Abstract
Background
Circular RNAs (circRNAs) exert important roles in carcinogenesis. Here, we aimed to uncover the working mechanism of circ_0006168 in esophageal squamous cell carcinoma (ESCC) development.
Methods
Western blot assay and real-time quantitative polymerase chain reaction (RT-qPCR) were used to determine protein and RNA expression, respectively. Wound healing assay and transwell migration assay were performed to assess cell migration ability, whereas cell invasion ability was evaluated by transwell invasion assay. 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and colony formation assay were utilized to analyze cell proliferation ability. Xenograft tumor model was utilized to assess the role of X-box binding protein 1 (XBP1) in xenograft tumor growth in vivo. Dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay and RNA pull down assay were used to verify intermolecular interactions.
Results
XBP1 silencing suppressed the migration, invasion and proliferation of ESCC cells in vitro and restrained the xenograft tumor growth in vivo. MicroRNA-516b-5p (miR-516b-5p) interacted with the 3ʹ untranslated region (3ʹUTR) of XBP1 in ESCC cells. MiR-516b-5p overexpression inhibited the proliferation and motility of ESCC cells. MiR-516b-5p was a molecular target of circ_0006168 in ESCC cells. The interference of circ_0006168 restrained the motility and proliferation of ESCC cells. Circ_0006168 acted as miR-516b-5p sponge to up-regulate XBP1 expression in ESCC cells. MiR-516b-5p silencing or the accumulation of XBP1 largely rescued the proliferation ability and motility in circ_0006168-silenced ESCC cells.
Conclusion
In conclusion, circ_0006168 contributed to ESCC development through promoting the proliferation and motility of ESCC cells via mediating miR-516b-5p/XBP1 axis.
Introduction
Esophageal squamous cell carcinoma (ESCC) is a common subtype of esophageal cancer (EC) with high incidence.Citation1,Citation2 ESCC patients are always diagnosed at late stage due to the lack of obvious early symptoms and diagnostic methods,Citation3 which results in poor outcome.Citation4 Furthermore, the molecular-targeted strategies for patients with ESCC are unsatisfactory due to the unclear molecular mechanism in ESCC development. Hence, identifying novel biomarkers for early diagnosis and therapy of ESCC is urgently needed to improve the prognosis of ESCC patients.
Circular RNAs (circRNAs) are stable non-coding RNA (ncRNA) molecules that are resistant to exonuclease due to their covalently closed circular structure.Citation5 CircRNAs could act as microRNA (miRNA) sponges to regulate cellular phenotypes.Citation6 For instance, circ_0006168 accelerated the malignant behaviors of ESCC cells through sponging miR-100.Citation7 Xie et al claimed that circ_0006168 facilitated EC progression through targeting miR-384/RBBP7 signaling.Citation8 However, the molecular mechanism behind the oncogenic role of circ_0006168 still needs to be illustrated. Based on bioinformatic analysis, miR-516b-5p was a potential target of circ_0006168. MiR–516b restrained the progression of several malignancies,Citation9–Citation11 including ESCC. In the current study, the interaction between miR-516b-5p and circ_0006168 and their functional association in ESCC progression were explored.
MicroRNAs (miRNAs) regulate protein coding through binding to messenger RNAs (mRNAs) to regulate cell cycle progression, cell apoptosis and motility.Citation12,Citation13 The target interaction between miR-516b–5p and XBP1 was predicted by bioinformatic database. XBP1 is an important transcription factor that is implicated in the process of unfolded protein response (UPR).Citation14 Recently, XBP1 was found to play an oncogenic role in many malignancies.Citation15–Citation17 Xia et al demonstrated that XBP1 accelerated the proliferation and motility in ESCC cells via up-regulating MMP9.Citation18 Jin et al proved that TUG1 contributed to ESCC development through sponging miR-498 and up-regulating XBP1.Citation19 Here, the interaction between XBP1 and miR-516b-5p was tested, and their functional relevance in regulating ESCC progression was analyzed.
In the current study, the expression pattern of XBP1 in ESCC cell lines and tissues was firstly analyzed. Loss-of-function experiments were then performed to analyze the biological functions of XBP1 in ESCC cells. Bioinformatic software and verification experiments were utilized to establish circRNA-miRNA-mRNA axis, and their functional association was tested by rescue experiments.
Materials and Methods
Clinical Samples
Thirty pairs of ESCC tissue samples and adjacent non-tumor tissues were collected from 30 ESCC patients at the First Affiliated Hospital of Nanchang University. These tissue samples were stored at −80°C within 30 min after surgical resection. Clinical experiments were performed with the approval of the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University and in accordance with the Declaration of Helsinki. Written informed consent was signed by every participant before the study. The association between clinicopathological features of ESCC patients and the expression level of circ_0006168 is shown in . RT-qPCR, Western blot assay and immunohistochemistry (IHC) assay were performed to determine RNA and protein expression. IHC assay was performed using the antibody against XBP1 (ab37152; Abcam, Cambridge, MA, USA) at the dilution of 1:300.Citation20
Table 1 Correlation Between Clinicopathological Features of ESCC Patients and the Expression Level of circ_0006168
Cell Lines
ESCC cell lines (Eca109, KYSE150, KYSE180 and KYSE450) and human esophageal epithelial cell line HET-1A were purchased from the Shanghai Institute of Chinese Academy of Sciences Cell Collection (Shanghai, China). All these five cell lines were cultured with Roswell Park Memorial Institute-1640 medium (RPMI-1460; Gibco, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS; Gibco), 1% 100 units/mL penicillin (Gibco) and 1% 100 μg/mL streptomycin (Gibco). All cell lines were maintained at 37°C incubator with 5% CO2.
Western Blot Assay
ESCC cells were collected and lysed using whole cell lysis buffer (Abcam). Protein quantification was conducted with the commercial bicinchoninic acid (BCA) protein assay kit (Invitrogen, Carlsbad, CA, USA). Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto the polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking, the membrane was incubated with primary antibodies, including anti-XBP1 (ab37152; Abcam) and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; ab9485; Abcam). After incubating with secondary antibody (Abcam) for 2 h, protein signals were determined using the commercial enhanced chemiluminescence kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Cell Transfection
XBP1 specific small interfering RNA (si-XBP1), circ_0006168 specific siRNA (si-circ_0006168), negative control siRNA (si-NC), short hairpin RNA targeting XBP1 (sh-XBP1), shRNA NC (sh-NC), XBP1 overexpression plasmid (XBP1), empty pcDNA vector (pcDNA), circ_0006168 overexpression plasmid (circ_0006168), empty pLCDH-cir vector (vector), miR-516b-5p mimics (miR-516b-5p), miRNA NC (miR-NC), miR-516b-5p inhibitor (inhibitor-miR-516b-5p) and inhibitor-NC were purchased from GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen) was used for transfection.
Wound Healing Assay
Cell monolayer in 6-well plates was scratched with the 200 μL pipette tip, and cell debris was removed through washing with phosphate buffer saline (PBS) buffer (Sangon Biotech, Shanghai, China). After creating the wound for 0 h and 24 h, cell images were obtained at the magnification of 40 ×.
Transwell Assays
ESCC cells were inoculated into the upper chambers with (in transwell invasion assay) or without Matrigel (BD Biosciences, San Jose, CA, USA; in transwell migration assay). The upper chambers were added with serum-free medium, while the lower chambers were added with culture medium plus 10% FBS (Gibco). After 24-h inoculation, ESCC cells that penetrated the membrane were fixed, stained and counted (Magnification: 100 ×).
3-(4,5-Dimethyl-Thiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
In brief, ESCC cells were inoculated into 96-well plates and transfected with plasmid or RNA for 0 h, 24 h, 48 h or 72 h. These transfected ESCC cells were incubated with 10 μL MTT reagent (Beyotime, Shanghai, China) for 4 h prior to incubation with 100 μL dimethyl sulfoxide (DMSO, Sigma, St. Louis, MO, USA). The absorbance at 490 nm was measured, and cell proliferation curve was generated.
Colony Formation Assay
ESCC cells were inoculated into 6-well plates at 300 cells per well. After 14-d culture at 37°C incubator with 5% CO2/95% air, the colonies were fixed with methanol (Sangon Biotech) for 15 min followed by staining using 0.1% crystal violet (Sangon Biotech) for 15 min. The number of colonies was counted.
Xenograft Tumor Model
Animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Nanchang University and in accordance with the National Guide for the Care and Use of Laboratory Animals. BALB/c nude mice from Vital River Laboratory Animal Technology (Beijing, China) were split into two groups (n=6). The mice in sh-XBP1 group were subcutaneously injected with Eca109 or KYSE150 cells (6 × 106 cells) stably transfected with sh-XBP1 in their right dorsal flank, while mice in sh-NC group were subcutaneously injected with Eca109 or KYSE150 cells (6 × 106 cells) stably transfected with sh-NC. The volume of tumors was monitored every week, and tumors were weighed after 4-week inoculation.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol reagent (Invitrogen) was used to isolate RNA samples from ESCC tissues and cell lines. The complementary DNA (cDNA) for circ_0006168, CCR4-NOT transcription complex subunit 6 like (CNOT6L) and XBP1 was synthesized using All-in-OneTM Synthesis Kit (FulenGen, Guangzhou, China). The cDNA for miR-516b-5p was obtained using miScript reverse transcription kit (Qiagen, Hilden, Germany). SYBR Green (Applied Biosystems, Foster City, CA, USA) was subsequently used to conduct PCR amplification reaction. U6 was used as the internal reference for miR-516b-5p while GAPDH was used as the internal reference for circ_0006168, CNOT6L and XBP1. The primers were listed in . The fold change was calculated using the 2−ΔΔCt method.
Table 2 Specific Primers in qRT-PCR Assay
Dual-Luciferase Reporter Assay
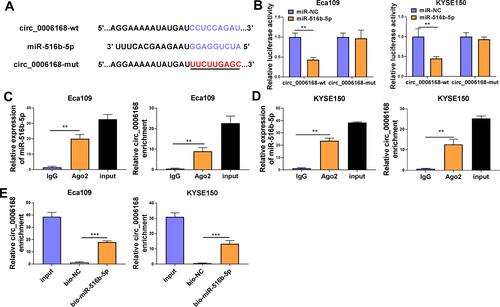
Dual-luciferase reporter assay was applied to test the interaction between miR-516b-5p and circ_0006168 or XBP1 mRNA 3ʹ untranslated region (3ʹUTR). Partial wild-type sequence of circ_0006168 or XBP1 mRNA 3ʹUTR was inserted into the psiCHECK2 vector (Promega, Madison, WI, USA) to obtain circ_0006168-wt or XBP1-wt. Meanwhile, circ_0006168-mut or XBP1-mut luciferase plasmid was constructed through inserting the mutant sequence of circ_0006168 or XBP1 3ʹUTR into the psiCHECK2 vector (Promega). ESCC cells were inoculated into 24-well plates and co-transfected with luciferase plasmid and miR-516b-5p or miR-NC. After transfection for 48 h, luciferase intensities were detected using the Dual-luciferase Reporter assay system (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using the EZMagna RIP kit (Merck, Darmstadt, Germany). ESCC cells were disrupted using RIP lysis buffer and then incubated with magnetic beads that pre-coated with Argonaute 2 (Ago2) antibody or Immunoglobulin G (IgG) antibody. After digesting with protease K, the levels of circ_0006168, miR-516b-5p and XBP1 mRNA were measured by RT-qPCR.
RNA Pull Down Assay
Cell lysate (2 μg) was incubated with biotinylated miR-516b-5p (bio-miR-516b-5p; 100 pmol) or its negative control (bio-NC; 100 pmol). 100 μL streptavidin agarose beads were added to incubate for 1 h. The levels of circ_0006168 and XBP1 mRNA were measured by RT-qPCR.
RNase R Digestion
A total of 2 μg RNA was incubated with 6 units of RNase R (Geneseed Biotech, Guangzhou, China) for 20 min at 37°C. The expression of circ_0006168 and CNOT6L was measured by RT-qPCR.
Statistical Analysis
The data from three independent experiments were represented as mean ± standard deviation. The differences in two groups or multiple groups were analyzed by Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test. P values <0.05 were regarded as statistically significant.
Results
XBP1 Interference Restrains Cell Migration and Invasion Abilities of ESCC Cells
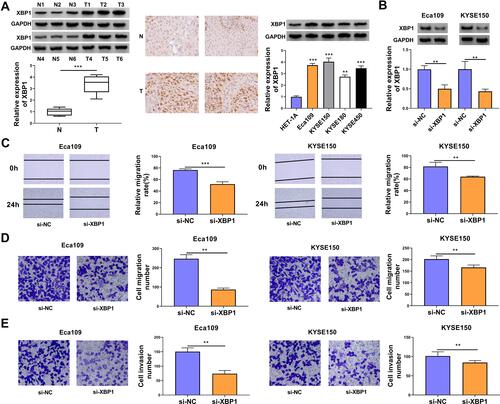
The expression characteristic of XBP1 in ESCC tissues and cell lines was initially explored. As shown in , XBP1 was highly expressed in ESCC tissues in comparison with that in para-cancerous normal tissues as verified by Western blot assay and IHC assay. Compared with human esophageal epithelial cell line HET-1A, XBP1 was observed to be notably up-regulated in four ESCC cell lines (), especially in Eca109 and KYSE150 cell lines. Thus, Eca109 and KYSE150 cell lines were chosen for loss-of-function experiments. High silencing efficiency of si-XBP1 was verified in both ESCC cell lines (). With the silencing of XBP1, the rate of wound healing was markedly reduced (), suggesting that XBP1 interference suppressed the migration ability of ESCC cells. Apart from wound healing assay, cell migration ability was further analyzed by transwell migration assay. The number of migrated ESCC cells was reduced in XBP1-silenced group compared with si-NC group (), which further demonstrated that XBP1 knockdown restrained the migration of ESCC cells. Based on the results of transwell invasion assay, we found that XBP1 interference notably decreased the number of invaded ESCC cells (), suggesting that XBP1 silencing markedly inhibited the invasion ability of ESCC cells. Taken together, XBP1 interference inhibited the migration and invasion of ESCC cells.
Figure 1 XBP1 interference restrains cell migration and invasion abilities of ESCC cells. (A) The relative protein expression of XBP1 was determined in ESCC tumor specimens and paired non-tumor specimens by Western blot assay and IHC assay. The relative level of XBP1 protein was examined in a panel of four ESCC cell lines (Eca109, KYSE150, KYSE180 and KYSE450) and normal esophageal cell line HET-1A by Western blot assay. (B–E) Eca109 and KYSE150 cells were transfected with si-NC or si-XBP1. (B) The protein expression of XBP1 was determined by Western blot assay. (C) Wound healing assay was performed to analyze the relative migration rate of transfected ESCC cells. (D) Transwell migration assay was conducted to analyze the number of migrated cells to assess cell migration ability. (E) Cell invasion number was analyzed in transwell invasion assay to assess cell invasion ability. **P<0.01, ***P<0.001.
XBP1 Silencing Suppresses Cell Proliferation Ability of ESCC Cells in vitro and in vivo
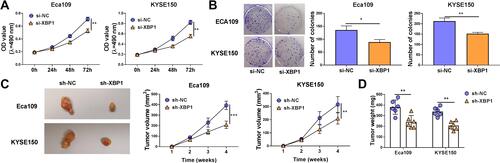
The effect of XBP1 silencing on the proliferation ability of ESCC cells was explored through MTT assay and colony formation assay. XBP1 interference significantly restrained the proliferation ability of ESCC cells (). With the silencing of XBP1, the number of colonies was notably decreased compared with that in si-NC group (). The results of MTT assay and colony formation assay together demonstrated that XBP1 silencing suppressed the proliferation ability of ESCC cells in vitro. Eca109 and KYSE150 cell lines stably transfected with sh-XBP1 or sh-NC were established to explore the role of XBP1 in xenograft tumor growth in vivo. As shown in and , XBP1 silencing significantly reduced tumor volume and weight, suggesting that XBP1 silencing suppressed ESCC progression in vivo. Overall, XBP1 interference restrained the proliferation of ESCC cells in vitro and in vivo.
Figure 2 XBP1 silencing suppresses cell proliferation ability of ESCC cells in vitro and in vivo. (A and B) Eca109 and KYSE150 cells were transfected with si-NC or si-XBP1. (A) Cell proliferation curve was generated via MTT assay to analyze cell proliferation ability. (B) Colony formation assay was performed to evaluate cell proliferation ability. (C) Eca109 or KYSE150 cells stably transfected with sh-NC or sh-XBP1 were injected into the nude mice to build xenograft tumor model. The tumor volume in two groups was measured and calculated every week. (D) Tumor weight in two groups was analyzed after four-week inoculation. *P<0.05, **P<0.01, ***P<0.001.
XBP1 is a Target of miR-516b-5p in ESCC Cells
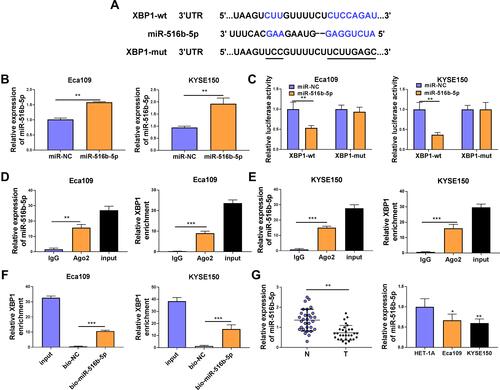
XBP1 was predicted as a downstream target of miR-516b-5p via TargetScan software based on their complementary sites shown in . High overexpression efficiency of miR-516b-5p mimics was verified in both ESCC cell lines (). The wild-type or mutant type 3ʹUTR fragment of XBP1 mRNA, including the putative or mutant binding sites with miR-516b-5p, was amplified and inserted into the luciferase plasmid, termed as XBP1-wt or XBP1-mut. Next, we tested the interaction between miR-516b-5p and XBP1 mRNA 3ʹUTR through co-transfecting ESCC cells with luciferase plasmid (XBP1-wt or XBP1-mut) and miR-516b-5p or miR-NC. MiR-516b-5p accumulation caused significant reduction in luciferase activity of wild-type reporter plasmid (XBP1-wt) (). MiR-516b-5p overexpression had almost no effect on the luciferase activity of mutant reporter plasmid (XBP1-mut) (), suggesting that miR-516b-5p interacted with XBP1 3ʹUTR in ESCC cells. RIP assay and RNA pull down assay were conducted to further verify the interaction between XBP1 and miR-516b-5p. MiR-516b-5p and XBP1 were simultaneously enriched when using Ago2 antibody ( and ), suggesting that there was spatial interaction between miR-516b-5p and XBP1 in RNA-induced silencing complex (RISC). As shown in , XBP1 was enriched using biotinylated miR-516b-5p, suggesting the interaction between miR-516b-5p and XBP1. MiR-516b-5p was down-regulated in ESCC tissue samples than that in adjacent normal tissues (). The level of miR-516b-5p was notably reduced in two ESCC cell lines compared with that in HET-1A cell line (). Taken together, miR-516b-5p interacted with XBP1 3ʹUTR, and miR-516b-5p was notably down-regulated in ESCC tissues and cell lines.
Figure 3 XBP1 is a target of miR-516b-5p in ESCC cells. (A) The complementary sequence between miR-516b-5p and XBP1 was predicted by TargetScan software. (B) The transfection efficiency of miR-516b-5p mimics was assessed by RT-qPCR. (C) Dual-luciferase reporter assay was applied to test if miR-516b-5p bound to XBP1 in ESCC cells. (D and E) RIP assay was conducted to test the interaction between miR-516b-5p and XBP1 in ESCC cells. (F) RNA pull down assay was used to verify if miR-516b-5p interacted with XBP1 using biotinylated miR-516b-5p. Bio-NC was used as the control group. (G) The relative expression of miR-516b-5p was examined in 30 pairs of ESCC tissues and adjacent non-tumor tissues via RT-qPCR. RT-qPCR was used to measure the expression of miR-516b-5p in Eca109, KYSE150 and normal esophageal cell line HET-1A. *P<0.05, **P<0.01, ***P<0.001.
MiR-516b-5p Accumulation Inhibits the Malignant Behaviors of ESCC Cells
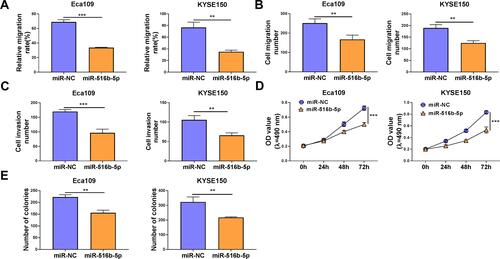
Gain-of-function experiments were performed to analyze the biological role of miR-516b-5p in ESCC cells. With the accumulation of miR-516b-5p, cell migration rate was significantly reduced compared with that in miR-NC group ( and Supplementary Figure 1A). Through performing transwell assays, we found that the numbers of migrated and invaded ESCC cells were significantly reduced in miR-516b-5p-overexpressed group ( and , Supplementary Figure 1B and C), demonstrating that miR-516b-5p overexpression suppressed the migration and invasion of ESCC cells. As verified by MTT assay, we found that miR-516b-5p overexpression notably hampered the proliferation of ESCC cells (). MiR-516b-5p overexpression also reduced the number of colonies in ESCC cells ( and Supplementary Figure 1D). These findings suggested that miR-516b-5p overexpression inhibited the motility and proliferation of ESCC cells.
Figure 4 MiR-516b-5p accumulation inhibits the malignant behaviors of ESCC cells. (A–E) Eca109 and KYSE150 cells were transfected with miR-NC or miR-516b-5p. (A) Cell migration ability was assessed by wound healing assay. (B and C) Cell migration and invasion abilities were analyzed by transwell migration assay and transwell invasion assay, respectively. (D) MTT assay was conducted to assess cell proliferation capacity. (E) The number of colonies was analyzed by colony formation assay. **P<0.01, ***P<0.001.
Circ_0006168 Interacts with miR-516b-5p in ESCC Cells
MiR-516b-5p-circRNAs interactions were explored using bioinformatic Circinteractome software, and miR-516b-5p was predicted to be a possible target of circ_0006168. The putative binding sequence between circ_0006168 and miR-516b-5p is shown in . There was a significant reduction in luciferase activity in circ_0006168-wt and miR-516b-5p co-transfected group compared with that in circ_0006168-wt and miR-NC co-transfected group (). Meanwhile, luciferase activity was unaffected in circ_0006168-mut group with the addition of miR-NC or miR-516b-5p (), demonstrating that miR-516b-5p interacted with circ_0006168 in ESCC cells. Compared with IgG antibody group, miR-516b-5p and circ_0006168 were both enriched in Ago2 antibody group ( and ), suggesting that miR-516b-5p interacted with circ_0006168 in RISC. Moreover, the interaction between miR-516b-5p and circ_0006168 was confirmed by RNA pull down assay (). Taken together, miR-516b-5p was a target of circ_0006168 in ESCC cells.
Figure 5 Circ_0006168 interacts with miR-516b-5p in ESCC cells. (A) The interaction between circ_0006168 and miR-516b-5p was predicted by Circinteractome software. (B) The target interaction between miR-516b-5p and circ_0006168 was tested via dual-luciferase reporter assay. (C and D) RIP assay was applied to confirm the target relationship between miR-516b-5p and circ_0006168. (E) RNA pull down assay was utilized to verify the interaction between miR-516b-5p and circ_0006168. **P<0.01, ***P<0.001.
Circ_0006168 is Highly Expressed in ESCC Tissues and Cell Lines
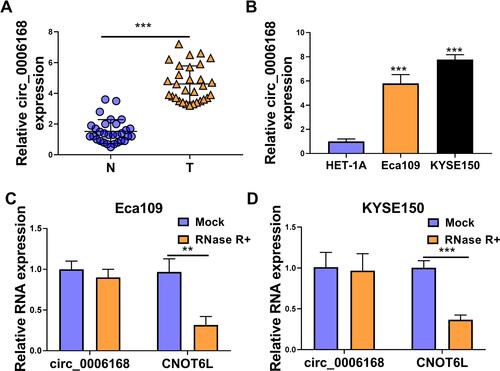
Circ_0006168 expression was up-regulated in ESCC tissues than that in adjacent non-tumor tissues (). High expression of circ_0006168 in tumor tissues was correlated with large tumor size, positive lymph node metastasis and advanced TNM stage in ESCC patients (). Compared with HET-1A cell line, circ_0006168 level was significantly up-regulated in both ESCC cell lines (). Subsequently, the stability of circ_0006168 was tested using exonuclease RNase R. Circ_0006168 was resistant to RNase R, whereas RNase R digestion markedly reduced the level of its linear counterpart CNOT6L ( and ). Taken together, circ_0006168 was a stable circular transcript, and it was aberrantly up-regulated in ESCC tissues and cell lines.
Figure 6 Circ_0006168 is highly expressed in ESCC tissues and cell lines. (A) The level of circ_0006168 was determined in a total of 30 ESCC tissues and paired normal tissues by RT-qPCR. (B) The level of circ_0006168 was examined in two ESCC cell lines and normal esophageal cell line HET-1A by RT-qPCR. (C and D) The relative levels of circ_0006168 and CNOT6L were examined in RNA samples isolated from ESCC cells treated with or without RNase R by RT-qPCR. **P<0.01, ***P<0.001.
Circ_0006168 Knockdown Hampers the Malignant Phenotypes of ESCC Cells in vitro
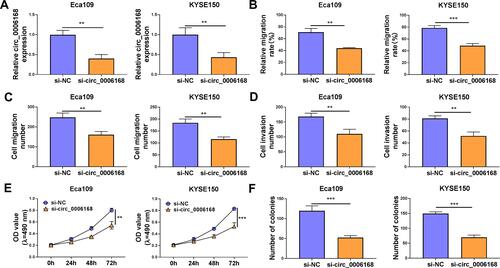
As verified by RT-qPCR, the interference efficiency of si-circ_0006168 was high in two ESCC cell lines (). Through performing wound healing assay and transwell migration assay, we found that circ_0006168 silencing notably suppressed the migration ability of ESCC cells ( and , Supplementary Figure 2A and B). As shown in and Supplementary Figure 2C, with the silencing of circ_0006168, the number of invaded ESCC cells was reduced, demonstrating that circ_0006168 knockdown suppressed the invasion ability of ESCC cells. The proliferation capacity of ESCC cells was blocked in circ_0006168-silenced group compared with si-NC group (). With the interference of circ_0006168, the number of colonies was decreased compared with that in si-NC group ( and Supplementary Figure 2D). Taken together, circ_0006168 interference blocked the migration, invasion and proliferation of ESCC cells.
Figure 7 Circ_0006168 knockdown hampers the malignant phenotypes of ESCC cells in vitro. (A–F) Eca109 and KYSE150 cells were transfected with si-NC or si-circ_0006168. (A) The expression of circ_0006168 was determined by RT-qPCR. (B) The relative migration rate was analyzed through conducting wound healing assay. (C and D) Transwell assays were used to analyze cell migration and invasion abilities. (E) The optical density at the wavelength of 490 nm was measured every 24 h via MTT assay, and cell proliferation curve was generated. (F) Colony formation assay was used to evaluate the number of colonies to assess cell proliferation ability. **P<0.01, ***P<0.001.
Circ_0006168 Acts as miR-516b-5p Sponge to Up-Regulate XBP1 in ESCC Cells
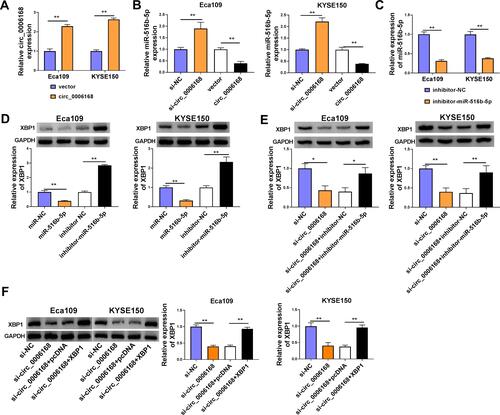
The overexpression efficiency of circ_0006168 plasmid was high in ESCC cells (). A negative regulatory relationship between circ_0006168 and miR-516b-5p was confirmed by RT-qPCR assay (). RT-qPCR assay verified the high silencing efficiency of inhibitor-miR-516b-5p in ESCC cells (). The negative regulatory relationship between miR-516b-5p and XBP1 in ESCC cells was verified by Western blot assay (). Circ_0006168 interference decreased the protein level of XBP1 in ESCC cells, and the addition of inhibitor-miR-516b-5p largely recovered the protein expression of XBP1 in ESCC cells (). These findings demonstrated that circ_0006168 silencing reduced the expression of XBP1 in ESCC cells partly through up-regulating miR-516b-5p. Circ_0006168 silencing reduced XBP1 protein level, while the addition of XBP1 plasmid largely regained the expression of XBP1 in ESCC cells (). Taken together, circ_0006168 positively regulated the expression of XBP1 partly through sponging miR-516b-5p in ESCC cells.
Figure 8 Circ_0006168 acts as miR-516b-5p sponge to up-regulate XBP1 in ESCC cells. (A) The overexpression efficiency of circ_0006168 overexpression plasmid was assessed by RT-qPCR. (B) The relative level of miR-516b-5p was measured in ESCC cells transfected with si-NC, si-circ_0006168, vector or circ_0006168 by RT-qPCR. (C) The knockdown efficiency of miR-516b-5p inhibitor (inhibitor-miR-516b-5p) in ESCC cells was assessed by RT-qPCR. (D) Western blot assay was applied to determine the expression of XBP1 in ESCC cells transfected with miR-NC, miR-516b-5p, inhibitor NC or inhibitor-miR-516b-5p. (E) Eca109 and KYSE150 cells were transfected with si-circ_0006168 alone or together with inhibitor-miR-516b-5p, and the protein level of XBP1 was examined by Western blot assay. (F) Western blot assay was implemented to measure the protein expression of XBP1 in ESCC cells transfected with si-NC, si-circ_0006168, si-circ_0006168 + pcDNA or si-circ_0006168 + XBP1. *P<0.05, **P<0.01.
Circ_0006168 Silencing-Mediated Effects are Largely Overturned by the Silencing of miR-516b-5p or the Overexpression of XBP1 in ESCC Cells
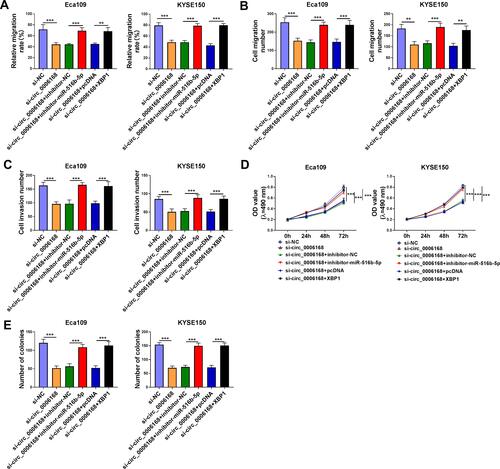
We performed rescue experiments to explore the functional relevance among circ_0006168, miR-516b-5p and XBP1 in ESCC cells. Circ_0006168 silencing-mediated suppressive effects on cell migration and invasion were largely counteracted by the silencing of miR-516b-5p or the overexpression of XBP1 via wound healing assay and transwell assays (–, Supplementary Figure 3A–C). Circ_0006168 knockdown suppressed the proliferation ability of ESCC cells, and this inhibitory effect was partly attenuated by the interference of miR-516b-5p or the overexpression of XBP1 (). Cell colony formation ability was suppressed with the silencing of circ_0006168, and the addition of inhibitor-miR-516b-5p or XBP1 plasmid largely recovered cell colony formation ability (, Supplementary Figure 3D). Overall, circ_0006168 silencing suppressed ESCC progression partly through up-regulating miR-516b-5p and reducing XBP1 expression.
Figure 9 Circ_0006168 silencing-mediated effects are largely overturned by the silencing of miR-516b-5p or the overexpression of XBP1 in ESCC cells. (A–E) Eca109 and KYSE150 cells were transfected with si-NC, si-circ_0006168, si-circ_0006168 + inhibitor-NC, si-circ_0006168 + inhibitor-miR-516b-5p, si-circ_0006168 + pcDNA or si-circ_0006168 + XBP1. (A) The migration rate of ESCC cells was analyzed by wound healing assay. (B and C) Transwell assays were performed to analyze cell migration and invasion abilities. (D) MTT assay was utilized to analyze cell proliferation ability. (E) Colony formation assay was used to analyze cell proliferation capacity. **P<0.01, ***P<0.001.
Discussion
Dysregulated circRNAs are found to be closely associated with the pathogenesis of many malignancies,Citation21 including ESCC.Citation22,Citation23 For example, Pan et al found that circ_0006948 was highly expressed in ESCC, and it accelerated ESCC progression through sponging miR-490-3p and up-regulating HMGA2.Citation24 Circ_0000337 was abnormally up-regulated in ESCC, and high level of circ_0000337 accelerated the proliferation and motility of ESCC cells.Citation25 We focused on the biological role of circ_0006168 in ESCC progression. Previous studies have identified circ_0006168 as a carcinogenic factor. Shi et al demonstrated that circ_0006168 aggravated ESCC development through enhancing mTOR expression via sponging miR-100.Citation7 Xie et al claimed that circ_0006168 contributed to ESCC development through targeting miR-384/RBBP7/S6K/S6 signaling cascade.Citation8 Here, we further investigated the working mechanism of circ_0006168 in ESCC progression. We found that circ_0006168 abundance was elevated in ESCC tissues and cell lines. Circ_0006168 silencing significantly hampered the migration, invasion and proliferation of ESCC cells, and the pro-tumor role of circ_0006168 in ESCC was in line with former articles.Citation7,Citation8
CircRNAs possess abundant miRNA binding sites, and they modulate the expression of downstream genes through competitively binding to miRNAs, which is also called competitive endogenous RNA (ceRNA) mechanism.Citation26 Based on the previous studies, circ_0006168 played an oncogenic role in ESCC through sponging miR-100Citation7 and miR-384.Citation8 Here, miR-516b-5p was identified as a novel miRNA target of circ_0006168 in our study. MiR-516b-5p functioned as a tumor suppressor in lung cancer and ESCC.Citation11,Citation27,Citation28 MiR-516b was reported to restrain the malignant potential of ESCC cells by directly regulating CCNG1.Citation11 We found that miR-516b-5p overexpression blocked the migration, invasion and proliferation of ESCC cells, and the anti-tumor role of miR-516b-5p was in agreement with the former study.Citation11 The results of rescue experiments revealed that circ_0006168 silencing-induced effects in ESCC cells were largely diminished by the knockdown of miR-516b-5p, suggesting that circ_0006168 silencing suppressed ESCC progression largely through up-regulating miR-516b-5p. Hereto, circ_0006168 was found to exert its pro-tumor role in ESCC through sponging miR-100,Citation7 miR-384Citation8 and miR-516b-5p, which provided novel insight in targeting circ_0006168-mediated carcinogenic effects in ESCC.
MiRNAs induce the translational repression or degradation of mRNAs through targeting their 3ʹUTR, leading to the changes in cell biological behaviors. For instance, miR-133b hampered the proliferation ability and motility of ESCC cells through down-regulating EGFR.Citation29 Liu et al found that miR-601 suppressed ESCC development through targeting HDAC6.Citation30 In our study, we identified XBP1 as a target of miR-516b-5p for the first time. XBP1 was identified as a pivotal regulator for cell fate determination during endoplasmic reticulum stress.Citation31 Recently, XBP1 was confirmed to play an oncogenic role in multiple malignancies, including breast cancer,Citation15 oral squamous cell carcinoma,Citation32 hepatocellular carcinomaCitation33 and ESCC.Citation18,Citation19 For instance, Xia et al found that XBP1 facilitated the proliferation ability and invasion ability of ESCC cells through inducing MMP9 expression.Citation18 Jin et al demonstrated that XBP1 acted as the downstream molecule of TUG1/miR-498 axis to contribute to ESCC development.Citation19 We found that XBP1 interference resulted in notable suppression in cell proliferation, migration and invasion abilities of ESCC cells, and the pro-tumor role of XBP1 in ESCC was consistent with former studies.Citation18,Citation19 Through performing rescue experiments, we found that XBP1 overexpression reversed circ_0006168 silencing-mediated inhibitory effects in the malignant behaviors of ESCC cells, suggesting that circ_0006168 silencing suppressed the migration, invasion and proliferation of ESCC cells largely through down-regulating XBP1. Circ_0006168 acted as a ceRNA to up-regulate the expression of XBP1 via absorbing miR-516b-5p in ESCC cells.
Our study demonstrated that circ_0006168 aggravated ESCC progression through acting as miRNA sponge to absorb miR-516b-5p to up-regulate XBP1 expression. Circ_0006168/miR-516b-5p/XBP1 signaling axis might be a novel treatment target for ESCC.
Disclosure
The authors declare that they have no conflicts of interest.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
- Takeno S, Noguchi T, Takahashi Y, Fumoto S, Shibata T, Kawahara K. Assessment of clinical outcome in patients with esophageal squamous cell carcinoma using TNM classification score and molecular biological classification. Ann Surg Oncol. 2007;14(4):1431–1438. doi:10.1245/s10434-006-9286-3
- Zhang Y, Zhang Y, Zhang L. Expression of cancer-testis antigens in esophageal cancer and their progress in immunotherapy. J Cancer Res Clin Oncol. 2019;145(2):281–291. doi:10.1007/s00432-019-02840-3
- Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi:10.1016/S0140-6736(12)60643-6
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;117:109151. doi:10.1016/j.biopha.2019.109151
- Xie ZF, Li HT, Xie SH, Ma M. Circular RNA hsa_circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):151–163. doi:10.26355/eurrev_202001_19906
- Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi:10.1016/j.bbrc.2018.08.152
- Zhu J, Zhang Y, Yang X, Jin L. Clinical significance and tumor-suppressive function of miR-516b in nonsmall cell lung cancer. Cancer Biother Radiopharm. 2017;32(4):115–123. doi:10.1089/cbr.2016.2163
- Zhao Y, Wang Y, Xing G. miR-516b functions as a tumor suppressor by directly modulating CCNG1 expression in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;106:1650–1660. doi:10.1016/j.biopha.2018.07.074
- Sun FB, Lin Y, Li SJ, Gao J, Han B, Zhang CS. MiR-210 knockdown promotes the development of pancreatic cancer via upregulating E2F3 expression. Eur Rev Med Pharmacol Sci. 2018;22(24):8640–8648. doi:10.26355/eurrev_201812_16628
- Lu L, Yue S, Jiang L, et al. Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology. 2018;67(3):1041–1055. doi:10.1002/hep.29593
- Andres SA, Wittliff JL. Relationships of ESR1 and XBP1 expression in human breast carcinoma and stromal cells isolated by laser capture microdissection compared to intact breast cancer tissue. Endocrine. 2011;40(2):212–221. doi:10.1007/s12020-011-9522-x
- Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107. doi:10.1038/nature13119
- Zhu H, Abulimiti M, Liu H, Su XJ, Liu CH, Pei HP. RITA enhances irradiation-induced apoptosis in p53-defective cervical cancer cells via upregulation of IRE1α/XBP1 signaling. Oncol Rep. 2015;34(3):1279–1288. doi:10.3892/or.2015.4083
- Bae J, Samur M, Munshi A, et al. Heteroclitic XBP1 peptides evoke tumor-specific memory cytotoxic T lymphocytes against breast cancer, colon cancer, and pancreatic cancer cells. Oncoimmunology. 2014;3(12):e970914. doi:10.4161/21624011.2014.970914
- Xia T, Tong S, Fan K, et al. XBP1 induces MMP-9 expression to promote proliferation and invasion in human esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6(9):2031–2040.
- Jin G, Yang Y, Tuo G, Wang W, Zhu Z. LncRNA TUG1 promotes tumor growth and metastasis of esophageal squamous cell carcinoma by regulating XBP1 via competitively binding to miR-498. Neoplasma. 2020;67(4):751–761. doi:10.4149/neo_2020_190805N717
- Gao D, Zhou Z, Huang H. miR-30b-3p inhibits proliferation and invasion of hepatocellular carcinoma cells via suppressing PI3K/Akt pathway. Front Genet. 2019;10:1274. doi:10.3389/fgene.2019.01274
- Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi:10.1016/j.canlet.2017.11.022
- Fan L, Cao Q, Liu J, Zhang J, Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18(1):16. doi:10.1186/s12943-018-0936-4
- Jiang C, Xu D, You Z, Xu K, Tian W. Dysregulated circRNAs and ceRNA network in esophageal squamous cell carcinoma. Front Biosci (Landmark Ed). 2019;24:277–290. doi:10.2741/4717
- Pan Z, Lin J, Wu D, et al. Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY). 2019;11(24):11937–11954. doi:10.18632/aging.102519
- Song H, Xu D, Shi P, et al. Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:1997–2006. doi:10.2147/CMAR.S195546
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
- Song H, Li H, Ding X, et al. Long non‑coding RNA FEZF1‑AS1 facilitates non‑small cell lung cancer progression via the ITGA11/miR‑516b‑5p axis. Int J Oncol. 2020;57(6):1333–1347. doi:10.3892/ijo.2020.5142
- Xu Y, Jiang T, Wu C, Zhang Y. CircAKT3 inhibits glycolysis balance in lung cancer cells by regulating miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol Lett. 2020;42(7):1123–1135. doi:10.1007/s10529-020-02846-9
- Zeng W, Zhu JF, Liu JY, et al. miR-133b inhibits cell proliferation, migration and invasion of esophageal squamous cell carcinoma by targeting EGFR. Biomed Pharmacother. 2019;111:476–484. doi:10.1016/j.biopha.2018.12.057
- Liu C, Tian X, Sun HB, Wang ZF, Jiang LF, Li ZX. MiR-601 inhibits the proliferation and metastasis of esophageal squamous cell carcinoma (ESCC) by targeting HDAC6. Eur Rev Med Pharmacol Sci. 2019;23(3):1069–1076. doi:10.26355/eurrev_201902_16995
- He Y, Sun S, Sha H, et al. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15(1):13–25. doi:10.3727/105221610X12819686555051
- Sun Y, Jiang F, Pan Y, et al. XBP1 promotes tumor invasion and is associated with poor prognosis in oral squamous cell carcinoma. Oncol Rep. 2018;40(2):988–998. doi:10.3892/or.2018.6498
- Zhou T, Lv X, Guo X, et al. RACK1 modulates apoptosis induced by sorafenib in HCC cells by interfering with the IRE1/XBP1 axis. Oncol Rep. 2015;33(6):3006–3014. doi:10.3892/or.2015.3920
