Abstract
Background
In the last few years, accumulating evidence has indicated that numerous long noncoding RNAs (lncRNAs) are abnormally expressed in gastric cancer (GC) and are associated with the survival of GC patients. This study aimed to conduct a meta-analysis on 19 lncRNAs (AFAP1 antisense RNA 1 [AFAP1-AS1], CDKN2B antisense RNA 1 [ANRIL], cancer susceptibility 15 [CASC15], colon cancer associated transcript 2 [CCAT2], gastric adenocarcinoma associated, positive CD44 regulator, long intergenic noncoding RNA [GAPLINC], H19, imprinted maternally expressed transcript [H19], HOX transcript antisense RNA [HOTAIR], HOXA distal transcript antisense RNA [HOTTIP], long intergenic non-protein coding RNA 673 [LINC00673], metastasis-associated lung adenocarcinoma transcript 1 [MALAT1], maternally expressed 3 [MEG3], promoter of CDKN1A antisense DNA damage activated RNA [PANDAR], Pvt1 oncogene [PVT1], SOX2 overlapping transcript [Sox2ot], SPRY4 intronic transcript 1 [SPRY4-IT1], urothelial cancer associated 1 [UCA1], X inactive specific transcript [XIST], ZEB1 antisense RNA 1 [ZEB1-AS1] and ZNFX1 antisense RNA 1 [ZFAS1]) to systematically estimate their prognostic value in GC.
Methods
The qualified literature was systematically searched in PubMed, Web of Science, Embase and Cochrane Database of Systematic Reviews (up to March 16, 2018), and one meta-analysis relating to the relationship between lncRNA expression and overall survival (OS) of GC patients was performed. The only evaluation criterion of survival results was OS.
Results
A total of 6,095 GC patients and 19 lncRNAs from 51 articles were included in the present study. Among the listed 19 lncRNAs, 18 lncRNAs (other than SPRY4-IT1) showed a significantly prognostic value (P<0.05).
Conclusion
This meta-analysis suggested that the abnormally expressed lncRNAs (AFAP1-AS1, ANRIL, CASC15, CCAT2, GAPLINC, H19, HOTAIR, HOTTIP, LINC00673, MALAT1, MEG3, PANDAR, PVT1, Sox2ot, UCA1, XIST, ZEB1-AS1 and ZFAS1) were significantly associated with the survival of GC patients, among which AFAP1-AS1, CCAT2, LINC00673, PANDAR, PVT1, Sox2ot, ZEB1-AS1 and ZFAS1 were strong candidates in predicting the prognosis of GC patients.
Introduction
In the last few years, accumulating evidence has indicated that numerous long noncoding RNAs (lncRNAs) are abnormally expressed in gastric cancer (GC) and are associated with the survival of GC patients.Citation1–Citation113 GC is the fourth most diagnosed tumor type and the third most common origin of tumor-related death all over the world.Citation114,Citation115 Although the incidence and mortality of GC are declining, >24,590 individuals are diagnosed with GC per year, of which 10,720 die from GC in the USA.Citation116 Although diagnosis and treatment strategies have been improved, the number of surviving cases remains low, since diagnosis often occurs in the late stages.Citation116,Citation117 Thus, the molecular characteristics about the carcinogenesis of GC and the recognition of new biomarkers for GC are urgently needed.
lncRNA is a new type of noncoding RNA that has a length of >200 nucleotides (nt) and lacks important open reading frameworks and can be divided into five main categories (sense, antisense, bidirectional, intronic and intergenic).Citation118 Abundant evidence has demonstrated that lncRNAs play significant regulatory roles in tumor biology via various mechanisms affecting transcriptional and posttranscriptional levels.Citation118–Citation120 Currently, for both cell behavior and clinicopathological factors, significant advances with respect to lncRNA effects on GC have been discovered.Citation121
On account of the obvious expression differences between normal and malignant tissues as well as causal roles of lncRNAs in cancer development, lncRNAs are now attracting increasing attention, which has led to numerous investigations of the correlation between lncRNA states and clinical results in GC. Nevertheless, most of these studies were performed with small samples, and there were inconsistently observed connections. Consequently, we conducted a meta-analysis to determine the accurate role of lncRNAs in the prognosis of GC patients, which possibly supplied us with new insights into the clinical value of combined detection in forecasting prognostic results and determining promising biomarkers in GC treatment strategies.
Methods
Literature search strategy
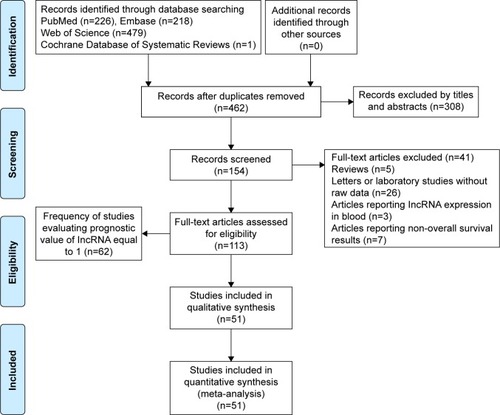
We basically performed a systematic selection of papers published in English from four databases (PubMed, Web of Science, Embase and Cochrane Database of Systematic Reviews). A comprehensive search was conducted using the subject term: lncRNA and gastric cancer. Two authors (Song Gao and Zhi-Ying Zhao) checked the titles and abstracts of the retrieved papers, and Yue Zhang reevaluated uncertain data. shows the flow diagram of the literature search and selection.
Inclusion criteria
We set up inclusion criteria for qualified papers, which were analyzed using our full-text assessment: 1) articles concerning the pertinence between lncRNA level in cancer tissues and prognosis of GC patients; 2) the survival results were estimated using overall survival (OS) and 3) full-text papers published in English.
Exclusion criteria
Articles that did not meet the abovementioned inclusion criteria, reviews, letters and laboratory studies without raw data were excluded. Articles of non-dichotomous lncRNA expression levels and frequency of studies evaluating prognostic value of lncRNAs equal to 1 were also excluded. If more than one paper had been published on the identical study cohort, only the most well-rounded investigation was selected for this research. In addition, if both of the univariate and multivariate outcomes were covered, only the latter were chosen, since they were adjusted for confounding factors.
Research frequency
gives the frequency of investigations reporting prognosis of GC patients, which included the lncRNA name, frequency of researched lncRNA and reference.
Table 1 Research frequency of lncRNAs in GC
Data extraction
The survival data were recovered from qualified articles independently by two authors (Song Gao and Zhi-Ying Zhao). Data extracted from them are as follows: researched lncRNA, first author’s name, paper publication year, reference, patient’s nationality, study design, histological type, patient number, neoplasm staging, cutoff value, detected method, follow-up period, survival analysis type, HRs and 95% CIs. The detailed data are shown in . If HR and 95% CI were not directly shown in the paper, data from survival curve were extracted. Disagreements were discussed with the third investigator (Yue Zhang).
Table 2 Basic information of included articles
Statistical analysis
Stata version 13.0 (StataCorp LP, College Station, TX, USA) was used for the whole meta-analysis. HR and 95% CI from GC patients were calculated on the basis of survival curve and patient number using Engauge Digitizer version 4.1 and Tierney’s method.Citation122 The random-effect model was used in the whole article because different histological type (frozen, formalin-fixed paraffin-embedded or undefined) from GC patients at different neoplasm staging, cutoff value and lncRNA detected method was used in the single study. The HR was considered significant if its P-value was <0.05 and 95% CI did not contain the value 1. Furthermore, the lncRNA was considered as a strong biomarker of prognosis, if its HR was >2. The Begg’s funnel plot was used to estimate publication bias, and a two-tailed P-value <0.05 was considered as significant. The sensitivity analysis was performed to examine how sensitive the merged HR was if the single study was removed, and if the point of evaluation was outside the 95% CI after it was removed from the whole analysis, a single research was considered as excessive influence.
Results
Meta-analysis
gives the basic information of the merged meta-analysis for researched lncRNAs.
Table 3 HR with 95% CI of lncRNA expression in GC
AFAP1 antisense RNA 1 (AFAP1-AS1), CDKN2B antisense RNA 1 (ANRIL), cancer susceptibility 15 (CASC15), colon cancer-associated transcript 2 (CCAT2), gastric adenocarcinoma associated, positive CD44 regulator, long intergenic noncoding RNA (GAPLINC) and H19, imprinted maternally expressed transcript (H19) demonstrated significantly prognostic value
Two articlesCitation3,Citation4 reported the relationship between high AFAP1-AS1 expression and OS, indicating that GC patients with its high expression had significantly worse OS than those with its low expression (HR=2.47, 95% CI=1.41–4.30, P<0.01).
Two researchesCitation9,Citation10 covered the connections between high ANRIL expression and OS, suggesting that GC patients with its high expression had significantly poorer OS than those with its low expression (HR=1.68, 95% CI=1.16–2.43, P<0.01).
Two investigationsCitation18,Citation19 analyzed the associations between high CASC15 expression and OS, showing that GC patients with its high expression had significantly shorter OS than those with its low expression (HR=1.99, 95% CI=1.21–3.28, P<0.01).
Two studiesCitation21,Citation22 focused on the correlation between high CCAT2 expression and OS, manifesting that GC patients with its high expression had significantly worse OS than those with its low expression (HR=2.17, 95% CI=1.53–3.09, P<0.01).
Two papersCitation32,Citation33 paid attention to the pertinence between high GAPLINC expression and OS, demonstrating that GC patients with its high expression had significantly poorer OS than those with its low expression (HR=1.49, 95% CI=1.18–1.89, P<0.01).
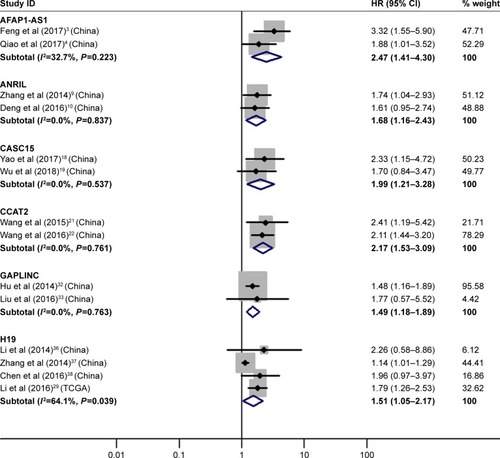
Four literatureCitation29,Citation36–Citation38 described the relativity between high H19 expression and OS, proving that GC patients with its high expression had significantly shorter OS than those with its low expression (HR=1.51, 95% CI=1.05–2.17, P=0.03; ).
Figure 2 Forest plot of pooled analyses of OS in association with high AFAP1-AS1, ANRIL, CASC15, CCAT2, GAPLINC and H19 expression levels.
Abbreviations: AFAP1-AS1, AFAP1 antisense RNA 1; ANRIL, CDKN2B antisense RNA 1; CASC15, cancer susceptibility 15; CCAT2, colon cancer associated transcript 2; GAPLINC, gastric adenocarcinoma associated, positive CD44 regulator, long intergenic noncoding RNA; H19, H19, imprinted maternally expressed transcript. OS, overall survival.
HOX transcript antisense RNA (HOTAIR) demonstrated significantly prognostic value
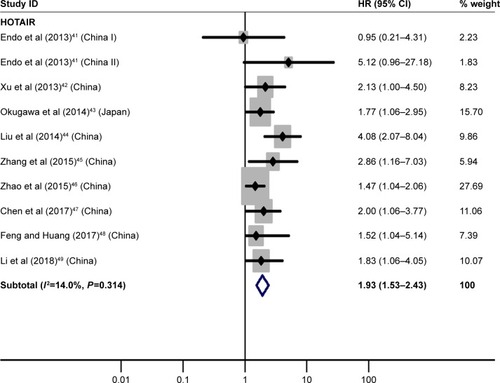
Nine essaysCitation41–Citation49 discussed the relation between high HOTAIR expression and OS, illuminating that GC patients with its high expression had significantly worse OS than those with its low expression (HR=1.93, 95% CI=1.53–2.43, P<0.01; ).
Publication bias
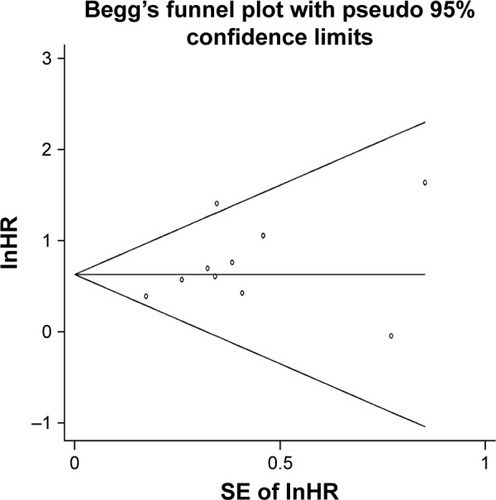
The Begg’s funnel plot was used to estimate publication bias, and its P-value was 0.20, so there was no significant publication bias in the pooled analysis of OS about high HOTAIR expression ().
Sensitivity analysis
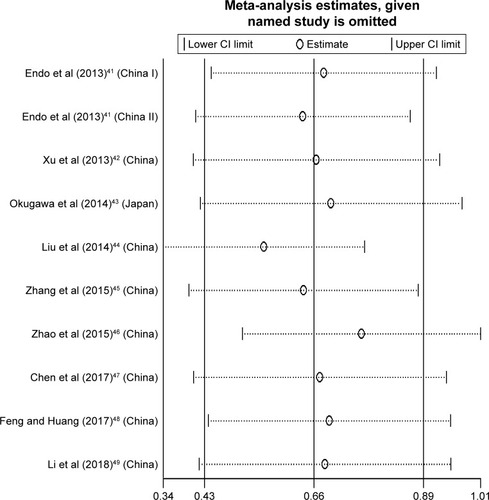
The sensitivity analysis was performed to examine how sensitive the merged HR was if the single study was removed. After this process, no individual study significantly affected the combined HR with 95% CI ().
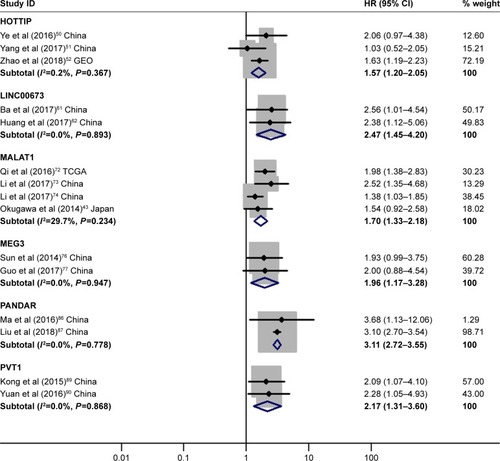
HOXA distal transcript antisense RNA (HOTTIP), long intergenic non-protein coding RNA 673 (LINC00673), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), maternally expressed 3 (MEG3), promoter of CDKN1A antisense DNA damage activated RNA (PANDAR) and Pvt1 oncogene (PVT1) demonstrated significantly prognostic value
The details are shown in and .
Figure 6 Forest plot of pooled analyses of OS in association with high HOTTIP, LINC00673, MALAT1, PANDAR, PVT1 expression levels, or low MEG3 expression levels.
Abbreviations: GEO, Gene Expression Omnibus; HOTTIP, HOXA distal transcript antisense RNA; LINC00673, long intergenic non-protein coding RNA 673; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MEG3, maternally expressed 3; OS, overall survival; PANDAR, promoter of CDKN1A antisense DNA damage activated RNA; PVT1, Pvt1 oncogene; TCGA, The Cancer Genome Atlas.
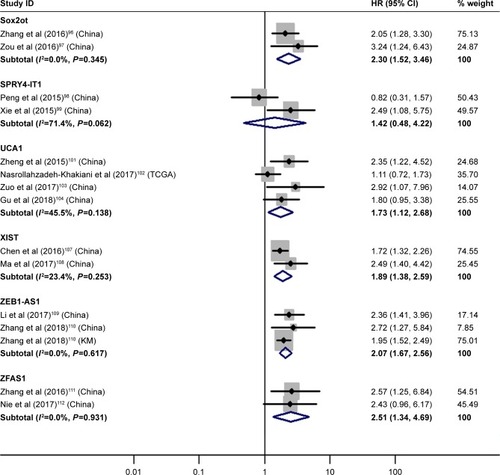
SOX2 overlapping transcript (Sox2ot), urothelial cancer-associated 1 (UCA1), X inactive specific transcript (XIST), ZEB1 antisense RNA 1 (ZEB1-AS1) and ZNFX1 antisense RNA 1 (ZFAS1) demonstrated significantly prognostic value
The details are shown in and .
Figure 7 Forest plot of pooled analyses of OS in association with high Sox2ot, UCA1, XIST, ZEB1-AS1, ZFAS1 expression levels, or low SPRY4-IT1 expression levels.
Abbreviations: OS, overall survival; Sox2ot, SOX2 overlapping transcript; SPRY4-IT1, SPRY4 intronic transcript 1; UCA1, urothelial cancer associated 1; XIST, X inactive specific transcript; ZEB1-AS1, ZEB1 antisense RNA 1; ZFAS1, ZNFX1 antisense RNA 1.
Discussion
Current situation
So far, the clinical treatment of GC remains limited. In the past score years, there has been little progress in both traditional and new treatment methods. Therefore, novel biomarkers that can improve the prognosis of GC patients are in need. Recently, there is an increasing evidence that lncRNAs can hinder the growth and metastasis of cancer. For example, Xu et alCitation123 reported that upregulating long stress-induced noncoding 5 (LSINCT5) significantly promoted the growth of the GC cell, while downregulating LSINCT5 suppressed its growth. Dan et alCitation124 conducted the cancer model experiments using mice, proving that MEG3 overexpression could suppress GC growth and metastasis in vivo by suppressing miR-21 expression. More importantly, several abnormally expressed lncRNAs have been discovered to touch upon the development of GC and perhaps possess prognostic potency in this illness. In view of the above consequences, we conducted this meta-analysis about the prognostic value of lncRNAs in GC.
Research finding
In the present research, a total of 51 articles reporting 19 lncRNAs, which were latent prognostic biomarkers and 6,095 GC patients were included, among which 18 lncRNAs (except SPRY4 intronic transcript 1 [SPRY4-IT1]) manifested a significantly prognostic value. Meanwhile, strong heterogeneity was only shown in two (H19 and SPRY4-IT1) analyses about lncRNAs, during which there was no significant associations between SPRY4-IT1 expression and OS. Further analysis suggested that AFAP1-AS1, CCAT2, LINC00673, PANDAR, PVT1, Sox2ot, ZEB1-AS1 and ZFAS1 were strong candidates in predicting prognosis of GC patients.
Molecular mechanisms
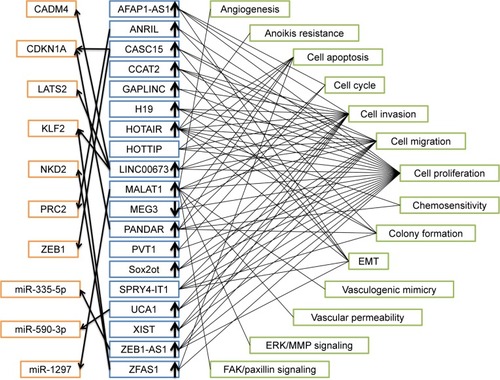
shows the summary of lncRNAs with aberrant expression, potential targets and pathways included in this study. It is noteworthy that there existed inconsistent outcomes about expression of HOTTIP and SPRY4-IT1 compared with normal controls, so these two lncRNAs were not shown to be up or down expressed. Unexpected results were findings that CDKN1A was target of both CASC15 and PANDAR and KLF2 was target of both LINC00673 and ZFAS1. In addition, cell proliferation was the most related cell function of these lncRNAs.
Figure 8 Summary of lncRNAs with altered expression, potential targets and pathways entered in this study.
Merits
The current study had several merits: 1) nearly all articles appraising the associations between OS of GC patients and lncRNA expression were searched and are clearly shown in ; 2) most of our meta-analyses revealed no or low heterogeneity (I2≤50.0%), indicating relatively consistent results of the meta-analyses and 3) all the included studies had a relatively large sample size (≥30), decreasing the error of low sample size to some degree.
Limitations
However, the limitations of this work could not be ignored: 1) only English papers were included in the present research, which may exclude potentially relevant articles; 2) most of the patients were from China, which cannot adequately represent the prognosis of global patients; 3) only the meta-analysis of HOTAIR was composed of nine articles,Citation41–Citation49 and other merged analyses about lncRNAs were from relatively small article number (two to four) and 4) the papers omitted due to no mention of OS may provide a lot of information on which lncRNAs hold promise for a prognostic value.
Inspirations
This study left several inspirations for us: 1) lncRNAs were arranged in an alphabetical order as shown in , via which the recently research frequency could be distinctly seen by clinical workers and scientific researchers; 2) the detailed outcomes of OS from the pooled analyses are shown in , through which combined detection of lncRNAs might better predict the survival time of GC patients and 3) for the molecular mechanisms of the included lncRNAs, their connections are shown in , which might play enlightening roles in future basic experiments on lncRNAs in GC.
Conclusion
This meta-analysis suggested that the abnormally expressed lncRNAs (AFAP1-AS1, ANRIL, CASC15, CCAT2, GAPLINC, H19, HOTAIR, HOTTIP, LINC00673, MALAT1, MEG3, PANDAR, PVT1, Sox2ot, UCA1, XIST, ZEB1-AS1 and ZFAS1) were significantly associated with the survival of GC patients, among which AFAP1-AS1, CCAT2, LINC00673, PANDAR, PVT1, Sox2ot, ZEB1-AS1 and ZFAS1 were strong candidates in predicting prognosis of GC patients.
Author contributions
Yue Zhang contributed toward study concept and design. Song Gao and Zhi-Ying Zhao were involved in acquisition of data. Song Gao, Zhi-Ying Zhao and Rong Wu carried out analysis and interpretation of data. Yue Zhang performed drafting of the manuscript. Song Gao, Zhi-Ying Zhao, Rong Wu, Yue Zhang and Zhen-Yong Zhang assisted with revision of manuscript. Yue Zhang and Zhen-Yong Zhang helped in supervision of work. All authors read and approved the final manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- LiFHuangCLiQWuXConstruction and comprehensive analysis for dysregulated long non-coding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in gastric cancerMed Sci Monit201824374929295970
- ChenXSunJSongYThe novel long noncoding RNA AC138128.1 may be a predictive biomarker in gastric cancerMed Oncol2014311126225260808
- FengYZhangQWangJLiuPIncreased lncRNA AFAP1-AS1 expression predicts poor prognosis and promotes malignant phenotypes in gastric cancerEur Rev Med Pharmacol Sci201721173842384928975981
- QiaoCFZhangYJinLDuXGQiaoZJHigh expression of lncRNA AFAP1-AS1 promotes cell proliferation and invasion by inducing epithelial-to-mesenchymal transition in gastric cancerInt J Clin Exp Pathol2017101393400
- QiFLiuXWuHLong noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancerJ Hematol Oncol20171014828209205
- HuangYZhangJHouLLncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathwayJ Exp Clin Cancer Res201736119429282102
- FanZYLiuWYanCIdentification of a five-lncRNA signature for the diagnosis and prognosis of gastric cancerTumour Biol20163710132651327727460075
- YangZWangRZhangTDongXHypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancerInt J Clin Exp Med2015811199541996826884908
- ZhangEBKongRYinDDLong noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449aOncotarget2014582276229224810364
- DengWWangJZhangJCaiJBaiZZhangZTET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cellsIUBMB Life201668535536427027260
- SaitoTKurashigeJNambaraSA long non-coding RNA activated by transforming growth factor-β is an independent prognostic marker of gastric cancerAnn Surg Oncol201522suppl 3S915S92225986864
- LiLZhangLZhangYZhouFIncreased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancerBiomed Pharmacother20157210911226054683
- LiuXWangYSunLLong noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxiaCancer Sci20181094988100029383777
- LüMHTangBZengSLong noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancerOncogene201635273524353426549025
- SunTTHeJLiangQLncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification patternCancer Discov20166778480127147598
- WangLChunyanQZhouYBCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patientsEur Rev Med Pharmacol Sci201721184064407029028095
- ZhouJHuangHTongSHuoROverexpression of long non-coding RNA cancer susceptibility 2 inhibits cell invasion and angiogenesis in gastric cancerMol Med Rep20171645235524028849111
- YaoXMTangJHZhuHJingYHigh expression of LncRNA CASC15 is a risk factor for gastric cancer prognosis and promote the proliferation of gastric cancerEur Rev Med Pharmacol Sci201721245661566729272000
- WuQXiangSMaJLong non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1Mol Oncol201812679981329489064
- LiuJNShangguanYMLong non-coding RNA CARLo-5 upregulation associates with poor prognosis in patients suffering gastric cancerEur Rev Med Pharmacol Sci201721353053428239816
- WangCYHuaLYaoKHChenJTZhangJJHuJHLong non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosisInt J Clin Exp Pathol20158177978525755774
- WangYJLiuJZLvPDangYGaoJYWangYLong non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the E-cadherin and LATS2Am J Cancer Res20166112651266027904778
- PeiYFZhangYJLeiYWuDWMaTHLiuXQHypermethylation of the CHRDL1 promoter induces proliferation and metastasis by activating Akt and Erk in gastric cancerOncotarget2017814231552316628423564
- HaoYPQiuJHZhangDBYuCGLong non-coding RNA DANCR, a prognostic indicator, promotes cell growth and tumorigenicity in gastric cancerTumour Biol2017396 1010428317699798
- XuTPWangYFXiongWLE2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axisCell Death Dis201786e283728569791
- PengWWuJFanHLuJFengJLncRNA EGOT promotes tumorigenesis Via Hedgehog Pathway in gastric cancerPathol Oncol Res2017
- XuTPHuangMDXiaRDecreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expressionJ Hematol Oncol201476325167886
- WuXZhangPZhuHLiSChenXShiLLong noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathwayBiomed Pharmacother2017961103110829239821
- LiCYLiangGYYaoWZIntegrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancerInt J Oncol20164851965197626935047
- ChongDQShanJLYangCSWangRDuZMClinical prognostic value of A FOXM1 related long non-coding RNA expression in gastric cancerEur Rev Med Pharmacol Sci201822241742129424899
- FengLZhuYZhangYRaoMLncRNA GACAT3 promotes gastric cancer progression by negatively regulating miR-497 expressionBiomed Pharmacother20189713614229091858
- HuYWangJQianJLong noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancerCancer Res201474236890690225277524
- LiuLZhaoXZouHBaiRYangKTianZHypoxia promotes gastric cancer malignancy partly through the HIF-1α dependent transcriptional activation of the long non-coding RNA GAPLINCFront Physiol2016742027729869
- SunMJinFYXiaRDecreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancerBMC Cancer20141431924884417
- YangFXueXZhengLLong non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stabilityFEBS J2014281380281324397586
- LiHYuBLiJOverexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancerOncotarget2014582318232924810858
- ZhangEBHanLYinDDKongRDeWChenJc-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancerMed Oncol201431591424671855
- ChenJSWangYFZhangXQH19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancerNeoplasma201663222323026774144
- ChenJFWuPXiaRSTAT3-induced lncRNA HAGLROS over-expression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagyMol Cancer2018171629329543
- ChenWMHuangMDKongRAntisense long noncoding RNA HIF1A-AS2 is upregulated in gastric cancer and associated with poor prognosisDig Dis Sci20156061655166225686741
- EndoHShirokiTNakagawaTEnhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancerPLoS One2013810e7707024130837
- XuZYYuQMDuYAKnockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancerInt J Biol Sci20139658759723847441
- OkugawaYToiyamaYHurKMetastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasisCarcinogenesis201435122731273925280565
- LiuXHSunMNieFQLnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancerMol Cancer2014139224775712
- ZhangZZShenZYShenYYHOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(C)-binding protein (PCBP) 1Mol Cancer Ther20151451162117025612617
- ZhaoWDongSDuanBHOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapyAm J Transl Res2015771295130226328013
- ChenWMChenWDJiangXMHOX transcript antisense intergenic RNA represses E-cadherin expression by binding to EZH2 in gastric cancerWorld J Gastroenterol201723336100611028970725
- FengXHuangSEffect and mechanism of lncRNA HOTAIR on occurrence and development of gastric cancerJ Cell Biochem2017
- LiHLiJZhangBZengHLong-chain non-coding RNA HOTAIR expression in tissue samples correlates with gastric cancer survivalInt J Clin Exp Med2018112856862
- YeHLiuKQianKOverexpression of long noncoding RNA HOTTIP promotes tumor invasion and predicts poor prognosis in gastric cancerOnco Targets Ther201692081208827103834
- YangYMaBYanYLong non-coding RNA HOXA transcript at the distal tip as a biomarker for gastric cancerOncol Lett20171411068107228693275
- ZhaoRZhangYZhangXExosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancerMol Cancer20181716829486794
- XieMSunMZhuYNLong noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expressionOncotarget2015632335873360126384350
- SunMNieFWangYLncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1Cancer Res201676216299631027651312
- MaBWangJSongYUpregulated long intergenic noncoding RNA KRT18P55 acts as a novel biomarker for the progression of intestinal-type gastric cancerOnco Targets Ther2016944545326855593
- ZhouBJingXYWuJQXiHFLuGJDown-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancerInt J Clin Exp Pathol20147128893889825674261
- ShanYYingRJiaZLINC00052 promotes gastric cancer cell proliferation and metastasis via activating the Wnt/β-catenin signaling pathwayOncol Res20172591589159928337962
- ChenWMHuangMDSunDPLong intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancerOncotarget2016799773978726799422
- ZhangZZZhaoGZhuangCLong non-coding RNA LINC00628 functions as a gastric cancer suppressor via long-range modulating the expression of cell cycle related genesSci Rep201662743527272474
- ZhangEYinDHanLE2F1-induced upregulation of long non-coding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIsOncotarget2016717232122322627036039
- BaMCLongHCuiSZLong noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancerOncotarget2017856955429555329221147
- HuangMHouJWangYLong noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancerMol Ther20172541014102628214253
- ZengSXieXXiaoYFLong noncoding RNA LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress gastric cancer progressionCancer Lett201841217918729107103
- FeiZHYuXJZhouMSuHFZhengZXieCYUpregulated expression of long non-coding RNA LINC00982 regulates cell proliferation and its clinical relevance in patients with gastric cancerTumour Biol20163721983199326334618
- MiaoYSuiJXuSYLiangGYPuYPYinLHComprehensive analysis of a novel four-lncRNA signature as a prognostic biomarker for human gastric cancerOncotarget2017843750077502429088841
- ChenXChenZYuSLong noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancerClin Cancer Res20182482002201429386218
- QinQHYinZQLiYWangBGZhangMFLong intergenic non-coding RNA 01296 aggravates gastric cancer cells progress through miR-122/MMP-9Biomed Pharmacother20189745045729091895
- ZouZDingQLiPOverexpression of lincRNA-ROR predicts poor prognosis in patients with gastric cancerInt J Clin Exp Pathol20169994679472
- HuYPanJWangYLiLHuangYLong noncoding RNA linc-UBC1 is negative prognostic factor and exhibits tumor pro-oncogenic activity in gastric cancerInt J Clin Exp Pathol20158159460025755750
- GuoWDongZShiYMethylation-mediated downregulation of long noncoding RNA LOC100130476 in gastric cardia adenocarcinomaClin Exp Metastasis201633549750827189370
- ZhaoYLiuYLinLThe lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1Mol Cancer20181716929510730
- QiYOoiHSWuJMALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10Oncotarget2016711126931270326871474
- LiJGaoJTianWLiYZhangJLong non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297Cancer Cell Int2017174428396617
- LiYWuZYuanJLong non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesisCancer Lett2017395314428268166
- ChenFTianYPangEJWangYLiLMALAT2-activated long non-coding RNA indicates a biomarker of poor prognosis in gastric cancerCancer Gene Ther2015
- SunMXiaRJinFDownregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancerTumour Biol20143521065107324006224
- GuoWDongZLiuSPromoter hypermethylation-mediated downregulation of miR-770 and its host gene MEG3, a long non-coding RNA, in the development of gastric cardia adenocarcinomaMol Carcinog20175681924193428345805
- NieFQMaSXieMLiuYWDeWLiuXHDecreased long non-coding RNA MIR31HG is correlated with poor prognosis and contributes to cell proliferation in gastric cancerTumour Biol20163767693770126692098
- QuanYZhangYLinWKnockdown of long non-coding RNA MAP3K20 antisense RNA 1 inhibits gastric cancer growth through epigenetically regulating miR-375Biochem Biophys Res Commun2018497252753429428732
- LaiYXuPLiuJDecreased expression of the long non-coding RNA MLLT4 antisense RNA 1 is a potential biomarker and an indicator of a poor prognosis for gastric cancerOncol Lett20171432629263428927028
- WangYZhangDWuKZhaoQNieYFanDLong noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublinesMol Cell Biol201434173182319324958102
- LinZLaiSHeXDecreased long non-coding RNA MTM contributes to gastric cancer cell migration and invasion via modulating MT1FOncotarget2017857973719738329228617
- FuJWKongYSunXLong noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancerJ Cancer Res Clin Oncol201614271571157927095450
- GuoXYangZZhiQLong noncoding RNA OR3A4 promotes metastasis and tumorigenicity in gastric cancerOncotarget2016721302763029426863570
- WangYQZhangQYWengWWUpregulation of the non-coding RNA OTUB1-isoform 2 contributes to gastric cancer cell proliferation and invasion and predicts poor gastric cancer prognosisInt J Biol Sci201612554555727019636
- MaPXuTHuangMShuYIncreased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancerBiomed Pharmacother20167817217626898439
- LiuJBenQLuELong noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancerCell Death Dis20189216829416011
- BiMYuHHuangBTangCLong non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1AGene201762633734328571676
- KongRZhangEBYinDDLong noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16Mol Cancer2015148225890171
- YuanCLLiHZhuLLiuZZhouJShuYAberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancerNeoplasma201663344244926925791
- SunJSongYChenXNovel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancerOncol Lett201510111512026170986
- SongWLiuYYPengJJIdentification of differentially expressed signatures of long non-coding RNAs associated with different metastatic potentials in gastric cancerJ Gastroenterol201651211912926045391
- HuYMaZHeYLiuWSuYTangZLncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1Biochem Biophys Res Commun2017491492693128754593
- YanKTianJShiWXiaHZhuYLncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3pCell Physiol Biochem2017423999101228683446
- ZhangHLuWLncRNA SNHG12 regulates gastric cancer progression by acting as a molecular sponge of miR-320Mol Med Rep20181722743274929207106
- ZhangYYangRLianJXuHLncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancerAm J Transl Res20168115035504327904704
- ZouJHLiCYBaoJZhengGQHigh expression of long noncoding RNA Sox2ot is associated with the aggressive progression and poor outcome of gastric cancerEur Rev Med Pharmacol Sci201620214482448627874951
- PengWWuGFanHWuJFengJLong noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancerTumour Biol20153696751675825835973
- XieMNieFQSunMDecreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transitionJ Transl Med20151325026238992
- ZhangEHeXYinDIncreased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57Cell Death Dis20167e2109
- ZhengQWuFDaiWYAberrant expression of UCA1 in gastric cancer and its clinical significanceClin Transl Oncol201517864064625903045
- Nasrollahzadeh-KhakianiMEmadi-BaygiMNikpourPAugmented expression levels of lncRNAs ecCEBPA and UCA1 in gastric cancer tissues and their clinical significanceIran J Basic Med Sci201720101149115829147491
- ZuoZKGongYChenXHTGFβ1-induced LncRNA UCA1 upregulation promotes gastric cancer invasion and migrationDNA Cell Biol201736215916728075173
- GuLLuLSZhouDLLiuZCUCA1 promotes cell proliferation and invasion of gastric cancer by targeting CREB1 sponging to miR-590-3pCancer Med2018741253126329516678
- ChenMWuXMaWDecreased expression of lncRNA VPS9D1-AS1 in gastric cancer and its clinical significanceCancer Biomark2017211232829036784
- CaiJWangDBaiZGYinJZhangJZhangZTThe long noncoding RNA XIAP-AS1 promotes XIAP transcription by XIAP-AS1 interacting with Sp1 in gastric cancer cellsPLoS One2017128e018243328792527
- ChenDLJuHQLuYXLong non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expressionJ Exp Clin Cancer Res201635114227620004
- MaLZhouYLuoXGaoHDengXJiangYLong non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancerOncotarget2017834125413527911852
- LiYWenXWangLLncRNA ZEB1-AS1 predicts unfavorable prognosis in gastric cancerSurg Oncol201726452753429113674
- ZhangLLZhangLFGuoXHZhangDZYangFFanYYDownregulation of miR-335-5p by long noncoding RNA ZEB1-AS1 in gastric cancer promotes tumor proliferation and invasionDNA Cell Biol2018371465229215918
- ZhangJJChenJTYaoKHHuaLWangCYHuJHUp-regulated expression of long non-coding RNA ZFAS1 associates with aggressive tumor progression and poor prognosis in gastric cancer patientsInt J Clin Exp Pathol20169220592063
- NieFYuXHuangMLong noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expressionOncotarget2017824382273823827246976
- LaiYXuPLiQDownregulation of long noncoding RNA ZMAT1 transcript variant 2 predicts a poor prognosis in patients with gastric cancerInt J Clin Exp Pathol2015855556556226191264
- McLeanMHEl-OmarEMGenetics of gastric cancerNat Rev Gastroenterol Hepatol2014111166467425134511
- TorreLABrayFSiegelRLFerlayJLortet-TieulentJJemalAGlobal cancer statistics, 2012CA Cancer J Clin20156528710825651787
- SiegelRLMillerKDJemalACancer statistics, 2015CA Cancer J Clin201565152925559415
- GBD 2013 Mortality and Causes of Death CollaboratorsGlobal, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013Lancet2015385996311717125530442
- PontingCPOliverPLReikWEvolution and functions of long noncoding RNAsCell2009136462964119239885
- CheethamSWGruhlFMattickJSDingerMELong noncoding RNAs and the genetics of cancerBr J Cancer2013108122419242523660942
- SpizzoRAlmeidaMIColombattiACalinGALong non-coding RNAs and cancer: a new frontier of translational research?Oncogene201231434577458722266873
- LiTMoXFuLXiaoBGuoJMolecular mechanisms of long non-coding RNAs on gastric cancerOncotarget2016788601861226788991
- ParmarMKTorriVStewartLExtracting summary statistics to perform meta-analyses of the published literature for survival endpointsStat Med19981724281528349921604
- XuMDQiPWengWWLong non-coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancerMedicine (Baltimore)20149328e30325526476
- DanJWangJWangYLncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancerBiomed Pharmacother20189993193829710493