Abstract
Introduction
Immune cell interactions and metabolic changes are crucial in determining the tumor microenvironment and affecting various clinical outcomes. However, the clinical significance of metabolism evolution of immune cell evolution in colorectal cancer (CRC) remains unexplored.
Methods
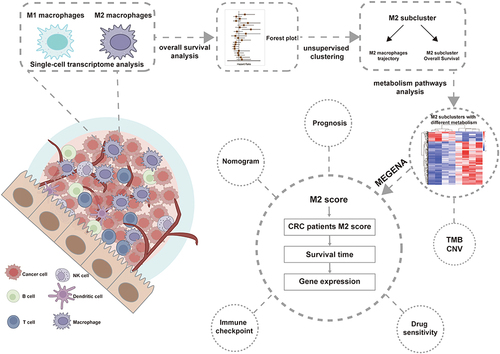
Single-cell RNA sequencing (scRNA-seq) and bulk RNA sequencing data were acquired from TCGA and GEO datasets. For the analysis of macrophage differentiation trajectories, we employed the R packages Seurat and Monocle. Consensus clustering was further applied to identify the molecular classification. Immunohistochemical results from AOM and AOM/DSS models were used to validate macrophage expression. Subsequently, GSEA, ESTIMATE scores, prognosis, clinical characteristics, mutational burden, immune cell infiltration, and the variance in gene expression among different clusters were compared. We constructed a prognostic model and nomograms based on metabolic gene signatures identified through the MEGENA framework.
Results
We found two heterogeneous groups of M2 macrophages with various clinical outcomes through the evolutionary process. The prognosis of Cluster 2 was poorer. Further investigation showed that Cluster 2 constituted a metabolically active group while Cluster 1 was comparatively metabolically inert. Metabolic variations in M2 macrophages during tumor development are related to tumor prognosis. Additionally, Cluster 2 showed the most pronounced genomic instability and had highly elevated metabolic pathways, notably those associated with the ECM. We identified eight metabolic genes (PRELP, NOTCH3, CNOT6, ASRGL1, SRSF1, PSMD4, RPL31, and CNOT7) to build a predictive model validated in CRC datasets. Then, a nomogram based on the M2 risk score improved predictive performance. Furthermore, our study demonstrated that immune checkpoint inhibitor therapy may benefit patients with low-risk.
Discussion
Our research reveals underlying relationships between metabolic phenotypes and immunological profiles and suggests a unique M2 classification technique for CRC. The identified gene signatures may be key factors linking immunity and tumor metabolism, warranting further investigations.
Data Sharing Statement
Publicly available datasets were analyzed in this study; these can be found in the UCSC Xena browser (https://xenabrowser.net/datapages), Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo), and GSEA database (https://www.gsea-msigdb.org/gsea/index.jsp).
Ethics Approval and Consent to Participate
Approval of human data: The public data of CRC patients used in the present study were approved by the Ethics Committee of Zhongnan Hospital of Wuhan University (Approval number: 2024062K). Approval of animal data: All animal experiments were approved by the Experimental Animal Welfare Ethics Committee, Zhongnan Hospital of Wuhan University (Wuhan, China) and performed in compliance with the Guidelines of Wuhan University for the Care and Use of Laboratory Animals (approval number: WP20230250).
Acknowledgments
Fengxing Huang, Youwei Wang and Yu Shao are co-first authors for this study. Qiu Zhao and are Lan Liu co-correspondence authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
All authors declare that they have no conflicts of interest in this work.