Abstract
Purpose:
To investigate whether the GLC3A locus harboring the CYP1B1 gene is associated with normal tension glaucoma (NTG) in Japanese patients.
Materials and Methods:
One hundred forty-two Japanese patients with NTG and 101 Japanese healthy controls were recruited. Patients exhibiting a comparatively early onset were selected as this suggests that genetic factors may show stronger involvement. Genotyping and assessment of allelic diversity was performed on 13 highly polymorphic microsatellite markers in and around the GLC3A locus.
Results:
There were decreased frequencies of the 444 allele of D2S0416i and the 258 allele of D2S0425i in cases compared to controls (P = 0.022 and P = 0.034, respectively). However, this statistical significance disappeared when corrected (Pc > 0.05). We did not find any significant association between the remaining 11 microsatellite markers, including D2S177, which may be associated with CYP1B1, and NTG (P > 0.05).
Conclusions:
Our study showed no association between the GLCA3 locus and NTG, suggesting that the CYP1B1 gene, which is reportedly involved in a range of glaucoma phenotypes, may not be an associated factor in the pathogenesis of NTG.
Introduction
Glaucoma is a progressive optic neuropathy leading to permanent visual loss that is often associated with elevated intraocular pressure (IOP). Primary open-angle glaucoma (POAG) is the most common type of glaucoma. Normal tension glaucoma (NTG) is an important subset of POAG; while many POAG patients have high IOP,Citation1 patients with NTG have statistically normal IOP.Citation2–Citation4 The prevalence of NTG is higher among the Japanese population than among Caucasians, and recent studies reported that 92% of POAG patients in Japan had NTG.Citation5–Citation8 The diagnosis of glaucoma is based on a combination of factors including optic nerve damage and specific field defects for which IOP is the only treatable risk factor. NTG, however, tends to be underdiagnosed because of an accompanying high myopia in many cases and the lack of elevated IOP. Although relatively higher IOP, myopia, and older age are known to be factors associated with the development of NTG,Citation9 they are not pathognomonic and there remains a growing interest in the identification of pathogenetic factors associated with NTG.
Glaucoma is genetically heterogeneous and the detection of susceptibility genes could provide useful information for early diagnosis of glaucoma. To date, over 30 genetic loci for glaucoma have been identified by linkage analysis in multiple pedigrees; Citation10–Citation12 14 loci of POAG, 3 loci of primary congenital glaucoma (PCG), and 1 locus of pigment dispersion syndrome have been designated GLC1A-GLC1N, GLC3A-GLC3C, and GPDS1 (with approval from the HUGO Genome Nomenclature Committee), respectively. Among them, GLC3A harbors cytochrome P450, family 1, subfamily B, polypeptide 1 (CYP1B1).Citation13 CYP1B1 sapans 8.5 kb on chromosome 2p21 with three exons, and mutation of this gene is a major cause of PCG.Citation10,Citation13 Most CYP1B1 mutations are genetic insertions, deletions, or missense mutations, indicating that CYP1B1 is relatively susceptible to recombination events.Citation10 CYP1B1 has also been found to be involved in the development of juvenile-onset glaucoma (juvenile open angle glaucoma [JOAG]).Citation14–Citation16 In addition, recent studies have shown that CYP1B1 mutations are related to POAG in several ethnic groups,Citation10 and there have been reports of delayed expression of a CYP1B1 mutation and coexistence of PCG and POAG in the same pedigree.Citation17,Citation18 These recent studies suggest that CYP1B1 mutations in the GLC3A locus may contribute to a broader range of glaucoma phenotypes than the PCG phenotype alone, including NTG.
In this study, with prospective SNP analysis of the CYP1B1 gene in NTG patients in mind, we performed comprehensive microsatellite mapping in and around the GLC3A locus and investigated the disease-susceptibility of this locus in NTG patients.
Materials and methods
Subjects
We recruited 243 Japanese subjects from the Yokohama City University, Yamanashi University, Gifu University, Kobe University, Yamaguchi University, Kumamoto University, Hokkaido University, Tokyo University, Niigata University, Kanazawa University, Hiroshima University, Tajimi Municipal Hospital, and Tokai University in Japan. Of these subjects, 142 were diagnosed with NTG, and 101 were control subjects. The control subjects were of the same age and sex as the NTG patients, and they were not affected by glaucoma or any ophthalmological or systemic diseases that could result in optic nerve or visual field changes. Furthermore, the control cases either had no myopia or had mild myopia with refractive errors of −3.00 D or less. All patients and control subjects were of Japanese ethnicity, with similar social backgrounds and residing in the same urban area. Informed consent was obtained from all patients, and the study was conducted in accordance with the Declaration of Helsinki and subsequent revisions thereof.
NTG patients enrolled in this study were diagnosed as such if the patient had the following conditions: glaucomatous changes in the optic nerve head with or without retinal nerve fiber layer defect and corresponding glaucomatous visual field defects; normal open angle with angle width greater than Shaffer grade 2; absence of intraocular pressure greater than 21 mmHg on repeated examination by Goldmann applanation tonometry without medication; neurologic, rhinologic, and general medical examinations including magnetic resonance imaging that failed to disclose any pathology responsible for optic nerve change. Glaucomatous optic nerve abnormality was diagnosed when the vertical cup/disc ratio of the optic nerve head was more than or equal to 0.7, or the rim width at the superior portion (11–1 hour) or inferior portion (5–7 hour) was less than or equal to 0.1 of disc diameter, or the difference of the vertical cup/disc ratio was greater than or equal to 0.2 between both eyes, or a nerve fiber layer defect was found. Glaucomatous visual field defect was defined on a hemifield basis using a reliable field data examined by the Humphrey® static visual field analyser (Carl Zeiss Meditec, Oberkochen, Germany,) C-30-2 program according to the Anderson and Patella’s criteria; Citation19 the hemifield was judged abnormal when the pattern deviation probability plot showed a cluster of three or more nonedge-contiguous points having sensitivity with a probability of less than 5% in the upper or lower hemifield, and in one of these with a probability of less than 1%.
The following inclusion and exclusion criteria were used to stringently categorize the patient groups in this study. We excluded individuals who were diagnosed under 20 or over 60 years of age and who had −8.0 D or higher myopic refractive error of spherical equivalence. The selection criteria of HFA mean deviation were stratified depending on the subjects’ age in order to minimize the effect of aging on retinal ganglion cell loss and subsequent visual field defect(s) (i) no limitation if the patient was diagnosed under 50 years of age (ii) −10.00 dB or worse in at least one eye if the patient was diagnosed between 50 and 55 years of age (iii) −15.00 dB or worse in at least one eye if the patient was diagnosed above 55 years of age. In this study, the cases exhibiting a comparatively early onset were selected as they suggest that genetic factors may show stronger involvement.
During diagnosis, patients whose refraction values had changed due to cataract surgery, refractive surgery, or other intervention were excluded from the study. In cases where a glaucomatous visual field defect was present only in one eye, the refraction value and glaucomatous visual field defect of the affected eye were adopted. In cases where a glaucomatous visual field defect was present in both eyes, the refraction value and glaucomatous visual field defect of the more severely affected eye were adopted.
Analysis of repeat polymorphisms for 13 microsatellite markers
Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) or the guanidine method. In this association study, we selected 13 highly polymorphic microsatellite markers that are located in and around the GLC3A locus as shown in Figure . The markers were determined based on the National Center for Biotechnology Information for fine mapping. Polymerase chain reaction (PCR) was performed in a reaction mixture with a total volume of 12.5 μl containing PCR buffer, genomic DNA, 0.2 mM dinucleotide triphosphates (dNTPs), 0.5 μM primers, and 0.35 U Taq polymerase. The PCR conditions were as follows: 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 sec, annealing at 56 °C for 30 sec, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 10 min. The reaction was carried out in a PCR thermal cycler (GeneAmp System 9700, Applied Biosystems, Foster City, CA, USA). The forward primer was labelled at the 5′ end with 6-FAM, VIC, PET, or NED (Sigma-Aldrich, St. Louis, MO, USA) (Table ). To determine the number of microsatellite repeats, the PCR products were denatured at 97 °C for 2 min, mixed with formamide, and electrophoresed using an ABI3130 Genetic Analyser (Applied Biosystems). The number of microsatellite repeats was estimated automatically using the GeneScan 672 software (Applied Biosystems) by the local Southern method with a size marker of GS500 TAMRA (Applied Biosystems).
Figure 1 Location of 13 microsatellite markers used in this study. The heavy bar above the chromosome represents the 2p22.1-p22.3 region including the GLC3A locus.
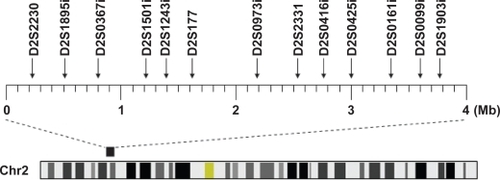
Table 1 Primer sequences of 13 microsatellite markers used in this study
Statistical analysis
Allelic frequencies were estimated by direct counting. The significance of allelic frequencies between the patient and control groups was evaluated by Fisher’s exact test. The probability of association was corrected by the Bonferroni inequality method, ie, by multiplying the obtained P values with the number of alleles compared. A corrected P (Pc) value of <0.05 was considered statistically significant. Statistical analyses were performed on a computer using the SPSS software (version 10.1; SPSS Inc., Chicago, IL, USA). Haplotype frequencies and linkage disequilibrium (LD) in the multi-locus analyses were calculated using PyPop.Citation20 Haplotype frequencies were estimated using the iterative Expectation-Maximization algorithm. LD was measured using Hedrick’s multiallelic D’ statistic.Citation21
Results
Patient age range was 21–58 years (mean 49.1 ± 9.7); 47.2% were male and 52.8% were female. The mean refraction value was −3.74 ± 3.02 diopters (D), and the mean deviation observed in the Humphrey® static visual field determination (Carl Zeiss Meditec, Oberkochen, Germany) was −10.14 ± 8.04 dB.
We genotyped 13 polymorphic microsatellite markers in and around the GLC3A locus in 142 patients and 101 controls (Figure ). The observed and expected frequencies of each genotype for the 13 markers in the case and control subjects were in Hardy–Weinberg equilibrium (data not shown). Only two adjacent markers, D2S0416i and D2S0425i, were significantly positive, as shown in Table , and the frequency of the 444 allele of D2S0416i and the 258 allele of D2S0425i were decreased in cases compared to controls (P = 0.022, OR = 0.59 and P = 0.034, OR = 0.42, respectively). However, this statistical significance disappeared (Pc > 0.05) when evaluated by Bonferroni correction. The magnitude of LD between these two markers was low, with pair-wise D’ = 0.25, and the comparison of haplotype consisting of two alleles (D2S0416i_444 and D2S0425i_258) rendered no significant difference between cases and controls (cases vs controls = 3.5% vs 7.3%, P = 0.055) (data not shown).
Table 2 Frequencies of 13 microsatellite markers in NTG cases and controls
Discussion
The purpose of this study was to investigate whether the GLC3A locus is associated with NTG in Japanese subjects, based on results from recent studies reporting that the CYP1B1 gene, located at the GLC3A locus on chromosome 2p21, could be a causative gene in POAG as well as PCG. To this end, we genotyped 13 microsatellite markers in and around the GLC3A locus. Here we report a lack of association between the GLC3A locus and NTG in Japanese patients, suggesting that genetic variation at this locus may not play an important role in the development of NTG.
Sarfarazi and colleagues mapped GLC3A as a putative PCG locus to 2p21 in 11 Turkish families, and the D2S177 microsatellite, located 270 kb telomeric to the CYP1B1 gene, showed a maximal LOD score of 9.40.Citation22 Bejjani and colleagues also reported that the maximal LOD score was 15.76 with D2S177 in 25 Saudi Arabian PCG families.Citation23 Recently, it has been reported that the D2S177 allele was associated with CYP1B1 mutations related to PCG.Citation24 Therefore, D2S177 may be potentially useful as a marker for genetic events associated with glaucoma, such as CYP1B1 mutations. In the present study, we did not find any significant association between D2S177 alleles and NTG, suggesting that CYP1B1 mutations may not be a risk factor for NTG. It has been hypothesized that mutations of CYP1B1 cause developmental abnormality in the structure and function of the anterior ocular segment.Citation25 However, NTG patients have statistically normal IOP with no abnormality in the structure of anterior ocular segment, suggesting that CYP1B1 is a potential disease susceptibility gene of PCG, POAG, and JOAG, but not NTG. On the other hand, we found a weak association between two adjacent markers, D2S0416i and D2S0425i, and NTG, although this link did not reach statistical significance when corrected. These two markers were located ∼860 kb centromeric to the CYP1B1 gene. The nearest gene of D2S0416i is the SOS1 (son of sevenless homolog 1 [Drosophila]) gene, which is about 40 kb from the marker. SOS1 encodes a protein that is a guanine nucleotide exchange factor for RAS proteins,Citation26 and mutations in this gene are associated with gingival fibromatosis and Noonan syndrome.Citation27–Citation29 Another marker, D2S0425i, is within the CDKL4 (cyclin-dependent kinase-like 4) gene, a member of the Ser/Thr protein kinase family. Currently, there are no reports suggesting any connection between these two genes and NTG.
Previous studies reported that mutations in genes such as optic atrophy 1 (OPA1),Citation30–Citation32 apolipoprotein E (APOE),Citation33,Citation34 and optineurin (OPTN)Citation35–Citation37 are implicated in NTG. Recently, WD repeat-domain 36 (WDR36),Citation38 endothelin receptor type A (EDNRA)Citation39,Citation40 methylenetetrahydrofolate reductase (MTHFR),Citation41 and β-1-adrenergic receptor (ADRB1)Citation42 have also been reported as risk factors for NTG. However, associations between these genes and NTG were not strong or not often replicated in other populations,Citation43–Citation47 suggesting that there may be other unknown genetic factors having more powerful effects on the development of NTG than those identified so far.
In conclusion, we performed an association analysis of the GLC3A locus including the CYP1B1 gene using microsatellite markers in NTG patients, but the marker involved in NTG was not detected in this locus. In NTG patients, diagnosis is usually made after a visual field defect has occurred, due to few subjective symptoms and normal IOP. Therefore it is necessary to identify a disease susceptibility gene and elucidate pathogenic mechanisms of NTG for early diagnosis, prevention, and therapeutic development.
Acknowledgements
This study was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan; a grant from the Ministry of Health, Labour, and Welfare, Japan; and a grant from the Johnson and Johnson KK Vision Care Company.
References
- QuigleyHAOpen-angle glaucomaN Engl J Med199332815109711068455668
- HitchingsRAAndersonSAA comparative study of visual field defects seen in patients with low-tension glaucoma and chronic simple glaucomaBr J Ophthalmol198367128188216671097
- HitchingsRALow-tension glaucoma – its place in modern glaucoma practiceBr J Ophthalmol19927684944961390534
- WernerEBNormal-tension glaucomaRitchRShieldsMBKrupinTThe Glaucomas2nd edSt. Louis, MOMosby1996769797
- ShioseYKitazawaYTsukaharaSEpidemiology of glaucoma in Japan- a nationwide glaucoma surveyJpn J Ophthalmol19913521331551779484
- KleinBEKleinRSponselWEPrevalence of glaucoma. The Beaver Dam Eye StudyOphthalmology19929910149915041454314
- BonomiLMarchiniGMarraffaMPrevalence of glaucoma and intraocular pressure distribution in a defined population. The Egna-Neumarkt StudyOphthalmology199810522092159479277
- IwaseASuzukiYAraieMThe prevalence of primary open-angle glaucoma in Japanese: the Tajimi StudyOphthalmology200411191641164815350316
- SuzukiYIwaseAAraieMRisk factors for open-angle glaucoma in a Japanese population. The Tajimi StudyOphthalmology200611391613161716828504
- VasiliouVGonzalezFJRole of CYP1B1 in glaucomaAnnu Rev Pharmacol Toxicol20084833335817914928
- FanBJWangDYLamDSPangCPGene mapping for primary open angle glaucomaClin Biochem200639324925816332362
- WiggsJLGenetic etiologies of glaucomaArch Ophthalmol20071251303717210849
- StoilovIAkarsuANSarfaraziMIdentification of three different truncating mutations in cytochrome P4501B01 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21Hum Mol Genet1997646416479097971
- VincentALBillingsleyGBuysYDigenic inheritance of early-onset glaucoma: CYP1B1, a potential modifier geneAm J Hum Genet200270244846011774072
- MelkiRColombELefortNBrézinAPGarchonHJCYP1B1 mutations in French patients with early-onset primary open-angle glaucomaJ Med Genet200441964765115342693
- AcharyaMMookherjeeSBhattacharjeeAPrimary role of CYP1B1 in Indian juvenile-onset POAG patientsMol Vis20061239940416688110
- PanickerSGReddyABMandalAKIdentification of novel mutations causing familial primary congenital glaucoma in Indian pedigreesInvest Ophthalmol Vis Sci20024351358136611980847
- SoleyGCBosseKAFlikierDPrimary congenital glaucoma: a novel single-nucleotide deletion and varying phenotypic expression for the 1546–1555dup mutation in the GLC3A (CYP1B1) gene in 2 families of different ethnic originJ Glaucoma2003121273012567107
- AndersonDRPatellaVMAutomated static perimetry2nd edSt. Louis, MOMosby1999121190
- LancasterANelsonMPMeyerDThomsonGSingleRMPyPop: a software framework for population genomics: analyzing large-scale multi-locus genotype dataPac Symp Biocomput2003851452512603054
- HedrickPWGametic disequilibrium measures: proceed with cautionGenetics198711723313413666445
- SarfaraziMAkarsuANHossainAAssignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneityGenomics19953021711778586416
- BejjaniBALewisRATomeyKFMutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi ArabiaAm J Hum Genet19986223253339463332
- BelmoudenAMelkiRHamdaniMA novel frameshift founder mutation in the cytochrome P450 1B1 (CYP1B1) gene is associated with primary congenital glaucoma in MoroccoClin Genet200262433433912372064
- HollanderDASarfaraziMStoilovIWoodISFredrickDRAlvaradoJAGenotype and phenotype correlations in congenital glaucoma: CYP1B1 mutations, goniodysgenesis, and clinical characteristicsAm J Ophthalmol20061426993100417157584
- Corbalan-GarciaSYangSSDegenhardtKRBar-SagiDIdentification of the mitogen-activated protein kinase phosphorylation sites on human Sos1 that regulate interaction with Grb2Mol Cell Biol19961610567456828816480
- HartTCZhangYGorryMCA mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1Am J Hum Genet200270494395411868160
- RobertsAEArakiTSwansonKDGermline gain-of-function mutations in SOS1 cause Noonan syndromeNat Genet2007391707417143285
- TartagliaMPennacchioLAZhaoCGain-of-function SOS1 mutations cause a distinctive form of Noonan syndromeNat Genet2007391757917143282
- AungTOcakaLEbenezerNDA major marker for normal tension glaucoma: association with polymorphisms in the OPA1 geneHum Genet20021101525611810296
- PowellBLToomesCScottSPolymorphisms in OPA1 are associated with normal tension glaucomaMol Vis2003946046414551537
- MabuchiFTangSKashiwagiKYamagataZIijimaHTsukaharaSThe OPA1 gene polymorphism is associated with normal tension and high tension glaucomaAm J Ophthalmol2007143112513017188046
- VickersJCCraigJEStankovichJThe apolipoprotein ɛ4 gene is associated with elevated risk of normal tension glaucomaMol Vis2002838939312379839
- LamCYFanBJWangDYAssociation of apolipoprotein E. polymorphisms with normal tension glaucoma in a Chinese populationJ Glaucoma200615321822216778644
- RezaieTChildAHitchingsRAdult-onset primary open-angle glaucoma caused by mutations in optineurinScience200229555571077107911834836
- FuseNTakahashiKAkiyamaHMolecular genetic analysis of optineurin gene for primary open-angle and normal tension glaucoma in the Japanese populationJ Glaucoma200413429930315226658
- SripriyaSNirmaladeviJGeorgeROPTN gene: profile of patients with glaucoma from IndiaMol Vis20061281682016885925
- WeisschuhNWolfCWissingerBGramerEVariations in the WDR36 gene in German patients with normal tension glaucomaMol Vis20071372472917563723
- IshikawaKFunayamaTOhtakeYAssociation between glaucoma and gene polymorphism of endothelin type A receptorMol Vis20051143143715988412
- KimSHKimJYKimDMInvestigations on the association between normal tension glaucoma and single nucleotide polymorphisms of the endothelin-1 and endothelin receptor genesMol Vis2006121016102116971893
- MabuchiFTangSKashiwagiKYamagataZIijimaHTsukaharaSMethylenetetrahydrofolate reductase gene polymorphisms c.677C/T and c.1298A/C are not associated with open angle glaucomaMol Vis20061273573916862068
- InagakiYMashimaYFuseNPolymorphism of beta-adrenergic receptors and susceptibility to open-angle glaucomaMol Vis20061267368016785856
- WooSJKimDMKimJYParkSSKoHSYooTInvestigation of the association between OPA1 polymorphisms and normal-tension glaucoma in KoreaJ Glaucoma200413649249515534475
- LiuYSchmidtSQinXNo association between OPA1 polymorphisms and primary open-angle glaucoma in three different populationsMol Vis2007132137214118079692
- LakeSLiveraniEDesaiMNormal tension glaucoma is not associated with the common apolipoprotein E gene polymorphismsBr J Ophthalmol200488449149315031162
- MiyazawaAFuseNMengkegaleMAssociation between primary open-angle glaucoma and WDR36 DNA sequence variants in JapaneseMol Vis2007131912191917960130
- WooSJKimJYKimDMParkSSKoHSYooTInvestigation of the association between 677C > T and 1298A > C 5,10-methylenetetra-hydrofolate reductase gene polymorphisms and normal-tension glaucomaEye200776 [Epub ahead of print]