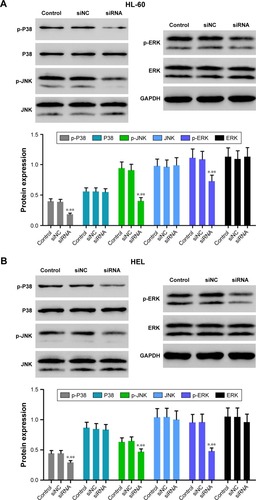
Accumulating evidence suggests that MAPK signaling plays a crucial role in carcinoma growth and metastasis. We then evaluated the phosphorylation (p-) of P38, JNK, and ERK by Western blotting. As shown in , PDIA3 siRNA significantly decreased the p-P38, p-JNK, and p-ERK expression compared with that in the control and siNC groups (P < 0.01). The results implied that PDIA3 siRNA inhibited cell proliferation, invasion, and migration, and induced cell apoptosis, possibly by suppressing MAPK phosphorylation.
Figure 6 Effect of PDIA3 siRNA on MAPK signaling.
Abbreviation: GADPH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
AML is the most common type of acute leukemia in adults, with over 20,000 new diagnoses each year in the US.Citation8,Citation9 AML usually results from a cooperative effect of several independent mutations that lead to cell malignant transformation. Several oncogenes that are most often activated in AML have been characterized.Citation10–Citation12 Interference of the involved critical genes or proteins is considered as a favorable strategy. Located at 15qn5, PDIA3 is reported to be a PDI. Increased PDIA3 expression was associated with an elevated Ki-67 index, indicating increased cancer cell proliferation, reduction in apoptotic cell death, and poor prognosis. Recent studies have shown that PDIA3 regulates cell invasiveness in cervical cancer and that a high level of PDIA3 is associated with a low patient survival rate.Citation13 As reported in our study, notable elevated expression of PDIA3 in bone marrow tissues of myeloid leukemia was found, proposing a new question about the role of PDIA3 in human AML development and progression. Consequently, the subsequent work was designed to explore the effect of PDIA3 siRNA on tumor migration and invasion in human AML.
The present study focused on the biological role of PDIA3 in AML. Notably, upregulated expression of PDIA3 was found in tissues of AML patients when compared with those of the healthy controls. HL-60 and HEL cell lines were further investigated for high expression of PDIA3. Our research revealed that PDIA3 siRNA significantly suppressed proliferation, invasion, and migration of AML HL-60 and HEL cells. PDIA3 siRNA also induced apoptosis of AML cells. Moreover, we demonstrated that the above effects may be mediated through the inhibition of PCNA, CDK1, RhoC, and MAT1 expression, and the promotion of caspase-9 expression at the protein level. The phosphorylation of P38, JNK, and ERK was also inhibited by PDIA3 siRNA in AML cells. Taken together, these findings indicated that PCNA, CDK1, MAT1, RhoC, and caspase-9, and MAPK signaling play a pivotal role in the process of antiproliferation, anti-invasion, antimigration, and apoptosis-induction triggered by PDIA3 siRNA.
We first found that siRNA of PDIA3 effectively inhibited cell proliferation and induced cell apoptosis. PCNA is a cyclin that is generally expressed in the S phase of the cell cycle.Citation14 CDK1 is one of the core regulatory proteins in cell cycle regulation.Citation14 In our results, siRNA of PDIA3 significantly suppressed the expression of PCNA and CDK1. Caspase-9 is a crucial element in cell apoptosisCitation15 and is upregulated, notably, by PDIA3 siRNA treatment. Tumor cell invasion and migration is the essential process in the metastasis of malignant tumor. MTA1 is a stress response protein that is upregulated in various stress-related situations, and cancer cells mostly live in a stressful environment. To cope with all these stresses, the expression of MTA1, which plays the role of a master regulator of gene expression, is upregulated and helps cancer cells to survive and migrate out of their original site.Citation16 It is reported that RhoC overexpression can give rise to cell invasion through regulation of cytoskeletal proteins and destruction of polarity in cancer cells. Our study showed that PDIA3 siRNA significantly inhibited the expression of MTA1 and RhoC.
To further explore the role of PDIA3 in AML, we performed GSEA of AML samples. In the present study, GSEA data indicated that higher PDIA3 expression was positively correlated with KEGG oxidative phosphorylation and amino sugar and nucleotide sugar metabolism pathways in AML samples. AML occurrence and progression depend upon oxidative phosphorylation and amino sugar and nucleotide sugar metabolism pathways.Citation17,Citation18
The expressions of oxidative phosphorylation-related factors (COX2 and COX5ACitation19) and amino sugar and nucleotide sugar metabolism pathway-related factors (UGP2 and UAP1Citation20) were significantly suppressed by PDIA3 knockdown. These data suggested the roles of PDIA3 on oxidative phosphorylation and amino sugar and nucleotide sugar metabolism, accounting for AML carcinogenesis. MAPK pathway is one of the most extensively studied protein kinase pathways, which can be subdivided into three subtypes, including ERK1/2, P38, and JNK.Citation21,Citation22 As an important signaling pathway in mammals, MAPK signaling pathway is stimulated by cytokines and involved in cell proliferation, differentiation, apoptosis, adhesion, invasion, metastasis, immunoregulation, and other important biological processes in AML.Citation23,Citation24
Collectively, we have demonstrated that PDIA3 siRNA effectively enhanced apoptosis and suppressed proliferation, invasion, and migration of AML HL-60 and HEL cells by regulating proliferation and metastasis-related proteins, oxidative phosphorylation and amino sugar, and nucleotide sugar metabolism pathways, and MAPK signaling. We will confirm these findings at in vivo level. These findings may elucidate the molecular mechanism underlying the effect of PDIA3 siRNA on cell growth, apoptosis, migration, and invasion in AML, and lead to the development of therapeutic approaches for the disease.
Acknowledgments
This work was supported by Scientific Research Program of Shanghai Municipal Commission of Health and Family Planning (program number 201540178).
Disclosure
The authors report no conflicts of interest in this work.
References
- IzziVLakkalaJDevarajanRExpression of a specific extracellular matrix signature is a favorable prognostic factor in acute myeloid leukemiaLeuk Res Rep2017991329270355
- GanapuleANemaniSKorulaAAllogeneic stem cell transplant for acute myeloid leukemia: evolution of an effective strategy in IndiaJ Glob Oncol20173677378129244983
- FontanaMCMarconiGFeenstraJDMChromothripsis in acute myeloid leukemia: biological features and impact on survivalLeukemia Epub20171218
- ChenJDoroudiMCheungJGrozierALSchwartzZBoyanBDPlasma membrane Pdia3 and VDR interact to elicit rapid responses to 1α,25(OH)(2)D(3)Cell Signal201325122362237323896121
- TakataHKudoMYamamotoTIncreased expression of PDIA3 and its association with cancer cell proliferation and poor prognosis in hepatocellular carcinomaOncol Lett20161264896490428101228
- ChayDChoHLimBJER-60 (PDIA3) is highly expressed in a newly established serous ovarian cancer cell line, YDOV-139Int J Oncol201037239941220596667
- RamosFSSerinoLTCarvalhoCMPDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancerGenet Mol Res20151426960696726125904
- SunYHuskeyRLTangLAdverse effects of intravenous vancomycin-based prophylaxis during therapy for pediatric acute myeloid leukemiaAntimicrob Agents Chemother2018623e01838e0191729229640
- SexauerANTasianSKTargeting FLT3 signaling in childhood acute myeloid leukemiaFront Pediatr2017524829209600
- SteinEMWalterRBErbaHPA phase 1 trial of vadastuximab talirine as monotherapy in patients with CD33-positive acute myeloid leukemiaBlood2018131438739629196412
- ZhouJDMaJCZhangTJHigh bone marrow ID2 expression predicts poor chemotherapy response and prognosis in acute myeloid leukemiaOncotarget2017854919799198929190891
- GuoQLuanJLiNMicroRNA-181 as a prognostic biomarker for survival in acute myeloid leukemia: a meta-analysisOncotarget2017851891308914129179505
- ChungHChoHPerryCSongJYlayaKLeeHKimJHDown-regulation of ERp57 expression is associated with poor prognosis in early-stage cervical cancerBiomarkers201318757357923957851
- JiaZZhaoWFanLShengWThe expression of PCNA, c-erbB-2, p53, ER and PR as well as atypical hyperplasia in tissues nearby the breast cancerJ Mol Histol201243111512022057681
- KimBSrivastavaSKKimSHCaspase-9 as a therapeutic target for treating cancerExpert Opin Ther Targets201519111312725256701
- SandhuCConnorMKislingerTSlingerlandJEmiliAGlobal protein shotgun expression profiling of proliferating mcf-7 breast cancer cellsJ Proteome Res20054367468915952714
- Whang-PengJHendersonESKnutsenTFreireichEJGartJJCyto-genetic studies in acute myelocytic leukemia with special emphasis on the occurrence of Ph1 chromosomeBlood19703644484575271809
- ZapfBCarpenterRJ3rdSnyderGGUlcerating tumor of tonsil and neck mass as occurrence of acute myelocytic leukemiaOtolaryngol Head Neck Surg19818945605636793965
- Corazao-RozasPGuerreschiPAndréFMitochondrial oxidative phosphorylation controls cancer cell’s life and death decisions upon exposure to MAPK inhibitorsOncotarget2016726394733948527250023
- MooltenFLTumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategyCancer Res19864610527652813019523
- ErikssonMPeña-MartínezPRamakrishnanRAgonistic targeting of TLR1/TLR2 induces p38 MAPK-dependent apoptosis and NFκB-dependent differentiation of AML cellsBlood Adv20171232046205729296851
- MakPYMakDHMuHApoptosis repressor with caspase recruitment domain is regulated by MAPK/PI3K and confers drug resistance and survival advantage to AMLApoptosis201419469870724337870
- ChoiCHXuHBarkHLeeTBYunJKangSIOhYKBalance of NF-kappaB and p38 MAPK is a determinant of radiosensitivity of the AML-2 and its doxorubicin-resistant cell linesLeuk Res20073191267127617218010
- LiQHuaiLZhangCIcaritin induces AML cell apoptosis via the MAPK/ERK and PI3K/AKT signal pathwaysInt J Hematol201397561762323550021
