Abstract
Metachromatic leukodystrophy (MLD) disorder is a rare lysosomal storage disorder that leads to severe neurological symptoms and an early death. MLD occurs due to the deficiency of enzyme arylsulfatase A (ARSA) in leukocytes, and patients with MLD excrete sulfatide in their urine. In this study, the ARSA gene in 12 non-consanguineous MLD patients and 40 healthy individuals was examined using polymerase chain reaction sequencing. Furthermore, the structural and functional effects of new mutations on ARSA were analyzed using SIFT (sorting intolerant from tolerant), I-Mutant 2, and PolyPhen bioinformatics software. Here, 4 new pathogenic homozygous mutations c.585G>T, c.661T>A, c.849C>G, and c.911A>G were detected. The consequence of this study has extended the genotypic spectrum of MLD patients, paving way to a more effective method for carrier detection and genetic counseling.
Introduction
Metachromatic leukodystrophy (MLD) is a rare lysosomal storage disorder, caused by deficiency of the enzyme arylsulfatase A (ARSA, E.C. 3.1.6.1) with a frequency of approximately 1 per 40,000 worldwide.Citation1 ARSA catalyzes initiative step of the metabolic pathway, sphingolipid 3′-o-sulfogalactosylceramide, known as sulfatide. Sulfatide is especially abundant in the myelin sheath of the nervous system.Citation2 Mutations in the ARSA gene () (GenBank accession number NP_000478) could lead to a deficiency in ARSA activity, leading to accumulation of sulfatide, especially in the nervous system.Citation3,Citation4 This phenomenon causes a progressive demyelination that leads to different neurological symptoms including ataxia, an initially flaccid and later spastic paresis, optic atrophy, and dementia.Citation5
To date, there is no effective treatment for MLD. However, bone marrow transplantation and stem cell therapy can be beneficial for patients with juvenile- and adult-onset forms in the early stages of the disease. In addition, gene and enzyme replacement therapies for the treatment of MLD have shown promising outcomes in mice models.Citation6–Citation11
ARSA deficiency is divided into 3 clinical subtypes: late-infantile (50%–60%), juvenile (20%–30%), and adult (15%–20%). The disorder course may range from 3 to 10 years or more in the late-infantile type and up to 20 years in the juvenile and adult types.Citation9
Typical magnetic resonance imaging (MRI) alterations in MLD have been explained in the literature. T2-weighted MRI (T2W MRI) of the brain in MLD patients typically indicates butterfly-shaped confluent white matter hyperintensities with early involvement of the corpus callosum.Citation12 In addition, there is elevated white matter involvement, including U-fibers and cerebellar white matter, as well as cerebral atrophy with progression of the MLD disease.Citation13–Citation16 In this study, the ARSA gene was examined in individuals who met the proposed clinical criteria for MLD in order to identify the pathogenic impact of the associated mutations in MLD.
Materials and methods
Patient collection and ethical statement
In this study, 12 Iranian non-consanguineous MLD patients, with a mean age of 3.5 years, were diagnosed between January 2009 and November 2012. Clinical characteristics of MLD patients are summarized in . Blood samples from 12 MLD patients and 40 healthy individuals were obtained from the Special Medical Center (SMC), Tehran, Iran. Written informed consent for genetic study and molecular analysis and consent to publish results was obtained from the healthy controls, patients, and parents on behalf of their children. The clinical ethics committee of SMC specifically approved this study (Approval No ML-41-1224) in December 2012. The exclusion criteria for healthy individuals were any history of familial and sporadic cancers, hereditary and non-hereditary metabolic disorders, and nuclear and mitochondrial DNA-associated disorders.
Table 1 Clinical and biochemical features of 12 Iranian MLD patients
Enzymatic ARSA activity assay
ARSA activity was determined in leukocytes using p-nitrocatechol sulfate as described previously by Molzer et al.Citation17
Neuroimaging (MRI) analysis
T2-weighted spin-echo sequences of the brain were carried out using a Siemens Magnetom Avanto 1.5 Tesla MRI (Munich, Germany). Images were evaluated in a blind fashion by a neuroradiologist.
DNA extraction and polymerase chain reaction (PCR)
The genomic DNA was extracted from the blood samples of MLD patients by the QIAamp DNA Micro Kit (Qiagen #56304). PCR primersCitation18 for amplification of exons 1–8 of the ARSA gene are as indicated in . Briefly, PCR was carried out in final volumes of 25 µL, containing 100–200 ng of total genomic DNA, 10 pmol of forward and reverse primers, 2.5 mM of MgCl2, 200 mM of each deoxyribonucleoside triphosphates (dNTP), and 1 U of super Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany). The PCR mixture was cycled for 35 times at 95°C for 1 minute; annealing temperature was based on temperature (TM) (°C) of forward and reverse primers (). The PCR products were examined on 2% agarose gel electrophoresis () in 0.5× Tris-borate-EDTA (TBE) buffer at 110 V for 50 minutes, and then stained with 0.002 mg/mL EtBr solution and visualized using ultraviolet light.
Figure 2 Agarose gel electrophoresis of PCR product. The presence of PCR products was confirmed by analyzing the products on a 2% agarose gel. From left: lane 1: exon 1 (405 bp), lane 2: exon 1–2 (737 bp), lane 3: exon 2*–4 (706 bp), lane 4: exon 5–7* (860 bp), lane 5: exon 7–8 (916 bp), lane 6: DNA ladder (Thermo Scientific Gene Ruler 100 bp #SM0241/2/3).
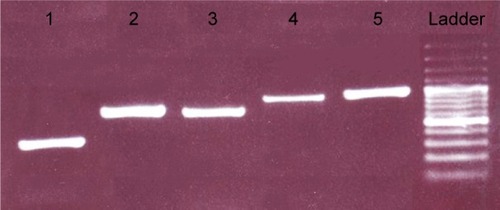
Table 2 PCR primers for amplification of ARSA gene in MLD patients
DNA sequencing and bioinformatics analysis
The PCR products were sequenced by forward or reverse primers on an ABI 3700 sequencer (Kosar Company, Tehran, Iran) and the results were compared using Finch TV program and were then analyzed on the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Moreover, the target sequence of patient was compared with normal reference sequence and ARSA gene mutations in exons and the splicing sites of the introns were identified. The functional and structural impacts of identified novel mutations in ARSA gene were assessed using in silico prediction algorithms including SIFT,Citation19 PolyPhen-2,Citation20 and I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html).
Statistical analysis
The chi-square test was used with Statistical Package for the Social Sciences (version 13) to examine the association between patient and healthy control samples, whereas P-value <0.05 was considered statistically significant.
Results
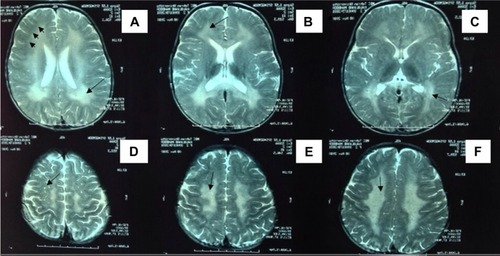
At T2W MRI, MLD patients demonstrated symmetric confluent areas of high signal intensity in the periventricular white matter (: arrows in A–C) with sparing of the subcortical U-fibers (: arrows in A). No enhancement is evident in computed tomography or MRI. The tigroid (: arrows in D) and leopard skin (: arrows in E and F) patterns of demyelination, suggesting sparing of the perivascular white matter, are identified in the periventricular white matter and centrum semiovale. The corpus callosum, corticospinal, and internal capsule tracts are also frequently involved. The cerebellar white matter may appear hyperintense at T2W MRI. In the later step of MLD, corticosubcortical atrophy is often ascertained, particularly after involvement of the subcortical white matter.
Figure 3 Axial T2-weighted brain MRI from patient P9.
Abbreviations: MRI, magnetic resonance imaging; P9, patient 9.
Eight exons of the ARSA gene were examined in 12 patients with MLD symptoms. The c.100G>A homozygous mutation in exon 1, c.661T>A homozygous mutation in exon 3, c.739G>A homozygous mutation in exon 4, c.827C>T homozygous mutation in exon 4, c.911A>G homozygous mutation in exon 5, c.931G>A homozygous mutation in exon 5, c.849C>G homozygous mutation in exon 4, A>G 96 relative to termination codon homozygous mutation, W195C homozygous mutation in exon 3, and c.978+1G>A homozygous mutation in intron 5 were detected as presented in . The 4 new mutations c.585G>T, c.661T>A, c.849C>G, and c.911A>G were significantly (P<0.05) identified in 4 patients ( and ). The possible structural and functional effects of identified new mutations in ARSA were examined using the bioinformatics SIFT, PolyPhen, and I-Mutant 2.0 software. Here, SIFT outcomes showed that W195C, F221I, D283E, and K340R mutations were determined as deleterious with scores of −0.734, −5.852, −3.908, and −2.931, respectively. I-Mutant analysis, based on the free energy change value (sign of DDG), demonstrated that p.W195C, p.F221I, p.D283E, and p.K340R mutations decreased protein stability. According to the PolyPhen score, the c.585G>T, c.661T>A, c.849C>G, and c.911A>G mutations were determined as probably damaging the protein structure and function with scores of 0.994, 1.000, 1.000 and 1.000, respectively ().
Figure 4 DNA sequencing result of the ARSA gene. (A) Sequence with a new G>T homo W195C mutation (patient 12). (B) Sequence with a new T>A homo F221I mutation (patient 2). (C) Sequence with a new C>G homo D283E mutation (patient 8). (D) Sequence with a new A>G homo K304R mutation (patient 6).
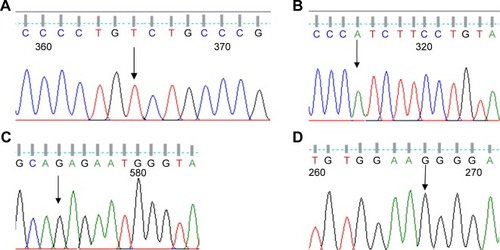
Table 3 Mutations in the ARSA gene in Iranian MLD patients
Table 4 Statistical and bioinformatics analysis of 4 novel pathogenic mutations
Discussion
In this study, 4 novel pathogenic mutations including c.585G>T, c.661T>A, c.849C>G, and c.911A>G in the ARSA gene were identified among 12 unrelated Iranian MLD patients. The previous reported mutations including the c.100G>A mutation in patient 1 (P1), c.736C>T mutation in P3, c.739G>A mutation in P4, c.827C>T mutation in P5, c.931G>A mutation in P7 and P9, A>G 96 relative to the termination codon in P15, and c.978+1G>A mutation in P11 that were reported by Gort et al,Citation21 Gieselmann et al,Citation22 Hasegawa et al,Citation23 Harvey et al,Citation24 Kreysing et al,Citation25 Gieselmann et al,Citation26 and Eng et al,Citation27 respectively.
Hence, additional research is required to affirm the biological role of these pathogenic mutations and confirm the in silico bioinformatics findings that have shown possible effects on protein structure and/or function in the absence of the ARSA enzyme.
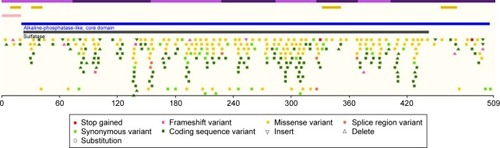
The ARSA gene has 8 exons, which are located on chromosome 22 (22q13.33).Citation25 At present, more than 150 mutations were identified in the ARSA gene according to the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=ARSA) ().
Figure 5 Mutations identified in the ARSA gene. (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=ARSA).
MLD is a heterogeneous disease with 3 frequent defective alleles including a missense mutation that leads to a Ilel79Ser substitution, a splice donor-site mutation at the exon 2/intron 2 border, and a missense mutation that causes a Pro246Leu substitution, which account for 12%, 25% (among European patients), and 25% of all defective alleles, respectively.Citation28,Citation29 Other mutant alleles were reported in only a few or single patients.Citation30
Conclusion
The result of this research has broadened the genotypic spectrum of Iranian patients with MLD, paving way to a more effective method for career detection, genetic diagnosis, and counseling of Iranian patients with MLD disorder.
Author contributions
MH conceived and designed the experiments. OA, EK, and SD performed the experiments and contributed to reagents/materials/analysis tools. FA, GYM, and MT analyzed the data. BK wrote the manuscript, contributed to the discussion, and reviewed the manuscript. All the authors read and approved the final manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Acknowledgments
The authors of this study would like to appreciate all Iranian patients with MLD disorder from the Medical Genetics Department of the Special Medical Center, Tehran, Iran, for blood donation. The authors would like to express their utmost gratitude to SMC for providing financial support (MLD-6102) to conduct this study. Additionally, the funders of this study had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
Disclosure
The authors report no conflicts of interest in this work.
References
- GustavsonKHHagbergBThe incidence and genetics of metachromatic leucodystrophy in northern SwedenActa Paediatr Scand19716055855905110535
- GieselmannVFrankenSKleinDMetachromatic leukodystrophy: consequences of sulphatide accumulationActa Paediatr Suppl2003924437479 discussion 4514989469
- van RappardDFBoelensJJWolfNIMetachromatic leukodystrophy: disease spectrum and approaches for treatmentBest Pract Res Clin Endocrinol Metab201529226127325987178
- GieselmannVKrägeloh-MannIMetachromatic leukodystrophy – an updateNeuropediatrics20104111620571983
- CostelloDJEichlerAFEichlerFSLeukodystrophies: classification, diagnosis, and treatmentNeurologist200915631932819901710
- BatziosSPZafeiriouDIDeveloping treatment options for metachromatic leukodystrophyMol Genet Metab20121051566322078456
- SoldersMMartinDAAnderssonCHematopoietic SCT: a useful treatment for late metachromatic leukodystrophyBone Marrow Transplant20144981046105124797185
- van EgmondMEPouwelsPJBoelensJJImprovement of white matter changes on neuroimaging modalities after stem cell transplant in metachromatic leukodystrophyJAMA Neurol201370677978223608771
- WangRYBodamerOAWatsonMSWilcoxWRACMG Work Group on Diagnostic Confirmation of Lysosomal Storage DiseasesLysosomal storage diseases: diagnostic confirmation and management of presymptomatic individualsGenet Med201113545748421502868
- BiffiANaldiniLNovel candidate disease for gene therapy: metachromatic leukodystrophyExpert Opin Biol Ther2007781193120517696818
- SevinCAubourgPCartierNEnzyme, cell and gene-based therapies for metachromatic leukodystrophyJ Inherit Metab Dis200730217518317347913
- CheonJEKimIOHwangYSLeukodystrophy in children: a pictorial review of MR imaging featuresRadiographics200222346147612006681
- Van der KnaapMSPouwelsPJWMagnetic resonance spectroscopy: basic principles and application in white matter disordersvan der KnaapMSValkJMagnetic resonance of myelination and myelin disordersBerlinSpringer2005859880
- FaerberENMelvinJSmergelEMMRI appearances of metachromatic leukodystrophyPediatr Radiol199929966967210460327
- KimTSKimIOKimWSMR of childhood metachromatic leukodystrophyAJNR Am J Neuroradiol19971847337389127040
- EichlerFGroddWGrantEMetachromatic leukodystrophy: a scoring system for brain MR imaging observationsAJNR Am J Neuroradiol200930101893189719797797
- MolzerBSundt-HellerRKainz-KorschinskyMZobelMElevated sulfatide excretion in heterozygotes of metachromatic leukodystrophy: dependence on reduction of arylsulfatase A activityAm J Med Genet19924445235261359786
- WangJZhangWPanHARSA gene mutations in five Chinese metachromatic leukodystrophy patientsPediatr Neurol200736639740117560502
- KumarPHenikoffSNgPCPredicting the effects of coding non-synonymous variants on protein function using the SIFT algorithmNat Protoc2009471073108119561590
- AdzhubeiIASchmidtSPeshkinLA method and server for predicting damaging missense mutationsNat Methods20107424824920354512
- GortLCollMJChabásAIdentification of 12 novel mutations and two new polymorphisms in the arylsulfatase A gene: haplotype and genotype-phenotype correlation studies in Spanish metachromatic leukodystrophy patientsHum Mutat199914324024810477432
- GieselmannVZlotogoraJHarrisAWengerDAMorrisCPMolecular genetics of metachromatic leukodystrophyHum Mutat1994442332427866401
- HasegawaYKawameHEtoYMutations in the arylsulfatase A gene of Japanese patients with metachromatic leukodystrophyDNA Cell Biol19931264934988101083
- HarveyJSNelsonPVCareyWFRobertsonEFMorrisCPAn arylsulfatase A (ARSA) missense mutation (T274M) causing late-infantile metachromatic leukodystrophyHum Mutat1993242612678104633
- KreysingJvon FiguraKGieselmannVStructure of the arylsulfatase A geneEur J Biochem199019136276311975241
- GieselmannVPoltenAKreysingJvon FiguraKArylsulfatase A pseudodeficiency: loss of a polyadenylation signal and N-glycosylation siteProc Natl Acad Sci U S A19898623943694402574462
- EngBNakamuraLNO’ReillyNIdentification of nine novel arylsulfatase a (ARSA) gene mutations in patients with metachromatic leukodystrophy (MLD)Hum Mutat2003225418419
- PoltenAFluhartyALFluhartyCBKapplerJvon FiguraKGieselmannVMolecular basis of different forms of metachromatic leukodystrophyN Engl J Med1991324118221670590
- BergerJLöschlBBernheimerHOccurrence, distribution, and phenotype of arylsulfatase A mutations in patients with metachromatic leukodystrophyAm J Med Genet19976933353409096767
- SchestagFYaghootfamAHabethaMThe functional consequences of mis-sense mutations affecting an intra-molecular salt bridge in arylsulphatase ABiochem J2002367Pt 249950412086582
- HGMD® [database on the Internet]Cardiff, UKCardiff University Available from: http://www.hgmd.cf.ac.uk/ac/gene.php?gene=ARSAAccessed April 4, 2017
