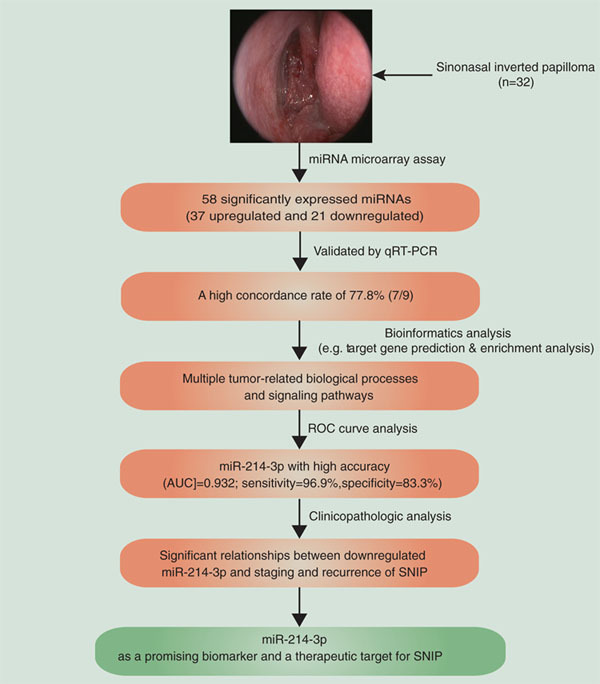
Abstract
Aim: To analyze the expression profile, diagnostic and clinicopathological significances of miRNAs in sinonasal inverted papilloma (SNIP). Materials & methods: The expression profile of miRNAs was analyzed using a miRNA microarray approach. The potential functions and clinical significances of specific miRNAs were further analyzed by bioinformatics and statistical methods. Results: The microarray assay identified 37 significantly upregulated and 21 downregulated miRNAs in SNIP. Of nine miRNAs randomly selected, the expression levels of seven miRNAs were confirmed by quantitative real-time PCR. The potential target genes of several candidate miRNAs were enriched in some biological processes and cellular signaling pathways related to tumorigenesis. Receiever operating characteristic curve analysis for miR-214-3p indicated an area under the curve of 0.932. Notably, its expression level was significantly decreased in SNIP tissues and associated with SNIP staging and recurrence. Conclusion: MiR-214-3p can possibly serve as a valuable biomarker and a therapeutic target for SNIP.
Graphical abstract
Sinonasal inverted papilloma (SNIP) is a benign epithelial neoplasm arising from the Schneiderian mucosa of nasal cavity and paranasal sinuses, accounting for 0.5–4% of all primary nasal tumors [Citation1]. The highest incidence of SNIP is observed in the fifth and sixth decades of life, with the male to female ratio of 2–5:1 [Citation2,Citation3]. Although SNIP is a histologically benign lesion, it is characterized by local invasiveness, a high recurrence rate and a tendency for malignant transformation [Citation4]. The exact etiologies of SNIP remain to be explored. Human papilloma virus (HPV) infection is generally recognized to be involved in the progression of SNIP. Other possible etiologies include allergy, chronic inflammation, smoking and occupational exposure [Citation5]. However, little is known about the underlying molecular genetic alterations and physiopathologic mechanism of this clinical entity.
MiRNAs are a class of small (approximately 22 nucleotides), single-stranded noncoding RNAs that negatively regulate post-transcriptional protein-coding gene expression through specifically binding to 3′ untranslated region of target mRNAs, leading to mRNA degradation or translation inhibition [Citation6,Citation7]. MiRNAs play important roles in various physiological and pathological processes, including individual development, tumorigenesis, immune responses and so on [Citation8–10]. Accumulating evidence indicates that some miRNAs affect the expression of genes and signaling pathways involved in the pathogenesis of head and neck tumors [Citation11–13]. Kakizaki et al. [Citation14] found that a total of 200 miRNAs exhibited a greater than twofold differential expression between SNIP and squamous cell carcinoma (SCC) arising from SNIP. Interestingly, miR-296-3p was thought to play a critical role in the malignant transformation of SNIP through the regulation of PTEN. However, genome-wide profiling, diagnostic and clinicopathological significances of miRNAs in SNIP appear to have received little attention.
In this study, to further elucidate the potential functions and clinical value of specific miRNAs, we characterized the miRNA expression profile of SNIP tissue with microarray analysis to distinguish the significantly upregulated and downregulated miRNAs. Based on the microarray data, we predicted the potential functions of the selected miRNAs and explored the associations between the altered miRNA expression and clinicopathological characteristics of SNIP by bioinformatics and statistical analysis. Together, our data lay the foundation for future functional and mechanism studies of SNIP-related miRNAs, and may provide novel targets for therapeutic intervention against SNIP.
Materials & methods
Patients & sample collection
SNIP tissue samples were obtained surgically from 32 patients with SNIP (26 males and six females; mean age 53.7 years; range: 32–85 years as the SNIP group). The diagnosis of SNIP was confirmed by histopathological examination. Detailed characteristics of these patients were summarized in . A total of 12 patients with only nasal septum deviation were selected to provide nasal mucosal tissue samples (nine males and three females; mean age 51.2 years; range: 28–64 years as the control group). Patients were admitted to the Departments of Otorhinolaryngology in Hangzhou First People’s Hospital, Nanjing Medical University and Second Affiliated Hospital, School of Medicine, Zhejiang University between 2012 and 2016. All tissue samples with clinical data were immediately preserved in RNAlater Solution (Ambion, TX, USA) after resection and then stored at -20°C until use. This study complied with the Declaration of Helsinki and was approved by the local ethics committee. Written informed consent was obtained at enrollment from each participating subject.
Table 1. Characteristics of 32 patients with sinonasal inverted papilloma.
RNA extraction
Total RNA was extracted from the tissue samples and purified using mirVana™ miRNA Isolation Kit without phenol (Ambion, TX, USA) following the manufacturer’s instructions. The RNA concentration was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, MA, USA). The integrity and quality of RNA was assessed by an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA), and the RNA samples with an RNA integrity number ≥6.0 and 28S/18S >0.7 were deemed acceptable to perform the miRNA microarray assay and reverse transcription.
MiRNA microarray assay
The RNA samples were analyzed at Shanghai Biotechnology Corporation (Shanghai, China) using the Agilent Human miRNA Microarray (8×60 K; version 21.0) with capture probes for a total of 2549 human miRNAs based on the Sanger miRBase database (release 21.0). Microarray assay was performed according to the manufacturer’s instructions. Briefly, the miRNA molecules in total RNA were labeled using miRNA Complete Labeling and Hyb Kit (Agilent Technologies). Each microarray slide was hybridized using 100 ng Cy3-labeled RNA in a hybridization oven (Agilent technologies, CA, USA). After hybridization, the slides were washed using Gene Expression Wash Buffer Kit (Agilent technologies), and then scanned using the Agilent Microarray Scanner (Agilent technologies) and Feature Extraction software program (version 10.7, Agilent technologies) with default settings. Raw data were normalized by Quantile algorithm, Gene Spring software program (version 12.6, Agilent technologies). A relative fold change >3 in the differential expression of miRNAs and a p-value < 0.01 were considered significant. The Gene Cluster (version 3.0) and Java Tree View software programs were used to perform the hierarchical cluster analysis of differentially expressed miRNAs and to visualize the miRNAs in the form of a heat map.
Quantitative real-time reverse transcription PCR
A cDNA synthesis was carried out using miScript II reverse transcription Kit (Qiagen, CA, USA). To validate the quantity of miRNA, quantitative real-time reverse transcription PCR (qRT-PCR) was performed on 7900 HT Sequence Detection System (ABI, CA, USA) with miRcute miRNA qPCR Detection Kit (SYBR Green, Tiangen, China) according to the manufacturer’s protocol. Forward primers specific to the mature miRNA from the Sanger miRNA database () were synthesized by Invitrogen (Shanghai, China). Expression analysis was carried out in triplicate for each sample. The endogenous control for normalization was U6 small RNA, and the expression levels of miRNA were calculated using the formula 2-ΔCT (ΔCt=CtmiRNA-CtU6).
Table 2. Primer sequences used for quantitative real-time reverse transcription PCR.
Enrichment analysis & network construction
The target geneswww.microrna.org for the significantly differentially expressed miRNAs were predicted using different algorithms, including TargetScan Human Release 7.1 (www.targetscan.org/), miRanda (www.microrna.org/) and PicTar (http://pictar.mdc-berlin.de/). As was previously described by Huang et al., the gene ontology (GO) and pathway analyses were conducted to annotate the functions of predicted miRNA targets using an online functional annotation tool (The Database for Annotation, Visualization and Integrated Discovery [DAVID] http://david.abcc.ncifcrf.gov/). GO terms and pathways with a p-value < 0.05 and a false discovery rate <0.05 were retained. The Cytoscape software was used to visualize and interpret the relationships between the GO, pathway and predicted target genes of interest for miRNA.
Statistical analysis
Statistical analyses were performed using the SPSS software for Windows (version 16.0; SPSS Inc., IL, USA). A two-tailed Student’s t-test was used to identify miRNAs that were differentially expressed between the SNIP and control groups. Pearson correlation analysis was performed to investigate the linear relationship between the microarray data and qRT-PCR results. Receiver operating characteristic (ROC) curve was established to evaluate diagnostic accuracy of miRNAs as biomarkers of SNIP. Fisher’s exact test was used to determine the relationships between clinicopathological characteristics and altered miRNA expression. p <0.05 was considered to indicate a statistically significant difference.
Results
MiRNA expression profiling in SNIP tissues
To identify differentially expressed miRNAs potentially involved in SNIP, we firstly examined the miRNAs expression profiles using the miRNA microarray technology and detected a total of 58 miRNAs that were significantly differentially expressed in SNIP tissues compared with the corresponding nontumorous tissues from the control group (fold change >3; p < 0.01). Of the 58 miRNAs, 37 were upregulated and 21 were downregulated in the SNIP group. Notably, miR-205-3p and miR-133a-3p were the most remarkably upregulated and downregulated miRNAs, respectively ( & ).
A hierarchical cluster analysis was performed with 58 differentially expressed miRNAs as compared the SNIP tissues from the SNIP group (n = 5) with nontumorous tissues from the control group (n = 5; fold change >3; p < 0.01). Each row indicates one miRNA, and each column indicates one sample. The miRNA expression levels are illustrated using a color key and histogram. The red and green bars denote high and low expression, respectively.
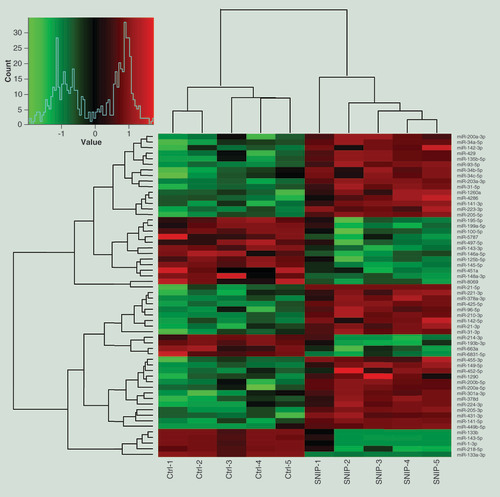
Table 3. List of differentially expressed miRNAs in sinonasal inverted papilloma tissues detected by miRNA microarray assay.
Validation of the candidate miRNAs by qRT-PCR
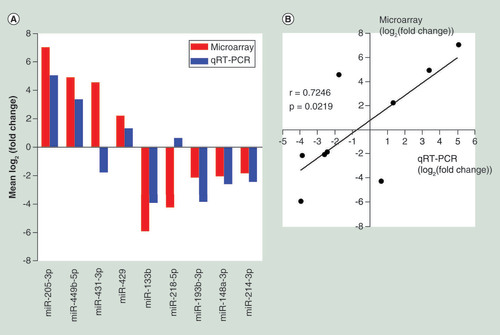
To confirm the microarray data, we performed qRT-PCR for nine candidate miRNAs randomly selected from the 58 differentially expressed miRNAs detected by the microarray experiment. A qRT-PCR was performed in two extend panel of the SNIP group (n = 32) and control group (n = 12). The qRT-PCR results for seven miRNAs (miR-205a-3p, miR-449b-5p, miR-429, miR-133b, miR-193b-3p, miR-148a-3p and miR-214-3p) were consistent with those from the microarray study, while the results for miR-431-3p and miR-218-5p were inconsistent, resulting in a concordance rate of 77.8% (7/9) (A). Pearson correlation analysis further indicated a positive correlation between the microarray data and qRT-PCR results (r = 0.7246, p = 0.0219) (B).
(A) Comparison of miRNA expression levels obtainedby miRNA microarray and qRT-PCR analysis. Upregulated and downregulated miRNAs are indicated by bars above and below the horizontal axis, respectively. (B) Pearson correlation analysis investigating the linear relationship between the microarray data and qRT-PCR results.
qRT-PCR: Quantitative real-time reverse transcription PCR.
Enrichment analysis of the potential targets of significantly upregulated and downregulated miRNAs
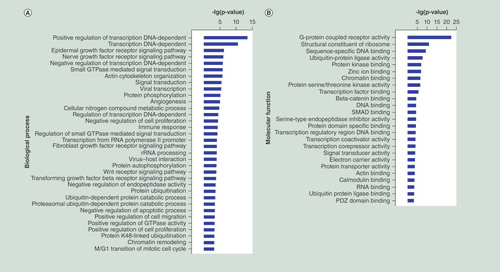
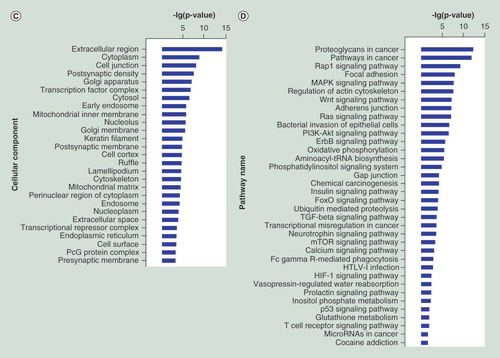
To explore the potential functions of the differentially expressed miRNAs in the progression of SNIP, we used three online databases (TargetScan, miRanda, and PicTar) to predict the target genes potentially regulated by these miRNAs. The functional enrichment (GO and pathway annotations) of these miRNA-regulated putative target genes were then analyzed. With GO analysis, we detected several significantly over-represented GO terms involved in biological process, molecular function and cellular component (p < 0.05; false discovery rate <0.05), including cell proliferation and apoptosis, such as positive regulation of cell proliferation and negative regulation of apoptotic process (A–C). Pathway analysis showed that a total of 36 significantly enriched pathways (p <0.05; false discovery rate <0.05). Many of these pathways were associated with the pathogenesis of tumors, such as Wnt signaling pathway, Rat sarcoma (Ras) signaling pathway and PI3K-Akt signaling pathway (D).
(A–C) GO functional enrichment of miRNA potential targets: (A) biological process, (B) molecular fuction, (C) cellular component. (D) Pathway annotation of the target genes of the miRNAs. Ordinate is the significant GO/pathway term (p < 0.05, FDR < 0.05).
Evaluation of miR-214-3p as a potential biomarker of SNIP
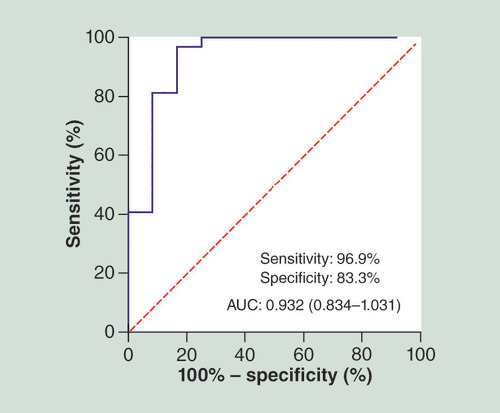
To verify whether miR-214-3p could discriminate the SNIP patients from controls, we performed qRT-PCR in two extended panel of the SNIP group (n = 32) and the control group (n = 12), and established ROC curve to determine its specificity and sensitivity as a diagnostic biomarker. ROC curve analysis revealed that miR-214-3p had a high accuracy in differentiating the SNIP patients from controls: area under the curve = 0.932; 95% CI: 0.834–1.031; sensitivity = 96.9%, specificity = 83.3% ().
Construction of a network reflecting the relationships between miR-214-3p, its predicated targets and associated GO & pathway terms
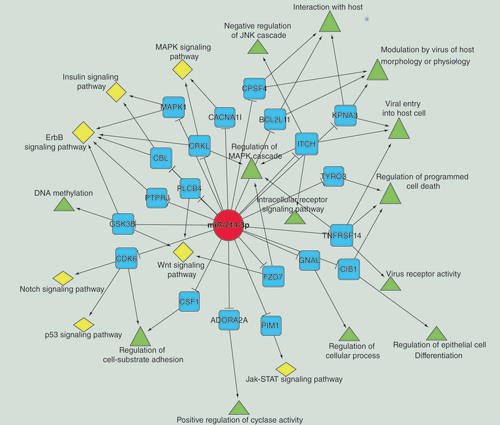
To further investigate the biological functions of significantly downregulated miR-214-3p in SNIP tissues, we constructed a network of miR-214-3P, its predicted targets and associated GO and pathway terms. As shown in , many genes, such as CDK6, BCL2L11, KPNA3 and ITCH, could be considered as the potential targets of miR-214-3p. These target genes were enriched in several GO and pathway terms linked to virus infection and pathogenesis of tumors, such as modulation by virus of host morphology or physiology, viral entry into host cell, p53 signaling pathway and Notch signaling pathway.
Relationships between clinicopathological characteristics & miR-214-3p expression
To determine the potential clinicopathological implications of the altered miR-214-3p expression, the real-time PCR results and clinicopathological characteristics from 32 patients with SNIP were analyzed using Fisher’s exact test. Clinicopathologic analysis revealed that the downregulation of miR-214-3p in SNIP was significantly related to tumor staging (p = 0.021) and recurrence (p = 0.038) ().
Table 4. Relationships between clinicopathological characteristics and miR-214-3p expression in patients with sinonasal inverted papilloma.
Discussion
Although SNIP is known to be a locally destructive, benign neoplasm of the nose and paranasal sinuses with a high tendency for recurrence and the possibility of malignant transformation, the underlying molecular mechanisms are still largely unknown. p53, DLEC1, gelsolin and caspase-3 have been reported to be abnormally expressed in SNIP tissue, some of which are possibly associated with the occurrence and development of SNIP [Citation15–17]. However, it has yet to be clarified whether and how the expression of these genes is fine-tuned at the post-transcriptional level. Such information will contribute to the development of preventative and therapeutic strategies against SNIP.
MiRNAs are a highly conserved class of short, noncoding RNAs that function as important post-transcriptional regulators, playing vital roles not only in regulating individual growth and development but also in tuning the expression of well-known genes involved in tumorigenesis [Citation9,Citation18]. Emerging evidence has indicated that several aberrantly expressed miRNAs are profoundly involved in the pathogenesis of head and neck neoplasms [Citation19]. As reported by Kakizaki et al. [Citation14], miR-296-3p was markedly upregulated in squamous cell carcinoma and might play a critical role in the malignant transformation of SNIP via the regulation of PTEN and the subsequent inhibition of the PI3K/Akt signaling pathway. To further elucidate the roles of miRNAs in the pathogenesis of SNIP, we studied the miRNA profiles in SNIP tissue using a microarray-based screening method and detected 58 candidate miRNAs (37 upregulated and 21 downregulated miRNAs) that had a threefold or greater change in expression. Nine of these miRNAs were then analyzed by qRT-PCR to confirm the reliability of the microarray data. We observed a high concordance rate of 77.8% and a positive correlation between the microarray data and qRT-PCR results.
The biological function of miRNAs is achieved mainly by manipulating the stability of mRNA and/or the initiation and progression of protein translation. Thus, the identification of miRNA target genes is considered to be a crucial step toward clarifying the miRNAs function. In our study, we intersected three prediction databases (TargetScan, miRanda and PicTar) in order to identify potential target genes of the differentially expressed miRNAs in SNIP tissues. These target genes can be classified into several functional categories by GO and pathway analyses based on the DAVID tools. DAVID is composed of an integrated biological knowledge base and analytic tools to understand biological meaning from large gene/protein lists. The procedure first requires uploading a gene list containing any number of common gene identifiers followed by analysis using one or more text and pathway-mining tools such as gene functional classification, functional annotation clustering, chart or table. Finally, investigators are able to obtain an in-depth understanding of the biological themes in lists of genes that are enriched in genome-scale studies. Interestingly, by following this protocol, we detected several genes that are involved in multiple tumor-related biological processes and signaling pathways, such as positive regulation of cell proliferation, Wnt signaling pathway and Ras signaling pathway. These findings suggest that the differentially expressed miRNAs detected in this study might participate in the pathogenic process of SNIP by regulating their target gene expression.
Recently, a growing number of studies have indicated the potential of using miRNAs as biomarkers in disease diagnoses and prognosis [Citation20–22]. For example, serum miR-378 has been suggested to be a noninvasive diagnostic biomarker for early detection of gastric cancer [Citation23]. Kovarikova et al. [Citation24] reported significantly upregulated miR-21 in sinonasal cancer tissue and sinonasal carcinoma patients and impaired survival of the patients with a high expression level of miR-21, suggesting that miR-21 could act as a valuable prognostic biomarker. Along with these clues, we investigated which of the differently expressed miRNAs in SNIP tissue could be used as diagnostic biomarkers. Our study showed that miR-214-3p could yield a ROC curve area of 0.932 with 96.9% sensitivity and 83.3% specificity in discriminating SNIP patients from controls. Thus, miR-214-3p probably provides great potential as a novel biomarker in molecular pathological diagnosis of SNIP. However, its specificity needs to be tested in depth between the SNIP and other nasal tumors. In addition, it remains to be further studied whether plasma/serum miR-214-3p can serve as a noninvasive diagnostic biomarker for early detection of SNIP.
As is well known, HPV infection is a definite risk factor for SNIP and plays an important role in the clinical evolution of SNIP [Citation25–27]. However, it is not yet clear whether miRNAs are involved in the pathogenesis of HPV-induced SNIP. Sannigrahi et al. [Citation28] demonstrated that HPV-16-mediated downregulation of miR-139-3p might promote oncogenesis in head and neck cancer by miR-139-3p targeting high-risk HPV-16 oncogenic proteins and revive major tumor suppressor proteins (p53, p21 and p16). Our bioinformatics data revealed that some of the putative target genes of miR-214-3p are related to the biological processes and mechanisms of virus infection, such as virus of host morphology or physiology and viral entry into host cells. Their relationships were visualized by the Cytoscape software, which is an open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data. In the working window of the Cytoscape software, you can import data files, such as expression profiles or GO annotations, generated by other applications or spreadsheet programs. These data can be mapped to node color, label, border thickness or border color, etc. according to user-configurable colors and visualization schemes.
In addition, we found that the characteristic expression of miR-214-3p was significantly associated with tumor staging and recurrence of SNIP. Given that SNIP is characterized by local invasiveness, tendency to recurrence, it is very necessary to detect the expression level of miR-214-3p in tissue samples from each patient with SNIP, which will contribute to evaluating the prognosis of SNIP patients. Therefore, our findings provided a possibility that miR-214-3p may serve as a gene expression regulator involved in HPV infection and oncogenesis of SNIP.
Conclusion
Our investigation, although preliminary, has revealed the potential biological functions of miRNAs in SNIP, providing new insight into the related molecular mechanism. We have further reported the strong correlation between the downregulation of miR-214-3p and tumor staging and recurrence of SNIP and identified miR-214-3p as a promising biomarker in SNIP. Such information has important clinical implications and would be helpful in identifying novel therapeutic targets for the treatment of SNIP patients.
Background
Sinonasal inverted papilloma (SNIP) is a histologically benign lesion. However, it is characterized by local invasiveness, a high recurrence rate and a tendency for malignant transformation.
Little is known about the underlying molecular genetic alterations and physiopathologic mechanism of this clinical entity.
Dysregulation of miRNAs has been implied in the progression of tumorigenesis. However, their role and clinicopathological significance in SNIP have yet to be identified.
MiRNA expression profiling in SNIP tissues
The miRNA microarray assay identified a total of 58 significantly expressed (37 upregulated and 21 downregulated) miRNAs in SNIP tissues.
The potential target genes of these significantly expressed miRNAs were involved in multiple tumor-related biological processes and signaling pathways, such as positive regulation of cell proliferation, Wnt signaling pathway, and Ras signaling pathway.
MiR-214-3p as a novel biomarker in SNIP
MiR-214-3p had high accuracy in differentiating the SNIP patients from controls: area under the curve = 0.932; 95% CI: 0.834–1.031; sensitivity = 96.9%, specificity = 83.3%.
Downregulation of miR-214-3p in SNIP was significantly related to tumor staging (p = 0.021) and recurrence (p = 0.038).
Conclusion
MiRNAs might participate in the pathogenic process of SNIP by regulating their target gene expression.
Our data might offer new ideas that miR-214-3p probably serves as a valuable biomarker and a therapeutic target for SNIP.
Ethical disclosure
This study complies with the Declaration of Helsinki and was approved by the Local Ethics Committee. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
YS Teng, ZH Lin and Y Li conceived and designed the experiments. ZH Lin, YQ Gao and XL Cao have made substantial contribution to collected tissue samples. YS Teng, FC Lin, XY Lou and YD Li performed the experiments and analyzed the data. YS Teng and YD Li prepared the figures and tables and drafted the manuscript. All authors read and approved the final manuscript.
Financial & competing interests disclosure
This work was supported by grants from Medical and Health Technology Program of Zhejiang (No: 2013RCA041), Science and Technology Development Project of Hangzhou (No: 20160533B09), Medical and Health Technology Program of Hangzhou (No: 2014A08), and Zhejiang Provincial Natural Science Foundation of China (No: LY17H130001). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Vrabec DP . The inverted Schneiderian papilloma: a 25-year study . Laryngoscope104 ( 5 Pt 1 ), 582 – 605 ( 1994 ).
- Lawson W , HoBT , ShaariCM , BillerHF . Inverted papilloma: a report of 112 cases . Laryngoscope105 ( 3 Pt 1 ), 282 – 288 ( 1995 ).
- Lawson W , PatelZM . The evolution of management for inverted papilloma: an analysis of 200 cases . Otolaryngol. Head Neck Surg.140 ( 3 ), 330 – 335 ( 2009 ).
- Krouse JH . Endoscopic treatment of inverted papilloma: safety and efficacy . Am. J. Otolaryngol.22 ( 2 ), 87 – 99 ( 2001 ).
- Govindaraj S , WangH . Does human papilloma virus play a role in sinonasal inverted papilloma?Curr. Opin. Otolaryngol.22 ( 1 ), 47 – 51 ( 2014 ).
- Ambros V . The functions of animal microRNAs . Nature431 ( 7006 ), 350 – 355 ( 2004 ).
- Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function . Cell116 ( 2 ), 281 – 297 ( 2004 ).
- Nannini M , RavegniniG , AngeliniSet al. miRNA profiling in gastrointestinal stromal tumors: implication as diagnostic and prognostic markers . Epigenomics7 ( 6 ), 1033 – 1049 ( 2015 ).
- Jansson MD , LundAH . MicroRNA and cancer . Mol. Oncol.6 ( 6 ), 590 – 610 ( 2012 ).
- Mattes J , CollisonA , FosterPS . Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation . Curr. Opin. Mol. Ther.10 ( 2 ), 150 – 157 ( 2008 ).
- Shen Z , QinX , YanMet al. Cancer-associated fibroblasts promote cancer cell growth through a miR-7-RASSF2-PAR-4 axis in the tumor microenvironment . Oncotarget8 ( 1 ), 1290 – 1303 ( 2017 ).
- Obayashi M , YoshidaM , TsunematsuTet al. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer . Oncotarget7 ( 7 ), 8223 – 8239 ( 2016 ).
- Mitra S , MukherjeeN , DasSet al. Anomalous altered expressions of downstream gene-targets in TP53-miRNA pathways in head and neck cancer . Sci. Rep.4 , 6280 ( 2014 ).
- Kakizaki T , HatakeyamaH , NakamaruYet al. Role of microRNA-296-3p in the malignant transformation of sinonasal inverted papilloma . Oncol. Lett.14 ( 1 ), 987 – 992 ( 2017 ).
- Altavilla G , StaffieriA , BusattoGet al. Expression of p53, p16INK4A, pRb, p21WAF1/CIP1, p27KIP1, cyclin D1, Ki-67 and HPV DNA in sinonasal endophytic Schneiderian (inverted) papilloma . Acta Otolaryngol.129 ( 11 ), 1242 – 1249 ( 2009 ).
- Chang PH , HuangCC , LeeTJet al. Downregulation of DLEC1 in sinonasal inverted papilloma and squamous cell carcinoma . J. Otolaryngol. Head Neck Surg.41 ( 2 ), 94 – 101 ( 2012 ).
- Cho JE , ParkW , KimDCet al. Down-regulation of gelsolin may play a role in the progression of inverted papilloma through an antiapoptotic mechanism . Am. J. Rhinol. Allergy26 ( 3 ), 177 – 182 ( 2012 ).
- Nohata N , HanazawaT , KinoshitaTet al. MicroRNAs function as tumor suppressors or oncogenes: aberrant expression of microRNAs in head and neck squamous cell carcinoma . Auris Nasus Larynx40 ( 2 ), 143 – 149 ( 2012 ).
- Koshizuka K , HanazawaT , AraiTet al. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma . Cancer Metastasis Rev.36 ( 3 ), 525 – 545 ( 2107 ).
- Shekari N , BaradaranB , ShanehbandiDet al. Circulating microRNAs: valuable biomarkers for diagnosis and prognosis of gastric cancer . Curr. Med. Chem.25 ( 6 ), 698 – 714 ( 2108 ).
- Yang Y , HuZ , ZhouYet al. The clinical use of circulating microRNAs as non-invasive diagnostic biomarkers for lung cancers . Oncotarget8 ( 52 ), 90197 – 90214 ( 2017 ).
- Ji X , TakahashiR , HiuraYet al. Plasma miR-208 as a biomarker of myocardial injury . Clin. Chem.55 ( 11 ), 1944 – 1949 ( 2009 ).
- Liu H , ZhuL , LiuBet al. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer . Cancer Lett.316 ( 2 ), 196 – 203 ( 2012 ).
- Kovarikova H , BubancovaI , LacoJet al. Deregulation of selected microRNAs in sinonasal carcinoma: value of miR-21 as prognostic biomarker in sinonasal squamous cell carcinoma . Head Neck39 ( 12 ), 2528 – 2536 ( 2017 ).
- Lin H , LinD , XiongX . Roles of HPV infection and Stathmin in the pathogenesis of sinonasal inverted papilloma . Head Neck38 ( 2 ), 220 – 224 ( 2014 ).
- Giotakis E , GomatosIP , AlevizosLet al. Apoptotic and proliferative status in HPV (+) and HPV (−) inverted papilloma patients. Correlation with local recurrence and clinicopathological variables . Pathol. Res. Pract.208 ( 6 ), 338 – 343 ( 2016 ).
- Scheel A , LinGC , McHughJBet al. Human papillomavirus infection and biomarkers in sinonasal inverted papillomas: clinical significance and molecular mechanisms . Int. Forum Allergy Rh.5 ( 8 ), 701 – 707 ( 2015 ).
- Sannigrahi MK , SharmaR , SinghVet al. Role of host miRNA Hsa-miR-139-3p in HPV-16-induced carcinomas . Clin. Cancer Res.23 ( 14 ), 3884 – 3895 ( 2017 ).