Abstract
Aim: Accurate noninvasive diagnostic aids for endometriosis are needed. We evaluated mitochondrial DNA deletions as potential biomarkers for endometriosis. Methods: The diagnostic accuracy of deletions was evaluated by quantitative polymerase chain reaction (QPCR) using well-characterized clinical specimens from all subtypes and stages of endometriosis in a case–control format (n = 182). Results: Deletions (1.2 and 3.7 kb) detected in blood differentiated between endometriosis and controls (area under the curve [AUC] 0.71–0.90). Differences in deletion levels were statistically significant (p < 0.05) for all disease subtypes and stages. Neither deletion was correlated with patient or specimen age or hormone status. The 1.2 kb deletion was not correlated with menstrual stage; the 3.7 kb deletion was significantly correlated between two of the groups. Conclusion: Biomarkers of the mitochondrial genome, including the deletions described here, offer a promising and largely unexplored avenue in the pursuit of diagnostic markers for endometriosis that can be effectively translated to clinical application.
Endometriosis is a burdensome disease that occurs in up to 5–10% of women of reproductive age and is a common cause of infertility [Citation1–7]. The disease is characterized by the presence of endometrial tissue (epithelial cells and stroma) outside of the uterus; endometrial tissue can be found on the pelvic peritoneum and Fallopian tubes, involve ovaries, the bowel and bladder and rarely more distal body sites [Citation8–11]. Women with endometriosis frequently suffer from symptoms including nonmenstrual pelvic pain, painful menstrual cramps, pain during intercourse, fatigue and infertility [Citation12], which can lead to a substantial reduction in quality of life (QoL) [Citation13]. Given the high prevalence and significant morbidity associated with the disease, the global endometriosis-related healthcare burden has been estimated to be in the hundreds of billions of Euros each year [Citation14].
The current gold standard for diagnosing endometriosis is laparoscopic surgery followed by histopathological confirmation [Citation5,Citation15]. Making a timely diagnosis is complicated by delayed reporting [Citation16] and misinterpretation of symptoms [Citation17], and can be further delayed if the patient is hesitant to undergo a costly and invasive laparoscopic procedure. Indeed, the delay in diagnosing endometriosis can be more than a decade [Citation16]. Due to these delays, the majority of women have developed moderate to severe symptoms by the time a definitive diagnosis is made, which can result in increased morbidity, treatment costs and decreased QoL [Citation14]. Clearly, there is a great need for reliable, noninvasive tests that can facilitate early detection of endometriosis and provide actionable real-time results.
Molecular biomarkers have been widely used as tools to measure, detect and predict human disease [Citation18–24]; however, the search for an endometriosis-specific biomarker has proven difficult [Citation25]. Some of the key challenges include nonstandardized sample collection, analysis methods and data interpretation and the lack of biomarker specificity [Citation17], though recent efforts have been made to harmonize methods of collecting and storage of biological specimen and reporting of endometriosis data, including the World Endometriosis Research Foundation EPHect Protocols [Citation26]. A variety of candidate biomarkers from blood, tissue and urine have been reported, but none have been successfully translated into clinical use. Many of these candidates have specific limitations on sample collection such as biopsy from diseased tissue, collection during a particular phase of menstruation or are dependent upon changes in regulatory patterns (e.g., gene expression and DNA methylation) induced by inflammation, which can overlap with other gynecological disorders [Citation10,Citation17] and increase the likelihood of false-positive detections. Thus, the ideal biomarker would be detectable from healthy cells or body fluids and independent of transient disease, inflammation-generated or cyclical physiological changes.
The mitochondrial genome represents a less-explored biomarker repository. Mitochondrial DNA (mtDNA) targets are attractive from a diagnostic perspective due to a high mutation frequency, limited DNA repair capability, presence in all nucleated cells and high copy number (thousands of genomes per cell) [Citation27]. As a result, even low-frequency mutations or deletion events can be amplified reliably from heteroplasmic mitochondrial populations. Indeed, mtDNA mutations have been well-described as biomarkers for several cancers across multiple body sites including bone, brain, breast, lung, colorectal, gastric, ovarian, prostate and endometrial tissues [Citation28–37]. Specifically, within the endometrium, mtDNA alterations were detected during multiple independent investigations of endometrial cancers [Citation37–40]. Although those studies did not reveal a consensus region within the mtDNA genome or a specific mtDNA alteration that correlated to endometrial disease, those data do support the tenet that changes in mtDNA could be used as a biomarker for detection of endometriosis.
In the current study, we investigated mtDNA deletions amplified from circulating blood as potential biomarkers for endometriosis. We identified 1.2 and 3.7 kb deletions and determined the diagnostic accuracy of the deletions as biomarkers using minimally invasive blood specimens collected from women of child-bearing potential with symptoms of endometriosis.
Methods
Participants & sample collection
This study utilized residual de-identified clinical specimens collected from prospectively enrolled patients as part of the EndOx study at Oxford Endometriosis CaRe Centre, John Radcliffe Hospital, University of Oxford (Oxford, UK). Briefly, specimens were collected from women scheduled to undergo laparoscopy for suspected endometriosis because of pelvic pain (symptomatic) or tubal ligation (asymptomatic). Study participants were female, aged 18 years or older (until menopause) and were confirmed as not pregnant. All specimens were obtained under a study protocol that received appropriate Ethics Committee approval from the National Research Ethics Service (Oxfordshire REC A, 09/H0604/58). All clinical specimens were anonymized to protect the identity of the source patient. The study was designed, implemented and reported in accordance with the International Council for Harmonisation, Harmonised Tripartite Guidelines for Good Clinical Practice, with applicable local regulations and the ethical principles laid down in the Declaration of Helsinki. All patients gave written informed consent prior to participation.
Blood specimens and extensive clinical phenotypic data were collected prior to surgery. Study specimens were collected, transported and stored in accordance with the standardized World Endometriosis Research Foundation EPHect procedures [Citation26,Citation41–44].
Patient populations/study cohorts
Clinical specimens used in this study were classified as asymptomatic controls, symptomatic controls of surgically confirmed absence of endometriosis or cases of surgically confirmed endometriosis. Asymptomatic controls were defined as specimens collected from a patient that underwent a scheduled tubal ligation without a clinical suspicion of endometriosis and surgically confirmed absence of endometriosis. Symptomatic controls were defined as specimens collected from patients having pain or other symptoms (excluding infertility) with a clinical suspicion of endometriosis, but no endometriosis lesions visualized by laparoscopy by experienced gynecological surgeons.
Endometriosis was scored by the operating surgeon using the revised American Society of Reproductive Medicine (rASRM) classification of endometriosis [Citation45]. Cases were grouped by disease subtype (peritoneal, ovarian, deep endometriosis) and rASRM stage, with stages I through IV representing minimal, mild, moderate and severe disease, respectively.
Sample handling, processing & mtDNA amplification
DNA extraction
Total DNA was extracted from 200 μl of plasma using the QIAamp 96 QIAcubeHT extraction kit (Qiagen, Crawley, UK), automated on a QIAcube HT system (Qiagen). Extracted DNA was eluted in 200 μl of AE buffer.
mtDNA deletion real-time QPCR
Amplification was performed in 20 μl reactions using a 96-well microplate (Bio-Rad, Hemel Hempstead, UK). Each well contained 5 μl of un-normalized DNA template, 1 × SYBR® Green master mix and 250 nM of each primer. Primer sequences and master mix are trade secrets of MDNA Life Sciences, Inc. PCR and SYBR Green I fluorescence were analyzed using a Chromo4 Real-time PCR Detection System (Bio-Rad). Cycling conditions for the 1.2 kb deletion were as follows: 3 min at 95°C, followed by 5 cycles of 30 s at 95°C, 30 s at 67°C and 30 s at 72°C; for each subsequent cycle, the annealing temperature was decreased by 0.5°C increments. Amplification conditions were: 45 cycles of 30 s at 95°C, 30 s at 65°C and 30 s at 72°C. All other deletions were amplified with a standard protocol of 45 cycles of 30 s at 95°C, 30 s at 58–65°C and 30 s at 72°C. Following amplification, melting curve analysis was performed from 70 to 90°C, reading every 0.5°C. Each plate of samples and controls was amplified in triplicate on three separate occasions.
Real-time QPCR normalization with 18S rRNA
Target amplicon quantity was normalized using the 18S rRNA nuclear DNA gene. Amplification reactions were performed as 20 μl reactions in a 96-well microplate. Each well contained 5 μl of un-normalized DNA template, 1 × SYBR Green master mix and 200 nM of each primer. Amplification and SYBR Green I fluorescence was analyzed using a Chromo4 Real-Time PCR Detection System. Amplification conditions were: 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 64.5°C and 30 s at 72°C. Following amplification, a melting curve analysis was performed from 70 to 90°C, reading every 0.5°C.
Quality control
The quantification cycle (Cq) was calculated using the CFX manager software regression model (Bio-Rad). The Cq of each deletion amplicon was normalized to the Cq of the multicopy nuclear target 18S rRNA gene amplicon. All samples were amplified in triplicate on separate plates and were considered to have passed if at least two of the three replicates were within 1.5 Cq and the melting temperature (Tm) was consistent with the target amplification product when present, (deletion Tm 81°C ± 2°C, 18S rRNA Tm 82°C± 2°C).
A total of two no template control samples were processed alongside each batch of DNA extractions and verified as negative for amplification of both the deletion target and the 18S rRNA gene. A total of two no template control reactions were included on each PCR plate and verified negative for amplification of both the deletion targets and the 18S rRNA gene. Deletion primer specificity was evaluated using rho 0 cellular DNA (to detect mitochondrial pseudogene amplification) as well as DNA from healthy male buccal swabs and DNA extracted from the rho 0 parental cell line (prior to depletion of mitochondria).
For the initial round of standard PCR reactions, extracted DNA from patients with confirmed endometriosis underwent whole genome amplification using the Repli-G mitochondrial DNA kit (Qiagen) to ensure sufficient DNA quantity during this phase.
Rho 0 cell preparation
Rho 0 cells were prepared as previously described [Citation46]. Briefly, cells from the human osteosarcoma cell line 143B (ATCC CRL-8303) were treated with ethidium bromide to deplete cytoplasmic mitochondrial DNA. Cells were grown to confluence in high glucose DMEM with pyruvate, L-glutamine, uridine (50 μg/ml) and 5% Fetal Bovine Serum.
Statistical analysis
No formal sample size calculation was performed; the number of clinical specimens used was deemed sufficient to satisfy the study objective.
For QPCR, targets were amplified from all specimens in triplicate and average Cq values were calculated. The normalized deletion value (ΔCq) was determined by quantifying the deletion amplicon relative to the 18S rRNA reference amplicon. Statistical analyses were performed using Graphpad Prism 5.0 (Graphpad Software, Inc., CA, USA) for receiver operating characteristic (ROC) curves and descriptive statistics. SPSS v17.0 (IBM Corp., NY, USA) was used to perform correlations and significance tests. Clinical characteristics were summarized using count and percentages for categorical data, and mean, standard deviation (SD) and range for continuous variables. The means of two groups were compared using the Student's t-test and the Mann–Whitney U-test for parametric and nonparametric distributions, respectively. Correlation between the two variables was assessed with the Pearson's correlation coefficient (r). With respect to the presence of endometriosis, ROC curves were constructed for all but the 6.5 kb deletion. The area under the curve (AUC) of the ROC and the sensitivity and specificity at selected cut-offs (described below) were calculated with 95% CI. The p-value <0.05 was considered statistically significant for all tests.
Results
Patient population & clinical specimens
Demographics and clinical characteristics for patients that provided the clinical specimens used to evaluate the 1.2 and 3.7 kb deletions are summarized in .
Table 1. Demographic and clinical characteristics.
One hundred eighty-two specimens were used in the 1.2 and 3.7 kb deletion evaluations. The mean (SD) age of patients that provided specimens was 34.4 (6.9) years. The mean ages were similar between the control and case groups with mean (SD) ages of 37.2 (6.8) and 33.8 (6.8) years, respectively and was statistically significantly different (p = 0.012). Of the 182 patient specimens used in the evaluation, 120 (65.9%) patients reported no hormone therapy within the 3 months prior to specimen collection, 55 (30.2%) reported having hormone therapy within 3 months prior to specimen collection and seven (3.8%) were undetermined. Menstrual cycle phase data were calculated using the last menstrual period (LMP) prior to the date of blood collection in relation to a patient's normal cycle length. Twenty-six (14.3%) patients reported no menstruation: 20 of whom were on hormones, 15 (8.2%) reported irregular menstruation, 31 (17.0%) were in the menstrual phase (between 1 and 5 days from the first day of LMP), 45 (24.7%) were in the follicular phase (5 to 14 days from LMP) and 65 (35.7%) were in the luteal + extended menstrual phase (>15 days from LMP).
The control group included a total of 32 specimens; 19 (59.4%) specimens collected from symptomatic patients (presenting with symptoms consistent with endometriosis other than infertility and surgical confirmed absence for the disease) and 13 (40.6%) specimens collected from asymptomatic patients scheduled for tubal ligation. The case group included 150 specimens from patients with three disease subtypes (peritoneal, ovarian and deep infiltrating endometriosis) that were classified into four stages (rASRM I through IV). A total of 52 (34.7%) specimen were collected from women with peritoneal, 47 (31.3%) were from women with ovarian and 51 (34.0%) were collected from women with deep endometriosis. A total of 65 (43.3%) specimens were from patients with stage I disease, 25 (16.7%) were stage II, 30 (20.0%) were stage III and 28 (18.7%) were stage IV. Two (1.3%) specimens had an unknown disease stage.
mtDNA deletions & preliminary evaluation: standard PCR
Seven candidate deletions were initially selected for evaluation based upon sequence composition, presence of a flanking repeat location within the major arc of the mitochondrial genome where proportionally more deletions are reported [Citation47] and observation previously in endometrial tissue (data not shown). Deletions were selected within the following genomic regions: CO2 to ATP6 (1.0 kb deletion); ATP6 to ND3 (1.2 kb deletion); ATP8 to ND4 (2.4 kb deletion); ATP6 to ND5 (3.7 kb deletion); ATP8 to ND5 (5.0 kb deletion); CO1 to ND5 (6.5 kb deletion); and CO2 to CytB (7.7 kb deletion). An initial round of standard (qualitative) PCR and visualization after gel electrophoresis was used to prequalify each deletion target and determine if each of the candidates were detectable, had sufficient copy number for reliable detection, had the predicted amplicon size, were specific and did not co-amplify nuclear pseudogenes or generate nonspecific amplification products.
All seven predicted deletions were detectable circulating in blood plasma. However, the 5.0 and the 6.5 kb deletions amplified the rho 0 cell DNA indicating potential co-amplification of nuclear mitochondrial pseudogenes (numts). Additionally, the 6.5 kb deletion had insufficient copy number and was not considered a viable candidate for further QPCR testing. The 7.7 and the 2.4 kb deletions were lower in copy number; however, still potentially detectable with QPCR so these were subject to further evaluation. The 5.0 kb deletion amplified DNA from the buccal swab of a healthy male indicating a potential lack of disease specificity. The 7.7 kb deletion had a low level of amplification from this specimen as well.
The remaining six deletions were further evaluated using QPCR to determine whether the targets were present in sufficient copy number in the absence of whole-genome amplification, of sufficient diagnostic accuracy, detectable in rho 0 cells using more sensitive QPCR and whether the assays’ precision was acceptable. Acceptable precision criteria were a maximum deviation of 1.5 Ct between a minimum of two out of three replicates for each target deletion.
Preliminary evaluation with clinical samples
As a preliminary assessment of the remaining six candidates, we evaluated the deletions using a set of 55 clinical specimens. No preselection criteria were applied prior to evaluating these specimens, which were the first group of specimens collected during the study. The specimens included 46 specimens from patients with confirmed endometriosis and nine specimens from symptomatic control patients. After initial QPCR testing, the 2.4 kb deletion was determined to have insufficient copy number and the 1.0 kb deletion amplified DNA extracted from rho 0 cells indicating co-amplification of numts and the 7.7 kb deletion amplified only at less stringent annealing temperatures meaning mispriming events would be more probable. These candidate deletions did not satisfy assay requirements as designed here but may still exist as biomarkers benefiting from further assay optimization to obtain better sequence specificity and assay sensitivity.
Of the seven deletions initially selected, the 1.2 and 3.7 kb deletions were present in sufficient copy number in plasma to facilitate easy and reliable detection. The assays were specific under the tested PCR conditions and accurately discriminated between symptomatic controls and endometrial disease cases (all subtypes and stages combined). The AUC (95% CI) for the 1.2 kb deletion was 0.8116 (0.6178–1.005), which was statistically significant (p = 0.0034). Similarly, the AUC (95% CI) for the 3.7 kb deletion was 0.8478 (0.6663–1.029), which was also significant (p = 0.0011; data not shown).
Diagnostic accuracy of the 1.2 & 3.7 kb deletions
To more fully evaluate the 1.2 and 3.7 kb deletions as clinically viable biomarkers of endometriosis, we determined the ability of these deletions to discriminate between symptomatic controls and all endometriosis types combined, between three subtypes, and four stages of endometriosis using a larger set of clinical specimens (). We also evaluated the frequency of the deletion in asymptomatic controls. Valid paired results (both target and 18S rRNA gene amplification) were obtained for 171 specimens with the 1.2 kb deletion and 181 specimens with the 3.7 kb deletion. The analyses were performed using only symptomatic control and confirmed disease specimens in order to more accurately reflect the clinically relevant patient populations, that is, women presenting with symptoms of endometriosis with surgical confirmation of disease status as an outcome. Importantly, the 1.2 and 3.7 kb deletions detected no difference between symptomatic and asymptomatic control specimens, p = 0.462 and p = 0.878, respectively.
Symptomatic controls versus all disease
Similar to the preliminary analysis using 55 clinical specimens, both the 1.2 and 3.7 kb deletions accurately discriminated between symptomatic control and endometrial disease specimens (peritoneal, ovarian and deep endometriosis specimens combined). The AUC (95% CI) for the 1.2 kb deletion was 0.7879 (0.6791–0.8967), which was statistically significant (p < 0.0001). The AUC (95% CI) for the 3.7 kb deletion was 0.807 (0.7063–0.9077), which was also significant (p < 0.0001; ). Coordinates of the ROC were examined and a threshold selected to optimize sensitivity. Applying a threshold of -4.43 for discrimination of symptomatic controls and all subtypes/stages of endometriosis using the 1.2 kb deletion results in sensitivity and specificity values of 81.8 and 72.2%, respectively. At a threshold of 10.51, sensitivity and specificity for the 3.7 kb deletion are 85.1 and 57.9%, respectively ().
The 1.2 and 3.7 kb deletions were evaluated for the ability to distinguish between symptomatic patient specimens and specimens from patients with confirmed endometriosis (all subtypes/stages combined). ROC curves were constructed and the AUCs were calculated.
AUC: Area under the curve; ROC: Receiver operating characteristic; Std: Standard.
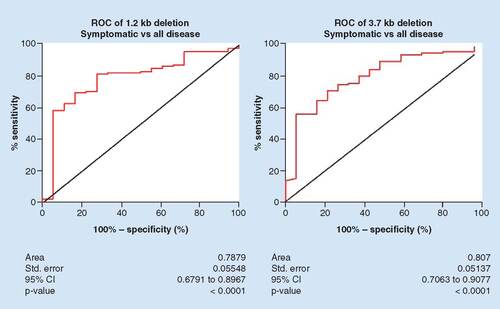
Table 2. Performance of the 1.2 and 3.7 kb deletions.
Combining the 1.2 kb deletion with the 3.7 kb deletion improved the diagnostic accuracy (AUC 0.827 [0.722–0.931]) between all symptomatic controls and all endometriosis and AUC 0.882 (0.784–0.980) between symptomatic controls and stage I/II disease (data not shown).
Disease by subtype – 1.2 kb deletion
An important feature of any diagnostic aid for endometriosis is the ability to accurately detect all disease subtypes. We evaluated the ability of the 1.2 kb deletion to differentiate between symptomatic control specimens and specimens from patients with confirmed peritoneal, ovarian and deep endometriosis. The distribution of the 1.2 kb deletion for each disease subtype is shown in . The mean (SD) ΔCt value was -4.89 (1.727) for asymptomatic controls, -4.312 (2.075), for symptomatic controls, -7.187 (2.581) for peritoneal disease, -6.291 (2.344) for ovarian disease and -6.193 (2.143) for deep endometriosis. The difference in normalized 1.2 kb deletion quantity between symptomatic controls was statistically significant for peritoneal (p < 0.0001), ovarian (p = 0.003) and deep endometriosis (p = 0.0012).
The 1.2 kb deletion was evaluated for the ability to distinguish between symptomatic patient specimens and specimens from patients stratified by subtype of endometriosis (peritoneal, ovarian, deep infiltrating). Top: The distribution of normalized 1.2 kb deletion for specimens from asymptomatic and symptomatic controls, patients with peritoneal, ovarian or deep infiltrating endometriosis or patients with low (I/II) or high (III/IV) stages of endometriosis. Box boundaries represent the 25th and 75th percentile, the line in the middle represents the median and the whiskers represent the 90th (top) and 10th (bottom) percentiles. Dots represent outlier values (left). Descriptive statistics are summarized for each group (right). Bottom: ROC curves for the 1.2 kb deletion were constructed and the areas under the curves were calculated, showing diagnostic accuracy.
Asympto: Asymptomatic control; Dev: Deviation; DIE: Deep infiltrating endometriosis; N: Number of specimens in each group; ROC: Receiver operating characteristic; Std: Standard; Sympto: Symptomatic control.
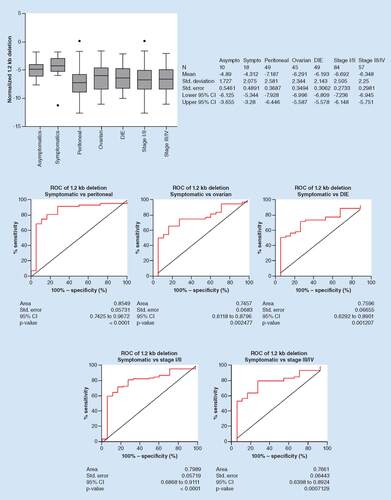
Diagnostic accuracy of the 1.2 kb deletion is shown in , with AUC (95% CI) values of 0.8549 (0.7425–0.9672), p < 0.0001 for detection of peritoneal, 0.7457 (0.6118–0.8796), p = 0.0025 for detection of ovarian and 0.7596 (0.6292–0.8901), p = 0.0012 for detection of deep endometriosis. Taken together, these data indicate that the 1.2 kb deletion was able to accurately distinguish between specimens collected from symptomatic controls and peritoneal, ovarian and deep endometriosis patients. Applying a threshold of -4.430 for discrimination of symptomatic controls and peritoneal endometriosis using the 1.2 kb deletion results in sensitivity and specificity values of 81.8 and 72.2%, respectively. At a threshold of -4.675, the sensitivity and specificity of the 1.2 kb deletion in discriminating between symptomatic controls and ovarian endometriosis is 75.6 and 72.2%, respectively. At a threshold of -4.350, the sensitivity and specificity of the 1.2 kb deletion in discriminating between symptomatic controls and deep endometriosis is 79.6 and 66.7%, respectively ().
Disease by subtype – 3.7 kb deletion
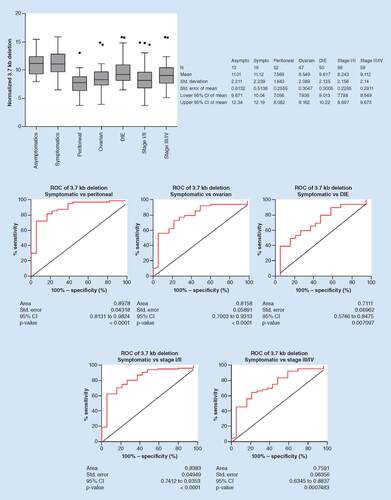
The distribution of the 3.7 kb deletion for each disease subtype is shown in . The mean (SD) ΔCt value was 11.01 (2.211) for asymptomatic controls, 11.12 (2.239) for symptomatic controls, 7.569 (1.843) for peritoneal, 8.549 (2.089) for ovarian and 9.617 (2.125) for deep endometriosis. The difference in deletion quantity between symptomatic controls was statistically significant for peritoneal (p < 0.0001), ovarian (p < 0.0001) and deep endometriosis (p = 0.0072). Diagnostic accuracy of the 3.7 kb deletion in detecting each of the three disease subtypes is shown in , with AUC (95% CI) values of 0.8978 (0.8131–0.9824), p < 0.0001 for detection of peritoneal, 0.8158 (0.7003–0.9313), p < 0.0001 for detection of ovarian and 0.7110 (0.5746–0.8475), p = 0.0071 for detection of deep endometriosis. Taken together, these data indicate that the 3.7 kb deletion was able to accurately distinguish between specimens collected from symptomatic controls and women with peritoneal, ovarian and deep endometriosis. Applying a threshold of 9.805 for discrimination of symptomatic controls and peritoneal endometriosis using the 3.7 kb deletion results in sensitivity and specificity values of 88.5 and 73.7%, respectively. At a threshold of 9.910, the sensitivity and specificity of the 3.7 kb deletion in discriminating between symptomatic controls and ovarian endometriosis is 80.9 and 68.4%, respectively. At a threshold of 11.01, the sensitivity and specificity of the 3.7 kb deletion in discriminating between symptomatic controls and deep endometriosis is 80.0 and 52.6%, respectively ().
The 3.7 kb deletion was evaluated for the ability to distinguish between symptomatic patient specimens and specimens from patients stratified by subtype of endometriosis (peritoneal, ovarian, deep infiltrating). Top: The distribution of normalized 3.7 kb deletion for specimens from asymptomatic and symptomatic controls, patients with peritoneal, ovarian or deep infiltrating endometriosis or patients with low (I/II) or high (III/IV) stages of endometriosis. Box boundaries represent the 25th and 75th percentile, the line in the middle represents the median and the whiskers represent the 90th (top) and 10th (bottom) percentiles. Dots represent outlier values (left). Descriptive statistics are summarized for each group (right). Bottom: ROC curves for the 3.7 kb deletion were constructed and the AUCs were calculated, showing the diagnostic accuracy.
Asympto: Asymptomatic control; AUC: Area under the curve; Dev: Deviation; DIE: Deep infiltrating endometriosis; N: Number of specimens in each group; ROC: Receiver operating characteristic; Sympto: Symptomatic control; Std: Standard.
Disease by stage – 1.2 kb deletion
Another important characteristic of a biomarker for endometriosis is the ability to detect both low and high stages of disease. We next evaluated the ability of the 1.2 kb deletion to differentiate between symptomatic control specimens and specimens from confirmed low (I/II) or high (III/IV) stages of disease. The distribution of the 1.2 kb deletion for stage I/II and stage III/IV disease is shown in . The mean (SD) ΔCt value was -4.312 (2.075) for symptomatic controls, -6.692 (2.505) for stage I/II and -6.348 (2.25) for stage III/IV. The difference between symptomatic controls was statistically significant for stage I/II (p < 0.0001) and stage III/IV (p = 0.001) disease groups. The difference between stage I/II and III/IV was not statistically significant (p = 0.406).
Diagnostic accuracy of the 1.2 kb deletion is shown in , with AUC (95% CI) values of 0.7989 (0.6868–0.9111), p < 0.0001 for detection of stage I/II and 0.7661 (0.6398–0.8924), p = 0.0007 for detection of stage III/IV disease. Thus, the 1.2 kb deletion was able to accurately distinguish between symptomatic controls and all stages of disease. At a threshold of -4.430, the sensitivity and specificity of the 1.2 kb deletion in discriminating between symptomatic controls and stage I/II endometriosis is 82.1 and 72.2%, respectively. At a threshold of -4.490, the sensitivity and specificity of the 1.2 kb deletion in discriminating between symptomatic controls and stage III/IV endometriosis is 80.7 and 72.2%, respectively ().
Disease by stage – 3.7 kb deletion
The distribution of the 3.7 kb deletion for stage I/II and stage III/IV disease is shown in . The mean (SD) ΔCt value was 11.12 (2.239), for symptomatic controls, 8.243 (2.156) for stage I/II and 9.112 (2.14) for stage III/IV. The difference between symptomatic controls was statistically significant for stage I/II (p < 0.0001) and stage III/IV (p = 0.0008) disease groups. Diagnostic accuracy of the 3.7 kb deletion is shown in , with AUC (95% CI) values of 0.8383 (0.7412–0.9353), p < 0.0001 for detection of stage I/II and 0.7591 (0.6354-0.8837), p = 0.0007 for detection of stage III/IV disease. The difference between stage I/II and III/IV was statistically significant for the 3.7 kb deletion (p = 0.016). These data indicate that the 3.7 kb deletion was able to accurately distinguish between symptomatic controls and all stages of disease. At a threshold of 10.17, the sensitivity and specificity of the 3.7 kb deletion in discriminating between symptomatic controls and stage I/II endometriosis is 87.6 and 63.2%, respectively. At a threshold of 11.00, the sensitivity and specificity of the 3.7 kb deletion in discriminating between symptomatic controls and stage III/IV endometriosis is 84.5 and 52.6%, respectively.
Correlation with patient age, specimen age, hormonal therapy & menstrual phase – 1.2 kb deletion
An ideal biomarker test would provide accurate results independent of patient and specimen age, treatment with hormonal therapy and timing of menstrual phase during specimen collection. The effect of these parameters on disease detection is summarized in . For the 1.2 kb deletion, we determined that there was no correlation with the detection of disease and patient age; the correlation coefficient (r) was r = 0.030 (p = 0.698). There was also no correlation with disease detection and specimen age by year of collection (r = 0.072; p = 0.353). When stratified by hormonal status (patients that received hormonal therapy or had no hormonal therapy within the 3 months prior to specimen collection), the difference in disease detection was not statistically significant (p = 0.120). When patients were stratified by menstrual phase (no menstruation, irregular menstruation, menstruation, follicular phase or luteal phase + extended menstruation), there was no statistically significant difference in disease detection with the 1.2 kb deletion (p = 0.228).
Table 3. Effect of patient and specimen age, hormonal therapy and menstrual cycle.
Similarly, disease detection based on the 3.7 kb deletion was not significantly correlated to patient age (r = 0.1034; p = 0.166) or specimen age (r = 0.0628; p = 0.4009) or significantly affected by hormonal therapy (p = 0.195). Detection of endometriosis with the 3.7 kb deletion was significantly correlated to menstrual phase (p = 0.036), which was driven specifically by the difference in detection between patients who reported no period and those that were in the follicular phase (p = 0.026) with a difference of 1.72. Combined, these data indicate that the accuracy of the 1.2 kb and is not significantly affected by these clinically relevant variables and accuracy of the 3.7 kb deletion is slightly affected by menstrual phase.
Discussion
Endometriosis is a highly prevalent disease in women of reproductive age that is associated with a large economic burden and results in a substantial reduction in the QoL of those affected. One of the key contributors to this clinical problem is the lack of diagnostic tools to facilitate early detection and intervention. The current diagnostic gold standard is a thorough laparoscopic inspection ideally followed by histologic confirmation of suspected lesions [Citation5,Citation15]. Because there are minimal objective data available, the diagnostic value of this process in largely unclear. The use of laparoscopic exams has been considered by some to be potentially inaccurate, and even paired with histologic confirmation accuracy reportedly ranges from 60 to 85% [Citation48–51]. The standard diagnostic process could be further complicated in cases where the disease presents itself atypically, or in early stages of disease that are not easily visualized and overlooked by inexperienced surgeons. Additionally, where medical intervention could be initiated in an effort to avoid or delay surgery, a presumptive diagnosis of endometriosis based upon an accurate biomarker test could provide needed evidence in support of this treatment. Thus, there is a clear need to improve upon the current standard, particularly in a way that could provide more routine results early in the course of disease.
In the current study, we identified and evaluated two novel mtDNA deletions as potential biomarkers for endometriosis. Assays targeting the 1.2 and 3.7 kb deletions met criteria for robust diagnostic tests, utilize a minimally invasive specimen, and if successfully translated into clinical use, could potentially help reduce the delay in time to diagnosis associated with current diagnostic practices and provide an opportunity for medical intervention prior to a surgical one. After setting a diagnostic threshold (as described above), the sensitivity of the 1.2 kb deletion assay was 81.8% and specificity was 72.2%. The diagnostic performance of the 3.7 kb deletion assay was similar with sensitivity and specificity of 85.1 and 57.9%, respectively. Thus, diagnostic assays based on either of these deletions have the potential to compliment the current standard of care. Of particular importance is the diagnostic accuracy of these deletions for early stage disease as later stage disease is more readily detected in current practice using ultrasound. In a primary care setting, a positive test result could support initiating first-line medical treatment for endometriosis such as oral contraceptives or trigger a specialist referral. Interestingly, as the difference in the means between the two control groups was very small and not significant (0.578 and 0.11 for the 1.2 kb and the 3.7 kb deletions, respectively), and thus diagnostic accuracy comparable, future development may explore utility in screening the healthy females beginning at menarche, perhaps to monitor for increases in the deletion(s) over time. An estimated 10% of women presenting with dysmenorrhea have secondary dysmenorrhea, with the majority caused by endometriosis [Citation52]. In this population, the 1.2 and 3.7 kb deletions would quite effectively rule out endometriosis with a negative predictive value of 97%. In a secondary care setting, a positive test could guide the decision to initiate treatment with second-line medication such as gonadotropin-releasing hormone antagonists or to proceed with laparoscopic surgery. Importantly, in the former setting a diagnostic cut-off could be selected to maximize test sensitivity as the risk associated with an incorrect false-positive result is less critical, whereas in the latter setting, a different diagnostic cut-off to maximize specificity and minimize the exposure of women without the disease to the risks associated with these interventions could be beneficial.
In addition to diagnostic accuracy, the ability of these two biomarkers to detect endometriosis was not correlated to patient age, specimen age or hormone status, and only the 3.7 kb deletion had a slight correlation to the patients’ phase of menstrual cycle at the time of specimen collection. Further study is needed to confirm whether this correlation with menstrual phase exists in a larger cohort of patients, or whether it is an artifact of the number of specimens used in this study. We demonstrated that both the 1.2 and 3.7 kb deletions accurately detect all subtypes and stages of disease. In contrast to the current diagnostic standard that involves visualization during surgery and possible excision of lesions for histological confirmation, assays based on mtDNA deletions require only a blood specimen and could potentially provide objective results before or instead of a surgical intervention. Thus, if successfully translated into clinical use, mtDNA-based assays have the potential to reduce the delays in diagnosis [Citation16] and provide actionable results earlier in the course of disease than currently possible.
From a practical standpoint, use of a blood-based biomarker assay has several advantages to effectively augment the current standard of care. The specimen is easy and inexpensive to collect via venipuncture, and there is a low likelihood to have comorbidities associated with collection. Blood specimens can be readily collected in an outpatient physicians’ office or clinic, eliminating the need for dedicated surgical space and equipment. As a result of the high copy number of mtDNA, standard DNA extraction methods are used without the need for enrichment techniques and ample DNA is recovered from a standard blood specimen so a low test failure rate can be anticipated. The assays use PCR-based technology that is cost-effective and widely used in clinical labs, and while the assays are quantitative, the output is easily interpreted, that is, a test result is either above or below a defined diagnostic cut-off which corresponds to either a positive or negative outcome. Finally, a lack of, or minimal correlation with menstrual stage ensures that sampling requirements are simplified and timing of menstruation need not be considered when scheduling venipuncture.
A key element in successful disease management is understanding disease epidemiology. Due, in part, to a relatively complex diagnostic process and symptoms that overlap with other gynecological disorders, the epidemiology of endometriosis is not well-characterized and varies across patient populations and geographic locations [Citation1,Citation2,Citation4,Citation6]. With the advent of molecular assays such as those described here, additional data could become more readily available and help fill in some of the gaps in our understanding of endometriosis epidemiology. Importantly, this study utilized publicly available standardized processes for specimen collection and processing, which will allow for more direct comparison of test results across different studies and patient populations [Citation41–44,Citation53].
Importantly, the location of the mtDNA deletions may also help shed light on the pathophysiological process of endometriosis. Both the 1.2 and 3.7 kb deletions affect all or part of the genes encoding for Complexes I and V (ATP synthase) of the respiratory chain and several tRNAs. Although these deletions likely result in abnormal mitochondrial ATP synthase and Complex I proteins, the heteroplasmic nature of mtDNA likely allows some degree of functional compensation within the population. Interestingly, the two best candidates out of the seven tested in this study are deletions within regions that overlap each other in the mitochondrial genome. Given that the 1.2 kb deletion region (ATP6 to ND3) resides within the larger 3.7 kb deletion (ATP6 to ND5), perhaps it is not surprising that the diagnostic accuracy of the two deletions is similar.
Based on the data presented here, the 1.2 and 3.7 kb mtDNA deletions are associated with endometriosis; however, an additional study is necessary to understand what mechanistic role this mitochondrial genomic region plays in the development of endometriosis.
Limitations of this study include the use of patient-reported hormone and menstrual status, which can be less accurate than taking study-specific data measurements. In addition, because the deletions were characterized using circulating cell-free DNA rather than directly from affected and unaffected tissue, the association with endometriosis is presumptive. These data, while encouraging, require replication and validation in a larger independent dataset and characterization in the tissue. These studies are currently underway.
Conclusion
Biomarkers derived from the mitochondrial genome, including the 1.2 and 3.7 kb deletions described here, offer a promising and largely unexplored avenue in the pursuit of diagnostic markers for endometriosis that can be effectively translated to clinical application. Based on a minimally invasive specimen, assays based on these markers could positively impact the diagnostic landscape for endometriosis by reducing the delay in diagnosis and providing rapid, actionable and objective test results. Further validation of these markers as diagnostic aids is ongoing.
Future perspective: mitochondrial biomarkers of disease
The mitochondrial genome, though well-characterized and first fully sequenced in 1981, has long been overlooked as a diagnostic tool outside of rare metabolic diseases such as Leber's Hereditary Optic Neuropathy. However, this genome is in many ways ideally suited to provide real-time disease data. The genome itself resides within the mitochondrial matrix, the site of most intracellular reactive oxygen species (ROS) production – ROS themselves are a potent DNA damaging agent. The increase in these mutagenic species is closely tied to the cellular stress response associated with disease progression. In many cases, the resulting mutated mitochondrial DNAs accumulate in parallel with this progression facilitating the identification of diagnostic cut-offs from a single mitochondrial biomarker, tailored to a specific clinically relevant end point. In cases, such as prostate cancer, where overdiagnosis and treatment is a concern and a repository of indolent disease exists within a population, a cut-off can be determined to select for more advanced disease. Alternatively, where earlier detection can positively impact patient outcomes, a cut-off to detect the initial accumulation of a disease specific mitochondrial deletion is of benefit. This quantitative accumulation may also provide meaningful information in post-treatment monitoring scenarios.
Importantly, owing to the proximity to ROS and the inability to effectively repair many types of mutations these mitochondrial mutations occur very early in the disease process, often well before nuclear genome alterations begin. As ongoing research continues to identify, report and commercialize novel disease biomarkers originating from the mitochondrial genome, the opportunity to enhance the current diagnostic and treatment pathways across a vast array of diseases will be significant.
Endometriosis is a significant health burden that affects up to 10% of women worldwide. Currently, diagnosis is based on surgical visualization followed by histological confirmation.
Diagnosis is often complicated due to variable clinical presentation and symptoms that overlap with other gynecological disorders. As a result, definitive diagnosis can be delayed up to a decade, which can result in higher morbidity and decreased quality of life for those affected.
Thus, there is a clear need for rapid, reliable diagnostic aids that can provide actionable results early in the course of disease.
Study specimens were collected from women scheduled to undergo laparoscopy for pelvic pain (symptomatic and cases) or tubal ligation (asymptomatic). Study participants were female, aged 18 years or older (until menopause) and were confirmed as not pregnant.
Seven candidate mitochondrial DNA deletions were identified and evaluated to determine whether each was detectable in plasma, had sufficient copy number for reliable detection, had the predicted amplicon size, were specific and did not co-amplify nuclear pseudogenes or generate nonspecific amplification products.
Six candidate deletions were further evaluated by QPCR and clinical specimens to determine whether each met the criteria for a robust diagnostic assay and evaluated accuracy in discriminating between endometriosis and control specimens. Two deletions were selected as potential biomarker candidates (1.2 and 3.7 kb deletions).
The 1.2 and 3.7 kb deletions accurately detected endometriosis, including all subtypes and disease stages, and detection was not correlated to patient or specimen age or hormone therapy. Only the 3.7 kb deletion was significantly correlated to menstrual phase, which was limited only to two phases.
Biomarkers derived from the mitochondrial genome, including the 1.2 and 3.7 kb deletions described here, offer a promising and largely unexplored avenue in the pursuit of diagnostic markers for endometriosis that can be effectively translated to clinical application.
Based on a minimally invasive specimen, assays based on these markers could positively impact the diagnostic landscape for endometriosis by reducing the delay in diagnosis and providing rapid, actionable and objective test results.
Authors contributions
All authors contributed to study design and data acquisition, analysis or interpretation and drafting of this manuscript. All the authors had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of data analysis.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. Written informed consent has been obtained from the participants involved.
Acknowledgements
The authors are grateful to all women who participated in the ENDOX study at the Oxford Endometriosis CaRe Centre, and thank the research nurses C Hubbard, K Barrett and E Shepherd for their dedication and hard work in collecting biological samples and clinical data as part of the studies. The authors are grateful to K. Zondervan and C. Becker for data and manuscript review and providing access to the invaluable ENDOX study collections.
Financial & competing interests disclosure
This study was funded by MDNA Life Sciences, Inc., J Creed, A Maggrah, B Reguly and A Harbottle are employed by or consult for MDNA Life Sciences, Inc. and are inventors of intellectual property including patent applications relating to the information disclosed herein. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
The authors acknowledge J Gilbreath of ICON, plc., who provided medical writing services in preparation of this manuscript. Funding for medical writing services was provided by MDNA Life Sciences, Inc.
Additional information
Funding
References
- Eskenazi B , WarnerML. Epidemiology of endometriosis. Obstet. Gynecol. Clin. North Am.24 (2), 235–258 (1997).
- Kjerulff KH , EricksonBA, LangenbergPW. Chronic gynecological conditions reported by US women: findings from the National Health Interview Survey, 1984 to 1992. Am. J. Public Health86 (2), 195–199 (1996).
- Houston DE , NollerKL, MeltonLJ, SelwynBJ, HardyRJ. Incidence of pelvic endometriosis in Rochester, Minnesota, 1970–1979. Am. J. Epidemiol.125 (6), 959–969 (1987).
- Wheeler JM . Epidemiology of endometriosis-associated infertility. J. Reprod. Med.34 (1), 41–46 (1989).
- Giudice LC , KaoLC. Endometriosis. Lancet364 (9447), 1789–1799 (2004).
- Vessey MP , Villard-MackintoshL, PainterR. Epidemiology of endometriosis in women attending family planning clinics. BMJ306 (6871), 182–184 (1993).
- Zondervan KT , BeckerCM, KogaK, MissmerSA, TaylorRN, ViganòP. Endometriosis. Nat. Rev. Dis. Primers.4 (1), 9 (2018).
- Barlow DH , KennedyS. Endometriosis: new genetic approaches and therapy. Annu. Rev. Med.56, 345–356 (2005).
- Giudice LC . Clinical practice. Endometriosis. N. Engl. J. Med.362 (25), 2389–2398 (2010).
- Fassbender A , VodolazkaiaA, SaundersPet al. Biomarkers of endometriosis. Fertil. Steril.99 (4), 1135–1145 (2013).
- Sonavane SK , KantawalaKP, MeniasCO. Beyond the boundaries-endometriosis: typical and atypical locations. Curr. Probl. Diagn. Radiol.40 (6), 219–232 (2011).
- Berkley KJ , RapkinAJ, PapkaRE. The pains of endometriosis. Science308 (5728), 1587–1589 (2005).
- Nnoaham KE , HummelshojL, WebsterPet al. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil. Steril.96 (2), 366–373.e8 (2011).
- Simoens S , DunselmanG, DirksenCet al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum. Reprod.27 (5), 1292–1299 (2012).
- Dunselman G a. J , VermeulenN, BeckerCet al. ESHRE guideline: management of women with endometriosis. Hum. Reprod.29 (3), 400–412 (2014).
- Hadfield R , MardonH, BarlowD, KennedyS. Delay in the diagnosis of endometriosis: a survey of women from the USA and the UK. Hum. Reprod.11 (4), 878–880 (1996).
- Ahn SH , SinghV, TayadeC. Biomarkers in endometriosis: challenges and opportunities. Fertil. Steril.107 (3), 523–532 (2017).
- Rifai N , GilletteMA, CarrSA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol.24 (8), 971–983 (2006).
- Hu C , van der HeijdenR, WangM, van der GreefJ, HankemeierT, XuG. Analytical strategies in lipidomics and applications in disease biomarker discovery. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.877 (26), 2836–2846 (2009).
- Liotta LA , PetricoinEF. Serum peptidome for cancer detection: spinning biologic trash into diagnostic gold. J. Clin. Invest.116 (1), 26–30 (2006).
- Petricoin EF , ArdekaniAM, HittBAet al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet359 (9306), 572–577 (2002).
- Petricoin EF , BellucoC, AraujoRP, LiottaLA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat. Rev. Cancer6 (12), 961–967 (2006).
- Zhang Z , BastRC, YuYet al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res.64 (16), 5882–5890 (2004).
- Anderson NL , AndersonNG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell Proteomics1 (11), 845–867 (2002).
- May KE , Conduit-HulbertSA, VillarJ, KirtleyS, KennedySH, BeckerCM. Peripheral biomarkers of endometriosis: a systematic review. Hum. Reprod. Update16 (6), 651–674 (2010).
- Rahmioglu N , FassbenderA, VitonisAFet al. Harmonization of clinical and laboratory data to improve biomarker discovery in endometriosis: WERF EPHect. In : D'Hooghe ThomasM ( Ed.). Biomarkers for Endometriosis. SpringerInternational, Cham, Switzerland, 199–226 (2017).
- Taylor RW , TurnbullDM. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet.6 (5), 389–402 (2005).
- Polyak K , LiY, ZhuHet al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet.20 (3), 291–293 (1998).
- Petros JA , BaumannAK, Ruiz-PesiniEet al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA102 (3), 719–724 (2005).
- Creed J , KlotzL, HarbottleAet al. A single mitochondrial DNA deletion accurately detects significant prostate cancer in men in the PSA ‘grey zone’. World J. Urol. 36 (3), 341–348 (2018).
- Parrella P , XiaoY, FlissMet al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res.61 (20), 7623–7626 (2001).
- Liang BC . Evidence for association of mitochondrial DNA sequence amplification and nuclear localization in human low-grade gliomas. Mutat. Res.354 (1), 27–33 (1996).
- Guo X-G , GuoQ-N. Mutations in the mitochondrial DNA D-Loop region occur frequently in human osteosarcoma. Cancer Lett.239 (1), 151–155 (2006).
- Alonso A , MartinP, AlbarranCet al. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis18 (5), 682–685 (1997).
- Liu VW , ShiHH, CheungANet al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res.61 (16), 5998–6001 (2001).
- Dai JG , XiaoYB, MinJX, ZhangGQ, YaoK, ZhouRJ. Mitochondrial DNA 4977 BP deletion mutations in lung carcinoma. Indian J. Cancer43 (1), 20–25 (2006).
- Liu VWS , YangHJ, WangYet al. High frequency of mitochondrial genome instability in human endometrial carcinomas. Br. J. Cancer89 (4), 697–701 (2003).
- Wang Y , XueWC, LiuVWS, NganHYS. Detection of mosaic pattern of mitochondrial DNA alterations in different populations of cells from the same endometrial tumor. Mitochondrion7 (1–2), 171–175 (2007).
- Semczuk A , LorencA, PutowskiLet al. Clinicoprognostical features of endometrial cancer patients with somatic mtDNA mutations. Oncol. Rep.16 (5), 1041–1045 (2006).
- Pejovic T , LadnerD, IntenganMet al. Somatic D-loop mitochondrial DNA mutations are frequent in uterine serous carcinoma. Eur. J. Cancer40 (16), 2519–2524 (2004).
- Rahmioglu N , FassbenderA, VitonisAFet al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: III. Fluid biospecimen collection, processing, and storage in endometriosis research. Fertil. Steril.102 (5), 1233–1243 (2014).
- Fassbender A , RahmiogluN, VitonisAFet al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: IV. Tissue collection, processing, and storage in endometriosis research. Fertil. Steril.102 (5), 1244–1253 (2014).
- Becker CM , LauferMR, StrattonPet al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: I. Surgical phenotype data collection in endometriosis research. Fertil. Steril.102 (5), 1213–1222 (2014).
- Vitonis AF , VincentK, RahmiogluNet al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: II. Clinical and covariate phenotype data collection in endometriosis research. Fertil. Steril.102 (5), 1223–1232 (2014).
- American Society for Reproductive Medicine . Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril.67 (5), 817–821.
- King MP , AttardiG. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science246 (4929), 500–503 (1989).
- Brandon MC , LottMT, NguyenKCet al. MITOMAP: a human mitochondrial genome database–2004 update. Nucleic Acids Res.33, D611–613 (2005).
- Stratton P , WinkelC, PremkumarAet al. Diagnostic accuracy of laparoscopy, magnetic resonance imaging, and histopathologic examination for the detection of endometriosis. Fertil. Steril.79 (5), 1078–1085 (2003).
- Mettler L , SchollmeyerT, Lehmann-WillenbrockEet al. Accuracy of laparoscopic diagnosis of endometriosis. JSLS7 (1), 15–18 (2003).
- Falcone T , MaschaE. The elusive diagnostic test for endometriosis. Fertil. Steril.80 (4), 886–888 (2003).
- Wykes CB , ClarkTJ, KhanKS. Accuracy of laparoscopy in the diagnosis of endometriosis: a systematic quantitative review. BJOG111 (11), 1204–1212 (2004).
- Harel Z . Dysmenorrhea in adolescents and young adults: etiology and management. J. Pediatr. Adolesc. Gynecol.19 (6), 363–371 (2006).
- Casper RF . Introduction: new tools for enhancing collaborative endometriosis research. Fertil. Steril.102 (5), 1211–1212 (2014).
