Abstract
Objectives:
To determine if histopathologic assessment of esophageal biopsies harvested for research study is justified due to the heterogeneity of tissues in the esophagus, and the consequent histopathologic mis-matches with the clinical histopathology of biopsies taken at the same level.
Methods:
Since 2004, patients undergoing upper endoscopy for a variety of clinical conditions were invited to provide additional esophageal biopsies; those were collected for research purpose at the same level as biopsies collected for clinical histopathology. Research biopsies were cut in two parts: one part was submitted to research histopathology and the other stored for molecular analysis. Results of clinical histopathology for each patient were summarized per biopsy level and compared to results obtained from research biopsies at the corresponding level.
Results:
A total of 377 level summaries were obtained from 137 patients. Clinical histopathology summaries classified 123 levels (32.6%) as squamous epithelium, 84 levels (22.3%) as metaplastic columnar-lined epithelium, 135 levels (35.8%) as columnar-lined epithelium with intestinal metaplasia, 30 levels (8%) as dysplasia, and 5 levels (1.3%) as adenocarcinoma. Research histopathology matched to clinical summaries on 120 of 123 (97.5%) levels for squamous epithelium, 52 of 84 (61.9%) for metaplastic columnar-lined epithelium, and 94 of 135 (69.5%) for columnar-lined epithelium with intestinal metaplasia. There were no matches for dysplasia between the groups; however, they agreed on all five cases of AC. On 59 (70.2%) metaplastic columnar-lined epithelium levels and on 62 (46%) columnar-lined epithelium with intestinal metaplasia levels, tissue heterogeneity was observed in clinical histopathology, with portions of squamous epithelium within the samples. Matches with pure tissue samples in both clinical and research histopathology levels were observed on 22 (26.2%) levels of metaplastic columnar-lined epithelium and in 55 (40.7%) levels of columnar-lined epithelium with intestinal metaplasia.
Conclusions:
The high proportion of mismatches and tissue heterogeneity observed, especially among columnar-lined epithelium with intestinal metaplasia and dysplasia, points to the necessity of determining the histopathology of the research samples to avoid sampling errors during molecular studies.
Introduction
In some individuals with chronic gastroesophageal reflux disease, the normal esophageal squamous epithelium can be replaced with a metaplastic columnar-lined epithelium which exhibits intestinal metaplasia. This condition is called Barrett’s esophagus. It can predispose to progression to dysplasia, and in some individuals adenocarcinoma can arise in this epithelium.Citation1 Currently, endoscopy with biopsy for histopathology is the gold standard for diagnosing Barrett’s esophagus, as well as the development of dysplasia and carcinoma. However, endoscopy findings, when compared to histopathologic diagnosis, have a sensitivity of only 82% and a positive predictive value of 34% for diagnosing columnar-lined epithelium with intestinal metaplasia. In part this might be related to focal distribution of intestinal metaplasia within columnar-lined epithelium due to heterogeneity of tissue types in the esophageal mucosa.Citation2 In addition, the identification of dysplasia is based on subjective histopathologic assessment, and there can be significant intra- and inter-observer variability between histopathologists when determining the presence of dysplastic changes.Citation3–Citation5 This highlights the need for new tools for the detection and follow up of various esophageal pathologies.Citation6,Citation7
Advances brought by high-throughput gene expression have made it possible to analyze the molecular characteristics of normal and abnormal tissues, including esophageal pathologies.Citation8–Citation11 The development of tissue biomarkers which correctly identify and the risk of progression to dysplasia and cancer would be of value for guiding clinical surveillance and early diagnosis of high risk lesions. This might facilitate more effective and timely therapy. Hence, research in this area might impact on future clinical practice if useful biomarkers can be identified.
However, to accurately identify specific biomarkers, it is necessary to be certain about the tissue type from which nucleic acids are extracted for biomarker discovery studies. Unfortunately, the heterogeneity of tissue types within the esophageal mucosa in patients who have developed Barrett’s esophagus complicates the identification of specific mucosal subtypes, and relying on collecting one biopsy for histopathology and an adjacent biopsy for nucleic acid extraction for research might lead to errors as adjacent tissue biopsies in such an individual may contain different pathologies.
With the aim of ensuring correct tissue classification for esophageal biopsy samples selected for subsequent molecular analysis and research, we developed a technique to identify tissue types. This entailed dividing stored biopsy samples into two pieces: one for histopathology, and the other for molecular study. In this study we determined the accuracy and utility of this method for classification of these tissue samples, and compared the outcomes with conventional histopathology assessment of biopsies obtained for clinical purposes from adjacent mucosa.
Materials and methods
Since 2004, patients undergoing upper gastrointestinal endoscopy for a variety of clinical conditions have been invited to provide additional endoscopic biopsy samples for laboratory research. Biopsies were collected for both clinical histopathology and research purposes. In individuals who had suspected Barrett’s esophagus, the biopsy collection protocol entailed the collection of a mucosal biopsy from four quadrants of the esophageal wall, 1 cm above the gastroesophageal junction, and then every 2 cm proximally for the full length of the columnar mucosa. These biopsy samples were placed in formalin and sent for conventional histopathology for clinical diagnostic purposes. A further three mucosal biopsies were obtained from the same levels in the esophagus, and three biopsies were also collected from the proximal stomach, and three from the proximal squamous epithelium, 5 cm above the upper limit of the columnar epithelium. These biopsy samples were processed for laboratory research as described below.
In individuals with suspected cancer, sufficient biopsies of the cancer were collected, placed in formalin and sent for conventional histopathologic assessment. Three to four extra biopsy samples were collected from the tumor for research purposes, as well as three biopsies from the proximal stomach, and three from the proximal squamous epithelium. In all other individuals three biopsies were also collected from the proximal stomach for research purposes, three from the esophageal squamous epithelium 2 cm proximal to the gastroesophageal junction and three from 8 cm more proximal. Some variation in the tissue collection protocol occurred according to clinical findings (eg, mucosal ulceration, and the presence of islands or tongues of columnar-lined epithelium with suspected intestinal metaplasia).
All biopsies collected for research were immediately placed in RNA-later® and stored at −20 °C until required for later study. When removed from storage, biopsies were thawed, and a small piece which comprised approximately 20% to 30% of each biopsy was cut off with a scalpel blade and placed in formalin. This piece was then embedded in paraffin, sliced and stained with hematoxylin and eosin, as well as ABPAS/D histochemical stain, and then examined using conventional light microscopy. The remainder of the biopsy was then stored at −80 °C for later nucleic acid extraction and molecular studies.
The histopathology slides prepared from the fragments of the research biopsy samples where examined by a single pathologist (DA) with expertise in esophageal pathology. The pathologist was unaware of the results of any conventional histopathology analysis of biopsy samples collected for clinical purposes from the same individuals. Intestinal metaplasia within columnar epithelium was confirmed by the presence of goblet cells, which were identified using ABPAS/D histochemical staining. The presence of dysplasia in any clinical biopsy samples was also determined.
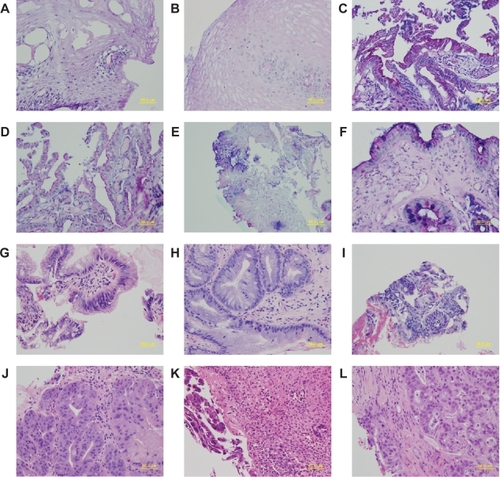
To assess the tissue type identified in the fragments of the research biopsies, we compared the diagnosis from biopsies collected for clinical histopathology with that for the histopathology from the research biopsy fragments, when both types of biopsy were obtained from the equivalent level in the esophagus; ie, the same distance from the gastroesophageal junction. In addition, to assess inter- and intra-observer variability, all slides from clinical samples reported to contain dysplastic tissues, and the corresponding slides from research samples were reviewed again by the pathologist. For this analysis the pathologist was blinded to the first diagnosis. Representative sections of each tissue type from matched clinical and research biopsies are shown in .
Figure 1 Sections of esophageal epithelium from matched research and clinical biopsies from the esophagus. Research biopsy sections are shown on the left panel and the matched clinical biopsy sections are on the right panel. A–F are sections stained with ABPAS/D. G–L are sections stained with H&E. A and B show stratified squamous epithelium, C and D show columnar lined epithelium without intestinal metaplasia, E and F show columnar-lined epithelium with intestinal metaplasia, G and H show changes consistent with low grade dysplasia, I and J represent carcinoma in-situ, and K and L show invasive cancer.
The tissue collection for this study was approved by the Flinders Clinical Research Ethics Committee and the Repatriation General Hospital Clinical Research Ethics Committee.
Results
One hundred thirty-seven individuals were included in this study, and endoscopic biopsies were obtained from 377 different levels (or sites). Clinical histopathology identified 123 levels (32.6%) as squamous epithelium, 84 levels (22.3%) as columnar epithelium without intestinal metaplasia, 135 levels (35.8%) as columnar epithelium with intestinal metaplasia, 30 levels (8%) as dysplasia (28, low grade dysplasia; 2, high grade dysplasia), and 5 levels (1.3%) as adenocarcinoma.
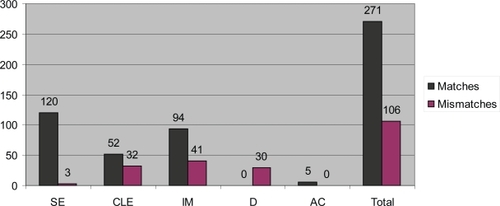
Two hundred seventy-one (71.9 %) of the diagnoses made by histopathology assessment of the research biopsy fragments, matched the clinical histopathology assessment, and in 108 (28.1%) the assessment differed (). The proportion of matches between assessment from corresponding research and clinical biopsies varied according to the pathological classification. There was a high level of concordance for squamous epithelium and adenocarcinoma, with all but three squamous epithelium and all adenocarcinoma research biopsies matching the classification of adjacent clinical samples. However, the tissue type identified in the clinical biopsies matched the type identified in the research biopsy fragments in only 52 of 84 (61.9%) columnar epithelium without intestinal metaplasia samples, and 94 of 135 (69.6%) the columnar epithelium with intestinal metaplasia samples (). Differences usually occurred when columnar epithelium without intestinal metaplasia was identified in one sample, and columnar epithelium with intestinal metaplasia in the matched samples, and vice versa.
Figure 2 Number of matches and mismatches between histopathology for research biopsy vs. clinical biopsy. The type of tissue present in the clinical biopsy is named on the X axis.
Abbreviations: AC, adenocarcinoma; CLE, columnar-lined epithelium; D, dysplasia; IM, columnar-lined epithelium with intestinal metaplasia; SE, squamous epithelium.
Heterogeneity within individual biopsy samples was identified when a portion of squamous epithelium was found alongside columnar epithelium with or without intestinal metaplasia, within the same biopsy sample. Although heterogeneity was observed in only 30 (7.6%) of the research biopsies, review of histopathology from the clinical biopsies showed a higher prevalence of heterogeneity in the clinical biopsy samples, 121 (32%). This might be due the larger size of the clinical biopsy samples, compared to the research samples, which were all split for histopathology and nucleic acid extraction. Matches where only one tissue type was observed in both clinical histopathology and research histopathology, were reported on 22 of 84 (26.2%) levels of metaplastic columnar-lined epithelium and in 55 of 135 (40.7%) levels of columnar-lined epithelium with intestinal metaplasia.
When research biopsy samples were obtained from areas adjacent to squamous mucosa identified by clinical histopathology, 120 (97.5%) research biopsies contained only squamous epithelium. In two biopsies (1.6%), research histopathology contained portions of columnar epithelium within squamous epithelium, and in one (0.9%) only columnar epithelium was identified.
On the 84 esophageal levels which contained metaplastic columnar epithelium without intestinal metaplasia on clinical histopathology assessment, 59 (70.2%) also had portions of squamous epithelium in at least one of the clinical biopsies. The histopathology findings from the research samples from the same level were identical in 52 (61.9%), whereas only squamous epithelium was identified in 22 biopsies (26.2%), and intestinal metaplasia within columnar epithelium in another 10 biopsies (11.9%). Histopathology identified columnar epithelium without intestinal metaplasia in both the research and clinical biopsy samples in 22 (26.2%) cases.
Table 1 Matches and mismatches between research histopathology results vs. clinical histopathology
Of the 135 esophageal levels which had columnar epithelium with intestinal metaplasia within the clinical biopsy samples, there were 62 (46%) levels which also contained portions of squamous epithelium. Histopathology from the research samples matched these findings in 94 (69.5%) biopsies. In eight (6%) biopsies only squamous epithelium was identified, in 29 (21.5%) only columnar epithelium without intestinal metaplasia, in two (1.5%) both squamous epithelium and columnar epithelium without intestinal metaplasia, and dysplasia was identified in two (1.5%).
There were few matches between the clinical and research pathology diagnoses for dysplasia. Of 30 clinical levels which had dysplasia in at least one of the four clinical biopsies (28, low grade dysplasia; 2, high grade dysplasia), the corresponding histopathology from the research biopsy fragment was columnar epithelium without intestinal metaplasia in nine (30%), columnar-lined epithelium with intestinal metaplasia in another 19 (63%), and low grade dysplasia in two (7%). The histopathology from the research biopsy fragments and the clinical biopsies matched in all five cases of adenocarcinoma.
Discussion
In this study we determined the proportion of matches between tissue types identified by histopathologic examination of fragments of esophageal mucosal biopsies collected for research purposes vs esophageal mucosal biopsies collected for clinical diagnosis. For clinical purposes, at each level multiple biopsies were usually collected from different aspects of the esophageal wall, and if Barrett’s esophagus was suspected clinically, four biopsies were collected at each level, one from each quadrant, using the Seattle biopsy collection protocol.Citation12 This meant that more than one biopsy sample was usually obtained for clinical diagnosis from appropriate levels within the esophagus. These biopsies were usually reported as a summary for each esophageal level rather than per biopsy, and this meant that in some individuals more than one tissue type was present at each esophageal level, whereas in others up to four biopsies from a level all identified a single tissue type.
When undertaking molecular biology research which seeks to identify changes in gene expression and other related changes within esophageal mucosal cells, it is important to accurately identify the mucosal tissue type from which nucleic acids are extracted. Whilst this might be done using laser capture microdissection, this technology is not suitable for the analysis of whole fresh esophageal biopsy specimens, and other strategies need to be considered. One option is to collect fresh tissue biopsies for research from the same level as biopsies that are collected for clinical diagnostic purposes, and then assume that the tissue type contained in the research biopsy is identical to that identified by histopathologic examination of the clinical biopsy specimens from the same level. Unfortunately, this assumption is not always correct, and the heterogeneity of tissue types found in the distal esophageal mucosa of individuals with Barrett’s esophagus means that more accurate identification of tissue type is required to avoid the errors which can arise if tissues are not correctly classified.
It was because of this problem that we developed the described technique of splitting the biopsies collected for research, with approximately 30% of the biopsy removed for histopathology. This was done to minimize the risk of erroneous classification of each individual biopsy due to heterogeneity. For such a method to be valid, however, the quality of the histopathology slides needs to be comparable to those obtained from conventional histopathology techniques. The research specimens were all initially stored and frozen in RNA later, then thawed and split before placing a fragment into formalin for fixation and subsequent embedding in paraffin, slicing, staining, and then examination under light microscopy. Our experience confirmed that the quality of the histopathology slides obtained by this process was good, and it allowed accurate assessment of each biopsy. Furthermore, we were still able to obtain good yields of DNA and RNA from the residual specimen (unpublished data). We have successfully used this for microarray and qPCR profiling of mRNA and microRNA expression in esophageal disease.Citation13,Citation14
Our results showed a high percentage of matches between clinical and research histopathology for both squamous epithelia and cancer. For squamous epithelia the endoscopic visual identification of the nature of the mucosa biopsied is usually accurate, and squamous mucosa is usually homogenous. This means that errors in classification of this mucosal type are unlikely to occur. Similarly, for cancer endoscopic visual identification is usually accurate, and biopsies can usually be obtained accurately.
On the other hand, a high proportion of mismatches were observed for columnar mucosa with versus without intestinal metaplasia, and for dysplasia. This highlights the importance of accurate identification of mucosal type, and confirms the risk of error if researchers rely on taking adjacent, but separate biopsies: one for clinical diagnosis and one for research purposes. This problem is partly due to the heterogeneity within metaplastic columnar mucosa in individuals with Barrett’s esophagus. It can also be due to difficulties with accurate biopsy collection in such individuals. Biopsy collection requires accurate manipulation of endoscopic biopsy forceps, and this can be difficult, particularly in individuals with short segments of Barrett’s esophagus, or tongues of metaplastic mucosa. Furthermore, as each biopsy is collected, there is usually a small amount of bleeding from the biopsy site, and this can obscure the endoscopist’s vision, rendering accurate collection of the next biopsy more difficult. This can contribute to errors in tissue identification, particularly when sampling close to the squamo-columnar junction. In our study there were a significant number of individuals in whom endoscopy identified tongues or islands of metaplastic columnar mucosa, highlighting this potential problem.
It is also possible, but perhaps less likely, that heterogeneity was less common in the research biopsies because they were smaller fragments than the clinical biopsies, and hence a different tissue might have been found if a different part of the research biopsy had been examined pathologically. For this reason, complete accuracy of diagnosis cannot be guaranteed by the method described, although the extent of misdiagnosis of tissues type has almost certainly been minimized by this method.
With dysplasia the problem of heterogeneity is also important, but in addition, inter-observer variability and subjectivity in the diagnosis and grading of dysplasia is likely to have further confounded the correct identification of mucosal type. In our study the inter-observer agreement between the original clinical diagnosis and the pathologist re-reviewing the pathology was 73%, which is in line with previous reports.Citation5 We found that intra-observer agreement when re-reporting the research biopsies was 93%. These findings reinforce the necessity of having access to an experienced pathologist, especially when dysplasia is considered. Correct characterization and grading of dysplasia is potentially of great benefit for research which is investigating the progression from Barrett’s esophagus to cancer. The identification of molecular biomarkers for normal and abnormal esophageal tissues is of considerable interest,Citation10–Citation12 and identification of consistent tissue signatures, in particular for dysplasia, have the potential to improve diagnosis and improve management.
We saw a high proportion of heterogeneity within different biopsies collected for clinical purposes, and if we relied on clinical pathology reports for tissue classification, significant errors could occur, and gene expression profiling of supposed esophageal mucosal types may not be correct. For this reason, the histopathology analysis of the fragments of the research biopsies is likely to minimize the risk of misclassification due to heterogeneity, as well as errors in biopsy collection, thereby improving the reliability of research data in this field.
Disclosure
The authors report no conflicts of interest in this work.
References
- JankowskiJAProvenzaleDMoayyediPEsophageal adenocarcinoma arising from Barrett’s metaplasia has regional variations in the westGastroenterology200212258859011845805
- EloubeidiMAProvenzaleDDoes this patient have Barrett’s esophagus? The utility of predicting Barrett’s esophagus at the index endoscopyAm J Gastroenterol19999493794310201460
- OrmsbyAHPetrasREHenricksWHObserver variation in the diagnosis of superficial oesophageal adenocarcinomaGut20025167167612377805
- AlikhanMRexDKhanARahmaniECummingsOUlbrightTMVariable pathologic interpretation of columnar lined esophagus by general pathologists in community practiceGastrointest Endosc199950232610385717
- SkacelMPetrasREGramlichTLSigelJERichterJEGoldblumJRThe diagnosis of low-grade dysplasia in Barrett’s esophagus and its implications for disease progressionAm J Gastroenterol2000953383338711151865
- JegoMVolantAFaycalJPrevalence and topography of intestinal metaplasia in columnar lined esophagusGastroenterol Clin Biol20073160160617646787
- EndlicherERummelePBeerSBarrett’s esophagus: a discrepancy between macroscopic and histological diagnosisEndoscopy2005371131113516281145
- WangKGanLJefferyEMonitoring gene expression profile changes in ovarian carcinomas using cDNA microarrayGene199922910110810095109
- MochHSchramlPBubendorfLHigh-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinomaAm J Pathol199915498198610233835
- SelaruFMZouTXuYGlobal gene expression profiling in Barrett’s esophagus and esophageal cancer: a comparative analysis using cDNA microarraysOncogene20022147547811821959
- VallbohmerDMarjoramPKuramochiHTowards the molecular characterization of disease: comparison of molecular and histological analysis of esophageal epitheliaJ Gastrointest Surg2007111095110417623264
- LevineDSHaggittRCBlountPLRabinovitchPSRuschVWReidBJAn endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in Barrett’s esophagusGastroenterology199310540508514061
- van RoonAHMayneGCWijnhovenBPImpact of gastroesophageal reflux on mucin mRNA expression in the esophageal mucosaJ Gastrointest Surg2008121331134018452049
- WatsonDIWijnhovenBPLMichaelMZMayneGCHusseyDJMicroRNA expression profiles in Barrett’s oesophagusANZ J Surg200777Suppl 1A45