Abstract
Antithrombotic therapy, including anticoagulants as well as antiplatelet drugs, is an important component in the treatment of cardiovascular disease. Variability in response to such medications, of which pharmacogenetic response is a major source, can decrease or enhance the benefits expected. This review is a comprehensive assessment of the literature published to date on the effects of genetic polymorphisms on the actions of a variety of antithrombotic medications, including warfarin, clopidogrel, prasugrel, and aspirin. Literature evaluating surrogate markers in addition to the impact of pharmacogenetics on clinical outcomes has been reviewed. The results of the studies are conflicting as to what degree pharmacogenetics will affect medication management in cardiovascular disease. Additional research is necessary to discover, characterize, and prospectively evaluate genetic and non-genetic factors that impact antithrombotic treatment in order to maximize the effectiveness and limit the harmful effects of these valuable agents.
Introduction
Cardiovascular disease continues to be the leading cause of death worldwide.Citation1 Antithrombotic therapies with both anticoagulants as well as antiplatelet drugs have been landmark accomplishments in the treatment of this disease state, and have been established as a cornerstone of therapy for a wide variety of ischemic vascular disease states, including acute coronary syndrome (ACS),Citation2 stroke,Citation3 peripheral vascular disease,Citation4 atrial fibrillation,Citation5 deep vein thrombosis,Citation6 and pulmonary embolism.Citation6 Interpatient variability has long been recognized with warfarin and its derivatives due to the narrow therapeutic index resulting in both bleeding events and inadequate therapeutic response, necessitating frequent International Normalized Ratio (INR) monitoring. Antiplatelet therapy has thus far been utilized with a population-based approach to treatment (eg, one dose for every patient). While this approach has been shown to be effective in clinical trials,Citation7,Citation8 concern has been growing over the inter-patient variability in antiplatelet response, and there is increasing investigation of individualized dosing of high-profile antiplatelet drugs such as clopidogrel.Citation9
While variability itself is recognized, the therapeutic implications and the source of this variability are not always completely defined and are likely multifactorial. Anticoagulation with warfarin, for example, is highly dependent on regulation of dietary vitamin K intake but is also strongly influenced by genetic variations. In addition, the measurement of platelet function as a surrogate for efficacy or safety is incompletely understood,Citation10 but such changes have been linked to both temporal variability as well as drug-drug interactions, the latter of which have not been consistently found to have a clinically significant impact on the efficacy of antiplatelet therapy.Citation11,Citation12
Pharmacogenetic variability is likely another major source of inconsistency in therapeutic response. Since the completion of the Human Genome Project, greater attention is turning to identifying genetic sources of variability in drug response, the clinical impact of variability in patients receiving the drug, and finally, to altering therapy at an individual level to achieve a consistent therapeutic response for each patient. Termed “personalized medicine”, this potentially represents a sea change in pharmacotherapeutics, where a genetic profile will determine the appropriate drug and/or dose the patient should receive for maximum therapeutic benefit with minimal risk of toxicity. There are several examples where pharmacogenetic tools already impact therapeutics. Perhaps the most promising example is in the field of oncology, where drugs have been developed for cancers with certain genetic expressions,Citation13 and a genetic profile may soon dominate the choice of chemotherapy a patient receives.Citation14 While this represents success in the clinical application of pharmacogenetics, the application of pharmacogenetics to cardiovascular disease is more varied.
This review is a comprehensive assessment of the literature published to date regarding the effects of genetic polymorphisms on the actions of a variety of antithrombotic medications, including warfarin, clopidogrel, prasugrel, and aspirin. Literature evaluating surrogate markers in addition to the impact of pharmacogenetics on clinical outcomes has been reviewed.
Warfarin
Warfarin is an anticoagulant, inhibiting the action of Vitamin K-dependant cofactors of the coagulation cascade, specifically, Factors II, VII, IX, and X. Warfarin use is indicated in prevention of thromboembolic events in several clinical settings, such as hypercoagulation disorders and atrial fibrillation. Appropriate response to warfarin is crucial due to the risk of adverse effects that occur at both ends of the spectrum. A low response to warfarin places the patient at risk of an embolic event. An increased response to warfarin can lead to intracranial hemorrhage or a gastrointestinal bleed. Due to the widespread variation in response to warfarin therapy, all patients receiving therapy are monitored closely using the INR. Experience has suggested that it is especially crucial to monitor therapy closely at initiation of warfarin in a naïve patient. Therefore, it is proposed that genetic testing be done before starting any patient on warfarin. Significant variation in the effects of warfarin has been attributed to polymorphisms of a major metabolic enzyme, cytochrome P450 (CYP2C9), and its pharmacologic target, the vitamin K epoxide reductase complex, subunit 1 (VKORC1). The most common variant alleles for the CYP2C9 enzyme, *2 and *3, are present in about 12% and 8%, respectively, of the Caucasian population, and are rarely expressed in African-American or Asian populations.Citation15,Citation16
In 2007, the United States Food and Drug Administration (FDA) approved additional labeling for the package insert recommending, but not requiring, genetic testing of the CYP2C9 and VKORC1 genes to help determine an appropriate warfarin dosing regimen. Key pharmacogenetic studies with warfarin, outlined below, have not only associated these polymorphisms with clinical outcomes, but also propose novel dosing algorithms that utilize these genetic polymorphisms in concert with traditional characteristics that affect warfarin response.
The CYP2C9 enzyme is important in the metabolism and inactivation of the active S-enantiomer of warfarin. Impaired function of this enzyme, possibly due to single nucleotide polymorphisms (SNP) in the CYP2C9 gene results in decreased warfarin clearance. As a result, patients with decreased CYP2C9 function are potentially at an increased risk of bleeding during the onset of therapy when receiving warfarin.
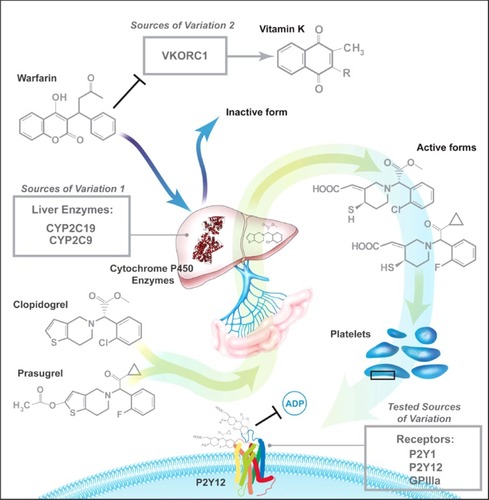
Vitamin K reductase (VKOR) is the target enzyme for warfarin. VKOR is responsible for regenerating vitamin K hydroquinone from the vitamin K cycle, the essential cofactor for the vitamin K-dependent coagulation factors. Warfarin use inhibits VKOR, resulting in decreased vitamin K hydroquinone production and resultant decreased coagulation cofactor production. Increased VKOR activity, such as that due to SNPs, leads to an increased response to warfarin inhibition of the vitamin K mediated cofactors ().Citation17
Figure 1 Previously-studied pharmacogenetic sources of variation in response to antithrombotic therapy.
Figure produced by Tom Dolan, University of Kentucky.
Higashi et al evaluated the association between variant CYP2C9 alleles and anticoagulation status as well as bleeding events.Citation18 They investigated whether carriers of CYP2C9 polymorphisms demonstrate an increased risk of extended time to therapeutic INR, out of range INR, and bleeding events. The study was conducted at a pharmacist-managed clinic and included any adult patient treated with warfarin. The primary outcome of this retrospective chart review was the overall anticoagulation status, as measured by time to therapeutic INR, incidence of supratherapeutic INR, and time taken to achieve stable dosing, with time to serious or life-threatening bleeding event as a secondary outcome. The investigators found that CYP2C9 genotype was significantly associated with the dose required to maintain therapeutic INR, with data suggesting a gene-dose effect. Regarding the primary endpoint, carriers of the CYP2C9 *2 and *3 polymorphisms had a lower rate of stable dosing (adjusted hazard ratio [HR] 0.65 (95% confidence interval [CI], 0.45–0.94)) and higher risk of supratherapeutic INR (adjusted HR 1.4 (1.03–1.90)). Additionally, patients with the polymorphisms were more likely to have bleeding events at initiation (HR 3.94 (1.29–12.06)) and over the entire study (HR 2.39 (1.18–4.86)).
The Warfarin Genetics (WARG) study has been the largest prospective, observational analysis of the clinical impact of pharmacogenetics on warfarin outcomes to date, evaluating 1,523 patients treated with warfarin in Sweden.Citation19 The goals of this national study were to evaluate the incidence of adverse bleeding events associated with warfarin treatment as well as to test the association between 29 different genotypes and warfarin dosing, specifically at the initiation of therapy. The VKORC1 polymorphism was the strongest predictor of dose, second was CYP2C9. Two VKORC1 SNPs, rs2359612 and rs9923231, were associated with 59.6% of the variance in maintenance dose, each SNP explaining 29.8%. VKORC1 SNPs were also associated with an increased risk of having an INR > 4 (HR 4.56 (95% CI, 2.85–7.30), P < 0.001). CYP2C9 SNPs also were significantly associated with dosing variance. The presence of the CYP2C9*2 allele explained 4.1% of variation, while the CYP2C9*3 allele explained 6.3%. Collectively, the presence of either CYP2C9 allele explained 11.8% of the variance in maintenance dose of warfarin. CYP2C9*3 homozygotes, which represented only 0.5% of the study population, had a significantly and excessively increased risk of an INR > 4 (HR 21.84 (9.46–50.42) P < 0.001). Additionally, 27% of patients either homozygous for any VKORC1 SNP or with CYP 2C9*3 had an INR > 4 during the first 5 weeks of therapy. The CYP2C9*2 SNPs did not have a significant association with having an INR > 4. Although there were no statistically significant differences found regarding bleeding events, CYP2C9 polymorphisms were associated with a numerical increase in bleeding within 5 weeks of warfarin initiation (P = 0.066). There were no differences in bleeding events between any of the groups over the entire 4-year observation period. Using the predictors identified as significant in their study, the authors developed a multiple regression model to predict a patient’s dose depending on specific genetic makeup.
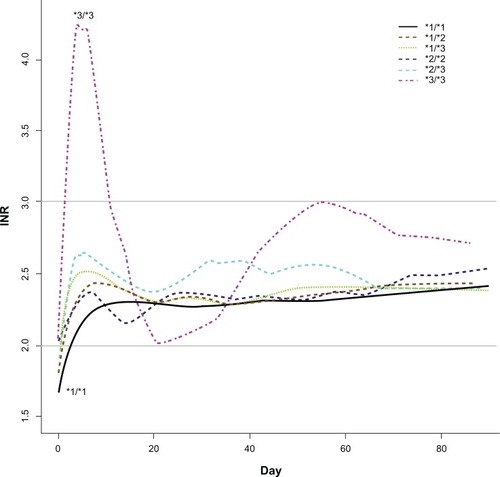
A prospective study evaluating the impact of CYP2C9 polymorphisms during the first three weeks of warfarin therapy in patients previously naïve to warfarin was completed by Lindh et al. The study population was a subpopulation (n = 219) of the WARG study that enrolled patients as they finished treatment, indicating that their warfarin therapy was a time-restricted therapy.Citation20 The primary endpoint was supratherapeutic INR during the first three weeks of treatment. Secondarily, the study also evaluated the time to reach a stable INR. Patients with either the CYP2C9*2 (CYP2C9*1/*2 and CYP2C9 *2/*2 carriers) or the CYP2C9*3 polymorphism (carriers of at least one CYP2C9*3 allele) had a significantly increased risk of having a supratherapeutic INR (INR > 3) during Week One (*2: RR 2.8 (95% CI, 1.2–6.7) and *3: RR 6.1 (95% CI, 2.7–13.6)) and Week Two (*2: RR 2.1 (95% CI, 1.2–3.7) and *3: RR 3.5 (95% CI, 2.1–5.8)) compared to CYP2C9*1/*1 homozygotes (). The risk was the similar during Week Three for both groups (*2: RR 1.0 (95% CI, 0.5–1.8) and *3: RR 1.1 (95% CI, 0.6–2.0)). These results do suggest an early association of warfarin dosing with CYP2C9 genotype. The authors suggested that this study may indicate that genotyping may be clinically useful when initiating warfarin treatment. Likewise, a systematic study of 188 patients receiving warfarin for six months or more with stable dosing regimens found CYP2C9 polymorphisms to be associated with decreased warfarin clearance.Citation21 Thus, such patients required lower doses to reach a therapeutic INR range.
Figure 2 Days taken to reach stable anticoagulation.
Reproduced with permission from Wadelius M, Chen L, Lindh J, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792.Citation19
© 2009 the American Society of Hematology.
Implementation of pharmacogenetic testing in warfarin treatment
While variant alleles were clearly shown to influence dosing with warfarin therapy, it was unclear how this data would aid clinicians in prescribing. To this end, several groups have attempted to formulate predictive dosing guidelines such that pharmacogenetic information could be of value at the onset of therapy. One prospective, randomized study compared a pharmacogenetic-guided dosing algorithm to a standard empiric dosing guide for 206 patients initiated on warfarin therapy ().Citation22 The main objective was to validate the predictive utility of this pharmacogenetic algorithm, the primary endpoint of the study being the average of the number of out-of-range INR standardized by the number of INRs obtained. The study failed to show a significant difference for the primary endpoint (out-of-range INRs: 30.7% (± 22.9) in the group dosed with the pharmacogenetic algorithm vs 33.1% (± 22.9) of the standard dosing group). However, the authors did show the pharmacogenetic algorithm slightly, but significantly, decreased the number of dose adjustments from 3.6 to 3.0 dose adjustments per patient (mean decrease of 0.62 adjustments (95% CI, 0.04–1.19), P = 0.035).
Table 1 Warfarin pharmacogenetic dosing algorithm and two examples of weekly doses in hypothetical patients
The International Warfarin Pharmacogenetics Consortium also has developed a dosing algorithm and performed a retrospective analysis of over 5000 patients, which evaluated whether these dosing recommendations were more predictive over the use of clinical variables alone.Citation23 The algorithm included the VKORC1 and CYP2C9 genotypes in addition to other variables to calculate a starting dose prediction. The investigators found the algorithm based on pharmacogenetic dosing more accurately estimated the warfarin dose when compared to traditional approaches, which included dosing based on clinical markers and a fixed-dosed regimen. The greatest predictive value was seen in patients receiving less than 21 mg per week to maintain a therapeutic INR range, with 35% of patients falling within 20% of the predicted dose, compared to 24% of patients using the clinical algorithm and 0% of the fixed-dose (P < 0.001). Of those prescribed less than 21 mg per week, the pharmacogenetically dosed group also experienced significantly fewer dose overestimations (59.7%) compared to the other two groups (clinical: 74.8%, fixed-dose: 100%; P < 0.001 for both comparisons).
The FDA’s action to recommend, not require, genetic testing before initiating warfarin therapy highlights the barriers to implementing routine pharmacogenetic testing prior to warfarin use. The studies described above present evidence supporting the value of genetically screening each patient. However, there is no prospective study that proves a significant effect of genetic testing on clinical outcomes. The clinical application of this information has yet to become universal, in part due to logistical issues with obtaining timely genotyping, but also due to an overall question of the cost-effectiveness of screening all patients to identify those with variant alleles.
Aspirin
Aspirin is an irreversible cyclooxygenase-1 (COX-1) inhibitor that decreases production of thromboxane A2, a potent platelet activator. The use of aspirin has been shown to decrease vascular events and mortality in a wide variety of cardiovascular settings.Citation24,Citation25 Variability in the ex vivo antiplatelet response to aspirin has been linked to many sources, including comorbidities and sex, among other parameters. Some studies have described a relationship between polymorphisms in genes encoding the COX-1 enzyme, such as the SNP prostaglandin-endoperoxide synthase 1 (PTGS1-1006A), and responsiveness, while others found an association with genetic variation in glycoprotein receptors, most consistently, GP IIIa’s PlA1/A2 polymorphism.Citation26 On the other hand, there is also evidence that the actual incidence of true clinical aspirin resistance is very low, and that aspirin failure has little to do with ex vivo-determined responsiveness.Citation27
Macchi et al investigated whether polymorphisms in genes encoding for platelet membrane glycoproteins affected the response to aspirin.Citation28 Ninety-eight patients with stable angina, not taking any antiplatelet or anti-inflammatory medications, were enrolled and given aspirin 160 mg/day. Aspirin response was assessed with a platelet function analyzer, PFA-100, which measures platelet function by simulating platelet-based hemostasis in vitro. The study investigated the effect of polymorphisms in platelet glycoproteins GP Ia/IIa (C807T), GP Ib alpha (C-5T Kozak), and GP IIIa (PlA1/A2) on platelet function following aspirin administration. Aspirin resistance was defined as a collagen epinephrine closure time (CEPI-CT) <186s. No significant relationship was found between the polymorphisms encoding GP Ia/IIa (C807T) or GP Ib alpha (C-5T Kozak). However, a significant relationship was found among patients with the platelet gene polymorphism PlA1/A1. Of the 29 patients with aspirin resistance, 25 (86.2%) had the A1/A1 SNP, and 4 (13.8%) had at least one PlA2 allele (P = 0.01). Carriers of the PlA1/A1 polymorphism had an increased risk of poor platelet response to aspirin therapy (odds ratio [OR] 4.4 [95% CI, 1.3–14.7]). The authors do admit the need for a larger study to validate these results, but also point out the potential clinical implications of tailoring therapy to a PIA genotype.
The hypothesis that variation in the COX-1 gene results in the failure of aspirin to adequately suppress platelet COX-1 was investigated by Maree et al in their evaluation of 144 stable patients with cardiovascular disease.Citation29 Aspirin response was evaluated with platelet aggregation studies utilizing light transmittance aggregometry (LTA). Aspirin resistance was defined as persistant arachidonic acid (AA)-induced platelet aggregation ≥20%. Polymorphisms in COX-1 were found to significantly affect response to AA-induced platelet aggregation and serum thromboxane A2 levels in patients receiving aspirin therapy (P = 0.004 for comparison among haplotypes). To date, there has been no assessment of the clinical impact of COX-1 polymorphisms in patients receiving aspirin therapy.
The relationship between genetic polymorphisms and decreased response to aspirin was again examined in 101 patients undergoing elective percutaneous coronary intervention (PCI).Citation30 The aim was to evaluate the association, if any, between the variable response to aspirin and platelet receptor polymorphisms (GP Iba, GP Ia, GP IIIa, GP VI) or COX-1 gene polymorphisms (A842G, C22T, C644A, C714A). This study used the PFA-100 and LTA to assess the effect of aspirin on platelet activity. Significant relationships were shown, but not consistently, between the two testing methods used. A significant association was identified between aspirin resistance and carriers of the SNP A842G in the COX-1 enzyme (OR 10.0 (95% CI, 1.2–87.0) P < 0.03) when assessed using LTA to measure the response to aspirin (defined as a slope greater than 12% / minute in AA-aggregation). However, there was not a significant association seen when using the PFA-100 method to assess platelet response to aspirin (decreased response defined as closure time <170s). On the other hand, carriers of the SNP C13254T in the GP VI platelet receptor had a significantly increased risk of decreased response to aspirin (OR 1.4 [95% CI, 1.4–22.2] P < 0.03) when response to aspirin was assessed using the PFA-100 method, but no significant association was shown when measuring platelet aggregation to assess the response. The authors did cite a small sample size as a limitation to their study, and called for larger studies in efforts to identify the specific patient populations that may be at risk of aspirin resistance due to genetic polymorphisms. Lev et al also found no significant association between the presence of the PlA1/A1 polymorphism and a decreased response to aspirin and clopidogrel in 120 patients undergoing elective PCI.Citation31 Platelet response was again measured using LTA, with a decreased response to aspirin defined as AA-induced aggregation ≥20%.
The potential causes, incidence and clinical impact of aspirin resistance are unclear. Measured variability in response to aspirin is multifactorial, with genetics playing what appears to be a small, undefined role. Given the mixed results shown in the above studies, as well as others not described, there is currently no defined role for pharmacogenetic testing to dose aspirin.
Thienopyridines
The thienopyridines ticlopidine, clopidogrel, and prasugrel irreversibly bind to the P2Y12 receptor, resulting in the inhibition of the platelet’s ability to activate in response to adenosine diphosphate (ADP). All thienopyridines are prodrugs that require absorption and activation by the cytochrome P450 system. Clopidogrel is metabolized to its active metabolite through a two-step process mediated by various cytochromes. Prasugrel is activated in a process mediated by esterases and by a single CYP450 step.
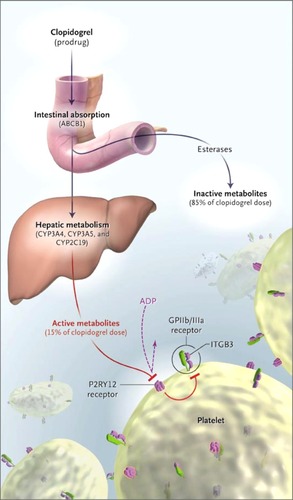
The exact sources of interpatient variability in response to clopidogrel are unknown but likely multifactorial. Pharmacogenetic sources of variability may involve pharmacokinetic, metabolic, or pharmacodynamic pathways (). SNPs in genes encoding the P-glycoprotein involved in intestinal absorption could cause a decreased response. SNPs in genes that encode the platelet receptor P2Y12, which serves as the target for the thienopyridines, could render the platelet less capable of responding to these medications. It is hypothesized that part of the cause of interpatient variability in response to the thienopyridines may lie in the CYP450 enzymatic system that metabolizes the drug to its active metabolite. Single nucleotide polymorphisms in genes encoding for the CYP3A4, CYP3A5, and CYP2C19 enzymes have been identified and evaluated in numerous studies with a variety of study designs as potential sources of variability The frequencies of the existence of these various alleles are not well defined. Reduced function alleles have been identified in 0%–40% of the population, depending on which population and which enzyme is evaluated. The results are conflicting, as some studies have found definitive associations with clinical outcomes such as cardiovascular events, and others have not. Regardless, in 2009, the FDA added information regarding the potential for pharmacogenetic variables to influence response to clopidogrel to the product labeling, although no guidance is given for how clinicians could utilize this information.
Figure 3 Sources of interpatient variability for clopidogrel.
Reproduced from Simon T, Verstuyft C, Mary-Krause M, et al Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375.Citation36
Copyright © 2009 Massachusetts Medical Society. All rights reserved.
Several pharmacodynamic estimates of the impact of genetic variation have been examined with clopidogrel. Brandt et al hypothesized that the loss-of-function polymorphisms of CYP enzymes (CYP2C19, CYP2C9, and CYP3A5) could contribute to decreased formation of the active metabolite of clopidogrel, thus affecting how well platelet activity is inhibited.Citation32 Investigators assessed the relationship between various SNPs and pharmacodynamic and pharmacokinetic responses to clopidogrel and prasugrel. They retrospectively examined the effect of loading doses of the respective thienopyridines on platelet function as measured by inhibition of platelet aggregation (IPA), where a poor responder was defined as <20% IPA in response to ADP. A significant association was demonstrated between the CYP2C19*2 allele and poor response to clopidogrel (of 18 subjects with the allele, 13 (72.2%) were poor responders (P = 0.030) compared to 41.1% of those subjects without the allele). Presence of a CYP2C9 SNP (*2/*2 or any *3) was also significantly associated with a poor response to clopidogrel (12 of 16 patients (75%) compared to 41.4% of patients without the SNP who were poor responders, P = 0.024). Additionally, the presence of these or any CYP allele polymorphism had no effect on the IPA response to prasugrel. Overall, the presence of either CYP2C19*2 or CYP2C9 (*2/*2 or *3) SNP was strongly associated with a poor response to clopidogrel (P < 0.001). The authors concluded that subjects with CYP2C19 and CYP2C9 SNPs were associated with a decreased exposure to the active metabolite of clopidogrel, not prasugrel. The group did not find an association between CYP3A5 polymorphisms and response to clopidogrel. The authors concluded that the polymorphisms of CYP2C9 and CYP2C19 provide an explanation for many cases of poor pharmacodynamic response to clopidogrel. Because this was a pharmacodynamic assessment only, the group did not evaluate clinical outcomes. Clinical evaluations of specific isoenzymes, in addition to the impact of genetic variability on clinical outcomes are detailed below.
CYP3A4
Lau et al investigated interindividual variability of 32 patients undergoing elective catheterization with stent placement receiving a clopidogrel loading dose of 300 mg followed by a maintenance dose of 75 mg daily. Platelet aggregation was measured at baseline and after 5 days of therapy. CYP3A4 activity was measured by the erythromycin breath test. An inverse correlation between platelet aggregation and CYP3A4 activity was seen in this study (r = −0.6, P = 0.003). CYP3A4 activity was shown to be responsible for one third of the varied response to clopidogrel (r = 0.36).Citation33 In addition to examining the effects of CYP3A4 on surrogate markers, an observational case-control study involving 186 patients receiving clopidogrel for ACS found only a trend toward significance of an association between the presence of a CYP3A4 polymorphism and the occurrence of a repeat cardiovascular event while receiving clopidogrel (16.8% vs 9.6%, P = 0.052).Citation34
CYP3A5
A multivariate analysis of 348 patients treated with clopidogrel after planned stent placement investigated the effect of a CYP3A5 polymorphism on the risk of atherothrombotic events at one and six months post-procedure.Citation35 The study identified CYP3A5 polymorphisms as an independent predictor of atherothrombotic events at six months follow-up. Patients with the CYP3A5 non-expressor allele (n = 193) had a significantly increased risk of an event over the group with the expresser allele (n = 155; adjusted OR 4.89 [1.89–18.7]). The study also found an increased risk at one month follow-up, but this difference was not significant (P = 0.16). A sub-group analysis of this study also found the number of concomitant CYP3A metabolizing medications to be an independent predictor of an event only in patients with the polymorphism. However, no association was found in a case-control study evaluating patients receiving clopidogrel for ACS.Citation34 The French Registry of Acute ST-elevation and Non-ST-elevation Myocardial Infarction (FAST-MI) trial investigated the effect of multiple polymorphisms on death or ischemic events during a one-year follow-up.Citation36 No association was found between the risk of clinical outcomes (death, nonfatal myocardial infarction [MI], stroke) in patients with a CYP3A5 polymorphism (P = 0.69).
CYP2C19
A pre-specified sub-group analysis of the EXCELSIOR study examined whether the loss-of-function CYP2C19 polymorphism is associated with increased platelet reactivity despite clopidogrel treatment in patients undergoing elective catheterization with stent placement.Citation37 The primary outcome was the proportion of patients with residual platelet aggregation (RPA) > 14% following a clopidogrel loading dose of 600 mg. In order to evaluate clinical outcomes, the association between RPA > 14% and risk of death or MI at one year were evaluated. The CYP2C19*2 allele was significantly associated with RPA > 14% before hospital discharge. Patients with RPA > 14% had a significant increase in the risk of death or MI (HR 3.0 [1.4–6.8], P = 0.004) at 1 year post-procedure. Considering that the CYP2C19 polymorphism accounted for a large proportion of patients with RPA > 14%, the authors estimated that the presence of this allele alone would be associated with a relative risk of death or MI at year 1 of 1.26, although the study was not adequately powered to determine the effect of variant alleles on clinical outcomes.
Shuldiner et al also investigated the association between CYP2C19 polymorphisms and clinical outcomes through a two-part study.Citation38 First, genes associated with poor response to clopidogrel were identified by assessing the Old Order Amish in the Amish Pharmacogenomics of Antiplatelet Intervention Study (Amish PAPI). The study replicated the findings in a time-to-event analysis of patients undergoing cardiac catheterization. Cardiovascular events included MI, ischemic stroke, stent thrombosis, unplanned revascularization, hospitalization for coronary ischemia, and death secondary to any cardiovascular cause. Patients with the CYP2C19 genotype had no baseline platelet aggregation difference (P = 0.58). After 1 year of follow-up, there was significant difference in cardiovascular event rates (20.9% versus 10.0%, HR 2.42 (1.18–4.99), P = 0.02). Patients still taking clopidogrel with the CYP2C19 variant at one year had a significantly increased risk (HR 3.40 (1.18–4.99), P = 0.02) versus those who did not have the variant. Additionally, the populations no longer taking clopidogrel at 1 year were compared. No increase was seen between carriers and non-carriers in the population that had discontinued clopidogrel therapy (HR 1.39 (0.39–4.88) P = 0.6). The authors determined that the CYP2C19 genotype variability accounts for 12% of the variation in clopidogrel response. Alternately, age, body mass index (BMI), and lipid levels accounted for 10% of the varied response. These data help confirm that the CYP2C19*2 variant is a determinant of ADP-stimulated platelet aggregation and that patients with this genotype may have reduced protection from ischemic events following PCI.
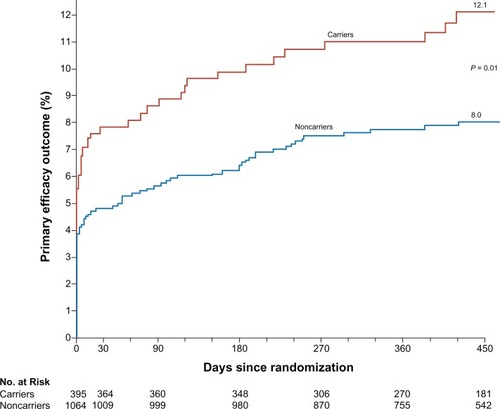
A sub-analysis of the TRITON-TIMI 38 study also examined the association between functional polymorphisms in CYP genes with plasma exposure to clopidogrel and platelet inhibition in healthy subjects.Citation39 Using a separate cohort of patients from the TRITON-TIMI 38 study treated with clopidogrel, it was tested whether the reduced-function CYP alleles were associated with a higher rate of adverse cardiovascular outcomes. The pharmacodynamic response was assessed with platelet aggregation in response to 20 μM of ADP and expressed as an absolute reduction in maximal platelet aggregation from baseline (tri-MPA). Outcomes were expressed as Kaplan-Meier estimates measured at 15 months and expressed as comparisons between carriers and noncarriers of at least one reduced-function allele (5 alleles were tested). Overall, a significant difference in outcomes between carriers and non-carriers of the CYP2C19 reduced-function allele was detected among patients randomized to clopidogrel (27.1% of the TRITON-TIMI study population). These patients were determined to be at a statistically significantly higher risk of death from cardiovascular (CV) causes, MI, and stroke (the primary efficacy outcome): 12.1% vs 8.0%, HR for carriers 1.53 (1.07–2.19), P = 0.01 (). The risk was also observed to be increased for each component of the primary outcome: death from CV causes (2.0% vs 0.4%, HR 4.79 [1.4–16.37]), non-fatal MI (10.1% vs 7.5%, HR 1.38 [0.94–2.02]), and nonfatal stroke (0.88% vs 0.24%, HR 3.93 [0.66–23.51]). Stent thrombosis was also increased for carriers of at least one CYP2C19 variant allele (2.6% vs 0.8%, HR 3.09 [1.19–8.0]). These data imply that CYP variation can lead to a reduced exposure to the active metabolite, less platelet inhibition, and less protection from ischemic events in patients receiving clopidogrel. Interestingly, no associations were found with any of the polymorphisms encoding for CYP enzymes among patients randomized to prasugrel therapy in TRITON.Citation40 It is unclear what contribution, if any, the reduced impact of genetic variability on prasugrel had on the overall efficacy of prasugrel in the TRITON trial.
Figure 4 TRITON TIMI primary efficacy outcome.
Copyright © 2009 Massachusetts Medical Society. All rights reserved.
In addition to the post hoc analysis of TRITON, an analysis of the FAST-MI registry (French registry of Acute ST-elevation or non-ST-elevation Myocardial Infarction) extensively studied the interaction of CYP2C19 polymorphisms and clopidogrel activity.Citation36 The investigators examined the effect of multiple SNPs of the enzyme. A total of 2,208 patients were analyzed for this study. The presence of any loss-of-function SNP of CYP2C19 (*2, *3, *4, or *5) was not associated with significantly increased risk of the composite endpoint (death, nonfatal MI, and stroke during a one-year follow-up period). However, carriers of two variant alleles did have an increased event risk (21.5% vs 13.3%, HR 1.98 (1.10–3.58)). This risk was increased even further in PCI as evidenced by a sub-group analysis. Patients had an increased event risk if they had PCI and were carriers of two of any of the CYP2C19 SNP alleles (HR 3.58 [1.71–7.51]). This observational study did not eliminate the possibility that associated polymorphisms may be direct contributors to atherothrombotic events, independent of clopidogrel use.
P2Y12 Receptor
In addition to metabolizing enzymes, potential sources of interpatient variability have been investigated. Specifically, polymorphisms in genes encoding for the platelet receptor that serves as the target for the thienopyridines have been evaluated. The effects of polymorphisms in the genes encoding platelet receptors related to aspirin and clopidogrel response were evaluated in an observational study by Lev et al.Citation31 Patients receiving an elective PCI were eligible for study enrollment. The authors investigated the effects of SNP on the genes encoding glycoprotein IIIa, P2Y12 and P2Y1 receptors. No significant associations were found between the various polymorphisms and response to clopidogrel as assessed by platelet aggregation studies. The FAST-MI analysis also investigated the effects of SNPs in receptors but found no association between the P2Y12 polymorphism and the risk of death, nonfatal MI, or stroke in patients treated with clopidogrel.Citation36
Despite the addition by the FDA recommending genetic testing for clopidogrel, there are studies that refute the clinical impact of polymorphisms on the effectiveness of clopidogrel. Most recently, the CHARISMA genetic sub-study, a placebo-controlled trial involving stable patients given aspirin and clopidogrel or aspirin and placebo, found that patients heterozygous for the CYP2C19*2 allele did not have increased CV events or bleeding events when taking clopidogrel or placebo.Citation41 Only homozygous carriers for the reduced-function CYP2C19 allele had significantly fewer bleeding events. The sub-study also showed an increased risk of ischemic events for patients homozygous for CYP2C19*2 (reduced-function) allele in both treatment and placebo arms, although this finding was not statistically significant.
The conflicting results with many different studies showcase the unanswered questions that linger regarding the clinical significance of pharmacogenetics in cardiovascular medicine. There are significant polymorphisms that result in interpatient variability in response to medications. However, being able to predict the specific response that an individual patient may or may not have based on his or her genetic code has yet to be defined with antithrombotic medications, especially clopidogrel. Despite this, the FDA has provided summarizing information to the product labeling regarding the potential for variant alleles, especially CYP2C19 polymorphisms, to influence clopidogrel effectiveness. In addition, some health systems have begun large-scale screening of clopidogrel pharmacogenetic information despite a lack of clear guidance in the literature for what therapeutic intervention to make in treating these patients.Citation42
Conclusion
The diagnostic and pharmacotherapeutic applications of genome-wide scanning are, without a doubt, a very promising avenue to achieving personalized medicine. In the cardiovascular field, the best-characterized example of the clinical implications of utilizing pharmacogenetic therapy is the impact of VKORC1 and CYP2C9 variants on warfarin response. These two gene products alone explain about 50% of the variability in response to warfarin, a potentially dangerous medication that is cumbersome to dose. Numerous prospective and retrospective studies establish the value of genetic variability to predict the appropriate warfarin dose for improving and easing the transition to a therapeutic INR level. In fact, the labeling for warfarin now includes a recommendation for genetic testing. Nonetheless, prospective studies confirming the value of genetic testing on clinical outcomes are still limited and inconclusive, and the pharmacoeconomic significance is certainly debated. In addition, there are current logistical concerns regarding genetic testing for warfarin therapy, including speed of testing, ethical and confidentiality issues regarding genetic information, and validation of appropriate genetically-guided dosing algorithms. The extensive previous and planned research in this area has verifies that 1) genetic testing will serve an invaluable role in tailored pharmacotherapy; 2) large-scale, prospective studies are required to confirm the usefulness of genetic testing, even when the impact of the variation is great; and 3) the logistics for moving genetic testing into routine clinical testing are not yet established.
Similar to the evidence for the anticoagulant warfarin, pharmacogenetic testing for antiplatelet agents also demonstrates strong potential for improving therapy, particularly for carriers of CYP2C19 variants taking clopidogrel. The path for clopidogrel, however, is even more arduous than that of warfarin for obtaining widespread application. There is a much smaller percentage of variability explained by the current paradigm, and no major prospective studies confirm the worthiness of genetic information for improving clinical outcomes. Likewise, any potential economic savings of this strategy have not been demonstrated. Most importantly, it is unclear how the pharmacogenetic information will be utilized in the setting of P2Y12 receptor antagonism. Although there are a variety of options for increasing the extent of inhibition (higher maintenance dose of clopidogrel, the recently approved prasugrel, and the potentially soon to be available ticagrelor), the appropriate use of these strategies, even in the absence of pharmacogenetic information, is difficult to assess in individual patients. Large-scale clinical trials for these agents establish a clinical benefit at the population level for reducing clinical events at the expense of increased bleeding.Citation43,Citation44 It certainly seems noble to alter therapy based on a single gene (CYP2C19) known to associate with clinical outcomes. Unfortunately, ignoring the rest of the genome, epigenetic mediators, proteome, etc, in addition to other potential predictors of adverse bleeding events, may prove risky when applying population-based strategies and incomplete genetic information to personalized treatment. Although the adverse cardiac events associated with an inappropriate amount of inhibition with clopidogrel are certainly devastating, the increased bleeding risk (sometimes even fatal bleeding) with alternative agents and doses should be evaluated with equal consideration. These murky waters are further complicated by the lack of a reliable and validated assay to measure platelet function. Currently, there are five tests available for genotyping for the CYP2C19 enzyme (ARUP Laboratories, Autogenomics [for research use only], Labcorp, Specialty Laboratories, and Quest Diagnostics). These tests generally involve a turnaround time of about five days and are only performed a few days per week. The currently available ex vivo assays marginally correlate with clinical outcomes, but do not yet possess the sensitivity, consistency, and standardization to tailor antiplatelet medications to individual patients with confidence. Without a monitoring tool, the effectiveness of genetic testing for antiplatelet agents is severely hindered.
Despite these current limitations, the promise of pharmacogenomics for antithrombotic medications is substantiated by the recent correlation of CYP2C19 variants with clopidogrel response and the storied, though not complete, path of warfarin. As time progresses, technology will continue to decrease the cost of whole-genome scans and other genetic tools, and allow for more efficient and secure transfer of information. These advances, along with a logistical platform for clinical utilization, will allow universal application of genetic information across all therapeutic areas. Then, personalized medicine, where genetic and other variations are measured and prospectively interpreted to improve outcomes and decrease costs, will be realized. In the meantime, additional research is necessary to discover, characterize, and prospectively evaluate genetic and non-genetic factors that impact antithrombotic treatment, in order to maximize the effectiveness and limit the harmful effects of these valuable agents.
Acknowledgements
The authors would like to acknowledge Tom Dolan, Medical Illustrator and Multimedia Developer, Teaching and Academic Support Center, University of Kentucky for his work on this document.
Disclosures
Dr Steinhubl is currently a full-time employee of The Medicines Company. The other authors report no conflicts of interest in this work.
References
- Lloyd-JonesDAdamsRCarnethonMHeart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics SubcommitteeCirculation20091193e21e18119075105
- AndersonJLAdamsCDAntmanEMACC/AHA 2007 guidelines for the management of patients with unstable angina/non ST-elevation myocardial infarction: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Writing Committee to Revise the 2002 Guidelines for the Management of Patients With Unstable Angina/Non ST-Elevation Myocardial Infarction): developed in collaboration with the American College of Emergency Physicians, the Society for Cardiovascular Angiography and Interventions, and the Society of Thoracic Surgeons: endorsed by the American Association of Cardiovascular and Pulmonary Rehabilitation and the Society for Academic Emergency MedicineCirculation20071167e148e30417679616
- SaccoRLAdamsRAlbersGGuidelines for prevention of stroke in patients with ischemic stroke or transient ischemic attack: a statement for healthcare professionals from the American Heart Association/American Stroke Association Council on Stroke: co-sponsored by the Council on Cardiovascular Radiology and Intervention: the American Academy of Neurology affirms the value of this guidelineCirculation200611310e409e4916534023
- HirschATHaskalZJHertzerNRACC/AHA 2005 Practice Guidelines for the management of patients with peripheral arterial disease (lower extremity, renal, mesenteric, and abdominal aortic): a collaborative report from the American Association for Vascular Surgery/Society for Vascular Surgery, Society for Cardiovascular Angiography and Interventions, Society for Vascular Medicine and Biology, Society of Interventional Radiology, and the ACC/AHA Task Force on Practice Guidelines (Writing Committee to Develop Guidelines for the Management of Patients With Peripheral Arterial Disease): endorsed by the American Association of Cardiovascular and Pulmonary Rehabilitation; National Heart, Lung, and Blood Institute; Society for Vascular Nursing; TransAtlantic Inter-Society Consensus; and Vascular Disease FoundationCirculation200611311e463e65416549646
- FusterVRydenLECannomDSACC/AHA/ESC 2006 Guidelines for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm SocietyCirculation20061147e257e35416908781
- HirshJGuyattGAlbersGWHarringtonRSchunemannHJExecutive summary: American College of Chest Physicians evidence-based clinical practice guidelines (8th Edition)Chest20081336 Suppl71S109S18574259
- SteinhublSRBergerPBMannJT3rdEarly and sustained dual oral antiplatelet therapy following percutaneous coronary intervention: a randomized controlled trialJAMA2002288192411242012435254
- YusufSZhaoFMehtaSRChrolaviciusSTognoniGFoxKKEffects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevationN Engl J Med2001345749450211519503
- BonelloLCamoin-JauLArquesSAdjusted clopidogrel loading doses according to vasodilator-stimulated phosphoprotein phosphorylation index decrease rate of major adverse cardiovascular events in patients with clopidogrel resistance: a multicenter randomized prospective studyJ Am Coll Cardiol200851141404141118387444
- GorogDASweenyJMFusterVAntiplatelet drug ‘resistance’. Part 2: laboratory resistance to antiplatelet drugs-fact or artifact?Nat Rev Cardiol20096536537319365406
- O’DonoghueMLBraunwaldEAntmanEMPharmacodynamic effect and clinical efficacy of clopidogrel and prasugrel with or without a proton-pump inhibitor: an analysis of two randomised trialsLancet2009374969498999719726078
- SawJSteinhublSRBergerPBLack of adverse clopidogrel-atorvastatin clinical interaction from secondary analysis of a randomized, placebo-controlled clopidogrel trialCirculation2003108892192412925453
- KroeseMZimmernRLPinderSEHER2 status in breast cancer--an example of pharmacogenetic testingJ R Soc Med2007100732632917606754
- HuangEIshidaSPittmanJGene expression phenotypic models that predict the activity of oncogenic pathwaysNat Genet200334222623012754511
- MoyerTPO’KaneDJBaudhuinLMWarfarin sensitivity genotyping: a review of the literature and summary of patient experienceMayo Clin Proc200984121079109419955245
- StehleSKirchheinerJLazarAFuhrUPharmacogenetics of oral anticoagulants: a basis for dose individualizationClin Pharmacokinet200847956559418698879
- RettieAEWienkersLCGonzalezFJTragerWFKorzekwaKRImpaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9Pharmacogenetics19944139428004131
- HigashiMKVeenstraDLKondoLMAssociation between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapyJAMA2002287131690169811926893
- WadeliusMChenLYLindhJDThe largest prospective warfarin-treated cohort supports genetic forecastingBlood2009113478479218574025
- LindhJDLundgrenSHolmLAlfredssonLRaneAseveral-fold increase in risk of overanticoagulation by CYP2C9 mutationsClin Pharmacol Ther200578554055016321620
- HermanDLocatelliIGrabnarIInfluence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dosePharmacogenomics J20055319320215824753
- AndersonJLHorneBDStevensSMRandomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulationCirculation2007116222563257017989110
- KleinTEAltmanRBErikssonNEstimation of the warfarin dose with clinical and pharmacogenetic dataN Engl J Med2009360875376419228618
- Randomised trial of intravenous streptokinase, oral aspirin, both, or neither among 17,187 cases of suspected acute myocardial infarction: ISIS-2. ISIS-2 (Second International Study of Infarct Survival) Collaborative GroupLancet1988286073493602899772
- Antiplatelet Trialists’ CollaborationCollaborative meta-analysis of randomised trials of antiplatelet therapy for prevention of death, myocardial infarction, and stroke in high risk patientsBMJ20023247329718611786451
- FeherGFeherAPuschGLupkovicsGSzaparyLPappEThe genetics of antiplatelet drug resistanceClin Genet200975111819067731
- CuissetTFrereCQuiliciJAspirin noncompliance is the major cause of “aspirin resistance” in patients undergoing coronary stentingAm Heart J2009157588989319376317
- MacchiLChristiaensLBrabantSResistance in vitro to low-dose aspirin is associated with platelet PlA1 (GP IIIa) polymorphism but not with C807T(GP Ia/IIa) and C-5T Kozak (GP Ibalpha) polymorphismsJ Am Coll Cardiol20034261115111913678940
- MareeAOCurtinRJChubbACyclooxygenase-1 haplotype modulates platelet response to aspirinJ Thromb Haemost20053102340234516150050
- LepantaloAMikkelssonJResendizJCPolymorphisms of COX-1 and GPVI associate with the antiplatelet effect of aspirin in coronary artery disease patientsThromb Haemost200695225325916493486
- LevEIPatelRTGuthikondaSLopezDBrayPFKleimanNSGenetic polymorphisms of the platelet receptors P2Y(12), P2Y(1) and GP IIIa and response to aspirin and clopidogrelThromb Res2007119335536016581111
- BrandtJTCloseSLIturriaSJCommon polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrelJ Thromb Haemost20075122429243617900275
- LauWCGurbelPAWatkinsPBContribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistanceCirculation2004109216617114707025
- BrackbillMLKiddRSAbdooADWarnerJGJrHarralsonAFFrequency of CYP3A4, CYP3A5, CYP2C9, and CYP2C19 variant alleles in patients receiving clopidogrel that experience repeat acute coronary syndromeHeart Vessels2009242737819337788
- SuhJWKooBKZhangSYIncreased risk of atherothrombotic events associated with cytochrome P450 3A5 polymorphism in patients taking clopidogrelCMAJ2006174121715172216754899
- SimonTVerstuyftCMary-KrauseMGenetic determinants of response to clopidogrel and cardiovascular eventsN Engl J Med2009360436337519106083
- TrenkDHochholzerWFrommMFCytochrome P450 2C19 681G > A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stentsJ Am Coll Cardiol200851201925193418482659
- ShuldinerARO’ConnellJRBlidenKPAssociation of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapyJAMA2009302884985719706858
- MegaJLCloseSLWiviottSDCytochrome p-450 polymorphisms and response to clopidogrelN Engl J Med2009360435436219106084
- MegaJLCloseSLWiviottSDCytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomesCirculation2009119192553256019414633
- BhattDLCHARISMA Genomics Substudy links loss-of-function allele and bleedingPresented at the Transcatheter Cardiovascular Therapeutics (TCT) Annual Meeting2009 [cited 2009 Dec 11]. URL: http://www.theheart.org/article/1008623.do
- Scripps HealthIndividualized therapies now available for anti-clogging medications [press release]1072009 URL:http://www.scripps.org/news_items/3521-scripps-becomes-first-in-u-s-to-offer-genetic-tests-to-stent-patients
- WallentinLBeckerRCBudajATicagrelor versus clopidogrel in patients with acute coronary syndromesN Engl J Med2009361111045105719717846
- WiviottSDBraunwaldEMcCabeCHPrasugrel versus clopidogrel in patients with acute coronary syndromesN Engl J Med2007357202001201517982182