Abstract
Epigenetic mechanisms work in an orchestrated fashion to control gene expression in both homeostasis and diseases. Among small noncoding RNAs, piRNAs seem to meet the necessary requirements to be included in this epigenetic network due to their role in both transcriptional and post-transcriptional regulation. piRNAs and PIWI proteins might play important roles in cancer occurrence, prognosis and treatment as reported previously. Nevertheless, the potential clinical relevance of these molecules has yet been elucidated. A brief overview of piRNA biogenesis and their potential roles as part of an epigenetic network that is possibly involved in cancer is provided. Moreover, potential strategies based on the use of piRNAs and PIWI proteins as diagnostic and prognostic biomarkers as well as for cancer therapeutics are discussed.
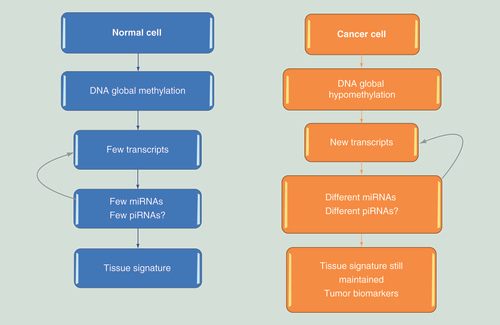
Small noncoding RNAs seem to work in epigenetic control by regulating mRNAs in a post-transcriptional fashion. The profile of miRNAs and piRNAs depends on the specificity of transcripts in each cell. In cancer cells, the shift to global hypomethylation modifies the transcripts and as a consequence, the piRNAs and miRNAs profiles. This specificity allows miRNA and piRNA tissue signatures to be identified, both in homeostasis and in diseases.
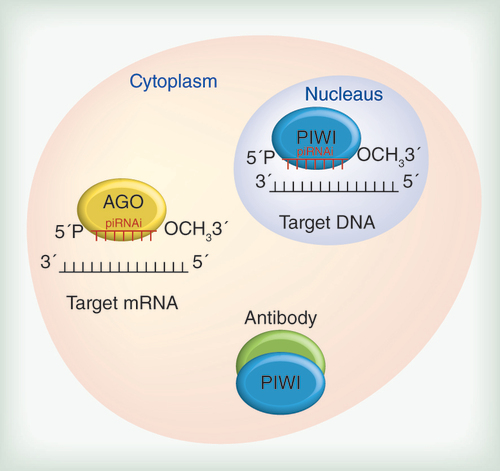
Based on the roles of piRNAs and PIWI proteins, these molecules may be excellent targets for cancer therapy. Antibodies against PIWI proteins could be useful at the post-translational level, whereas synthetic piRNAs could be applied in transcriptional and post-transcriptional approaches. When targeting DNA, the ‘piRNAi’ should contain the specific sequence to allow them to associate with the PIWI proteins. Conversely, when targeting mRNA, the ‘piRNAi’ should contain the specific sequence to allow them to associate with proteins of the Argonaute family.
Epigenetics as reversible controllers of gene expression patterns
In oncology, epigenetic alterations can serve as prognostic or diagnostic biomarkers and may play a role in therapy in the future [Citation1–3].
In contrast to genetics, epigenetics does not involve modifications of the genomic code but rather influence whether a specific gene is expressed. Every normal human cell contains the complete genomic information available for transcription, but instead of being expressed and resulting in proteins, the majority of genes in each cell are ‘turned off.’ Only a small set of genes is ‘turned on,’ which determines the specificity of differentiated tissues [Citation4–6].
Even when every gene is present, somatic cells express only a restricted, specific set of genes. In other words, a neuronal cell specifically expresses the transcripts necessary and sufficient to allow it to appear and function as a neuron, whereas a hepatocyte expresses a different set of genes that causes it to differentiate as a liver cell.
Importantly, the epigenetic marks, or codes, of each gene can be mitotically transmitted through generations, maintaining its expression profile, and these epigenetic marks are generationally transmitted, making these marks hereditable in a highly precise manner [Citation4,Citation7–9].
An important feature of epigenetic status is its reversibility. For most cells, epigenetics can be modified, and consequently, expressed genes can be silenced and vice versa [Citation10,Citation11]. Despite the complexity of the mechanisms involved in these fine regulations, under some circumstances, this process is reversible with various biological potential and clinical implications [Citation2].
Complex machinery is involved in epigenetic control, defining the accessibility to the transcription of each gene. Chemical modifications in DNA and histones together with noncoding RNAs establish a complex regulatory network that controls genome function. These epigenetic mechanisms modulate gene expression at both transcriptional and post-transcriptional levels [Citation12–16]. DNA methylation patterns, chromatin modifications and noncoding RNAs seem to work together in a redundant fashion of epigenetic regulation to create a complex regulatory network, including bidirectional or even multidirectional interactions among them [Citation13,Citation17].
Epigenetic dysregulation has important consequences for cell homeostasis and can contribute to the development of diseases. However, considering the reversibility of most epigenetic events, in the setting of diseases including cancer, each of these epigenetic regulators could be considered to be therapeutic targets that can alter pathological gene regulation and re-establish health status.
Cancer & epigenetics
In oncology, epigenetics have been shown to be involved in the development of cancer; they influence prognoses and therapeutic outcomes and represent a potential therapeutic option for many cancer types [Citation18–20].
Considering the classical concepts of genetic cancer mechanisms, such as increased oncogene activity and decreased tumor suppressor gene function, both could result from epigenetic abnormalities rather than ‘genetic alterations.’
Even with more recent identification of cancer cell hallmarks and capabilities [Citation21], it is currently recognized that each of these cancer features can also be achieved via epigenetic pathways. Thus, cancer epigenetics might lead to a myriad of possible clinical applications.
Nevertheless, these great ‘windows of opportunities’ remain almost unexplored. Except for DNA methylation, which usually represents an epigenetic silencer mark that has recently been added to the collection of medical information, including its recognition as a cause of some subtypes of cancers and a prognostic biomarker, little is known and much less is clinically implemented in the epigenetic field of cancer [Citation22].
Despite the current scenario of rare clinical applications of epigenetics in the cancer field, knowledge is increasing exponentially, although not in an integrative manner; that is, much scientific advancement is achieved as isolated views of the entire complex process. Although it is welcome because many patients will benefit, an integrative interpretation of the epigenetic network players seems to be much more promising.
Epigenetic alterations in cancer seem to be more accessible for medical intervention in many cancer scenarios than genetic alterations. In the last decade, drugs that modify chromatin or DNA methylation status have been used alone or in combination to affect cancer therapeutic outcomes [Citation23–26].
Recently, some strategies based on noncoding RNAs as cancer biomarkers and new creative therapeutic interventions have entered the clinical field and appear to have great potential for becoming part of routine clinical practice in the near future.
In this field, an almost unimaginable strategy for treating a cancer-related infection, hepatitis C, is most likely going to be utilized in clinical practice in the next few years. It was discovered that a synthetic small noncoding RNA seems to be highly efficient at treating hepatitis C, a leading cause of liver cancer [Citation27]. This opens new avenues of future investigations of many other cancer-related infections that are responsible for a broad spectrum of highly common human cancers.
Noncoding RNAs
Instead of coding for protein synthesis, noncoding RNAs exert the opposite function, either in the cytoplasm by regulating mRNAs usually in a negative manner that blocks protein synthesis or in the nucleus by binding to DNA, which can also result in gene silencing through a fascinating new mechanism. According to their size, these regulatory ncRNAs can be further classified as long ncRNAs (lncRNAs) (>200 bp) and small noncoding RNAs (sncRNAs) (<200 bp; miRNAs, siRNAs, piRNAs, scnRNAs, PASRs, TASRs, tiRNAs and diRNAs) [Citation17,Citation28–37].
Previously, lncRNAs have emerged as important players in biological regulation. These ncRNAs orchestrate the regulation of genetic information from DNA to RNA to protein by different mechanisms [Citation17]. More than 100,000 human lncRNA transcripts have been identified, but the majority of them remain almost unknown, and scientific information regarding their functions and roles in human diseases and biological phenomena is lacking [Citation38].
miRNAs are the most well-studied ncRNAs. Their mechanism of action is post-transcriptional; they interact with Argonaut proteins to form an RNA-induced silencing complex (RISC) and bind to complementary sequences of mRNAs, causing their inability to translate the protein or, less frequently, resulting in mRNA degradation [Citation39,Citation40].
Moreover, in vitro synthesis of miRNAs (interfering RNAs) allows for their utilization as therapeutic bullets to shut down target mRNAs, thus blocking the synthesis of specific proteins involved in disease. Similarly, if a specific required protein is scarce, in vitro sequences can block miRNAs that silence the scarce protein, thus allowing its translation resulting in the production of the desired protein by the target cell. Due to these and many other direct clinical applications, miRNAs are rapidly becoming familiar to both researchers and physicians [Citation41,Citation42].
Notably, there is a class of ncRNAs that direct de novo cytosine DNA methylation at the loci from which they are produced, in a process known as RNA-directed DNA methylation. In the RNA-directed DNA methylation pathway, transcripts from transposons and other repetitive elements are produced, presumably by Pol IV. These transcripts serve as templates for an RNA-dependent RNA polymerase to generate double-stranded RNAs that are processed into 24 nt siRNAs. The siRNAs are then associated with AGO4 to guide DNA methylation, resulting in transcriptional silencing of transposons as well as some genes that are adjacent to repeats [Citation43–46].
Another recently described ncRNA, piRNA, is a single-stranded (23–36 nt) sncRNA with a proposed specific function of interacting with PIWI proteins in early embryogenesis in germ cells and stem cells to silence transposable elements in the genome at the transcriptional level [Citation46,Citation47]. Nevertheless, the name “PIWI-interacting RNAs” does not define the complete set of activities of these small RNAs because piRNAs have recently been reported to play an important role in the control of genomic expression through different mechanisms [Citation47].
In this paper, a brief overview of piRNAs biogenesis and their potential roles as part of an epigenetic network that is possibly involved in cancer is provided. Moreover, potential strategies using piRNAs and PIWI proteins as diagnostic and prognostic biomarkers as well as for cancer therapeutics are discussed.
piRNAs
Studies on the biological function and possible clinical relevance of piRNAs are still in the beginning stages. There are many gaps to be filled regarding the understanding of biogenesis, and it is necessary to define the roles of piRNAs in epigenetic control.
Based on their origins, piRNAs can be divided into three groups: transposon-derived piRNAs, which are typically transcribed from both genomic strands and produce both sense and antisense piRNAs; mRNA-derived piRNAs, which are always sense to the mRNA from which they are processed and often originate from 39 UTRs; and lncRNAs-derived piRNAs, which produce piRNAs from the entire transcript. piRNA function is only well understood for transposon-derived piRNAs [Citation42,Citation48].
After transcription, piRNA primary transcripts (pri-piRNAs) are processed to mature piRNAs. It is not very clear how the putative precursors are processed into mature piRNAs, but two main routes have been described: the primary synthesis mechanism and the ‘ping-pong’ amplification mechanism [Citation42].
The primary synthesis relies on the transcription of small nucleotide sequences from clusters of piRNA genes by RNA polymerase II. After export to the cytoplasm, these transcripts are processed in smaller sequences and reach their main partner, the PIWI protein, to form a piRNA+PIWI complex. This complex migrates back to the nucleus and through complementary base pairing of piRNAs and DNA, it reaches its target gene and mobilizes silencer machinery to block the transcription of that target gene. In this way, piRNAs are transcriptional regulators that act mainly on transposable element sequences [Citation49,Citation50].
The second mechanism, known as ‘ping-pong,’ allows the production of many piRNAs in the cytoplasm. Instead of associating with PIWI proteins, piRNAs join with AGO3 or AUB proteins. piRNAs+Ago3 and piRNAs+Aub contain sequences that are complementary to each other. In this way, a piRNA+Ago complex targets and cuts a sequence of RNA that will result in a new RNA sequence that will function as a substrate for the formation of a new piRNA that is able to load an Aub protein. In the same way, the resulting piRNA+Aub protein complex will cut a complementary RNA sequence, resulting in the production of additional RNA substrates that form new piRNA+Ago3 complexes. Thus, the product of the piRNA cytoplasmic function is the substrate for an additional functional piRNA molecule, in a process based on an amplification mechanism [Citation47].
Similar to the formation of the miRNA silencer complex (RISC), piRNA post-transcriptional regulation (piRISC) occurs in the cytoplasm. The piRISC protects the integrity of the genome from invasion by ‘genomic parasites’ – transposable elements – by silencing them. Besides, transposons elements, mRNA and lncRNA also are targets of these piRNA complexes [Citation51].
It has become evident that the interaction of piRNAs with PIWI proteins in germ cells and stem cells during early development does not encompass the entire function of piRNAs. In brief, PIWI proteins are known to be mainly expressed in early development and in germ and stem cells, whereas piRNAs are expressed in many adult differentiated cells [Citation49,Citation52].
Currently, it is clear that piRNAs participate in post-transcriptional activities similar to miRNAs by interacting with other Argonaut proteins and silencing mRNAs in the cytoplasm [Citation47,Citation53]. It could explain the expression of piRNA in the soma but also raises new important issues about the possible interactions between these two classes of sncRNAs.
The new data being generated by many different approaches, including robust sequence technology, are helping to answer many questions about piRNAs. However, the data should be carefully analyzed to allow macro interpretation in addition to the specific goal of each investigation.
Profile of miRNAs & piRNAs as tissue signatures
The pattern of expression of miRNA and piRNAs seems to be part of an epigenetic network for controlling diverse biologic phenomena, including cancer. In summary, differentiated normal cells are characterized by global methylation, and specific areas are unmethylated, resulting in a restricted set of transcripts [Citation5,Citation46]. As a possible consequence, a small and specific set of miRNAs and piRNAs would be necessary to effect epigenetic regulation ().
According to this hypothesis, for a specific cell, only a restricted set of genes is active and is post-transcriptionally regulated by a few miRNAs and piRNAs [Citation54,Citation55]. This allows a definition of unique tissue signatures, based on the profile of miRNA and piRNAs expressed in a given tissue.
In cancer tissues, the shift to global hypomethylation and focal hypermethylation could imply different profiles of miRNA and piRNA expression, which also seem to potentially constitute cancer-specific signatures.
Strengthening these assertions, repetitive regions of the genome, including transposable elements that are usually methylated and therefore not transcribed in normal cells because they represent threats to genome integrity, become unmethylated in cancer cells [Citation15,Citation56], thus permitting their transcription. Based on this finding, additional piRNAs might be needed to exert epigenetic control to avoid dangerous expression of these repetitive elements.
Generally, the process of demethylation cited above occurs by inhibition of DNMTs or their targets, leading to a gradual loss of methylation during successive cell divisions in passive demethylation. Moreover, the active demethylation mechanisms include base excision repair and other companion proteins [Citation57].
As previously discussed, the miRNA and piRNA expression profiles appear to have organ and even tissue signatures, which seem to be relevant to the epigenetic control of differentiated cells [Citation54,Citation58]. Additionally, the profile of piRNAs in human saliva as well as miRNA profile data demonstrated robust reproducibility across individuals [Citation59]. In this scenario, bioinformatic analyses of piRNA profiles in diverse tissues could shed light on the role of piRNA as post-transcriptional regulators.
Despite the gaps in knowledge about piRNAs, these molecules seem to meet the requirements for being part of the epigenetic network, in addition to miRNAs, DNA methylation, chromatin modifications, lncRNAs and possibly many others.
Preliminary data from our team demonstrate that the diversity of expressed piRNAs increases from normal gastric mucosa to tumor samples, reaching an intermediate value in samples that are adjacent to tumor tissue, which may be as a consequence of new transcripts generated in the cancer process. Additionally, the highest expressed piRNAs in normal gastric mucosa are downregulated in cancer and in samples that are adjacent to cancer tissue, allowing the identification of cancer signatures based on piRNA profiles.
piRNAs as cancer biomarkers
piRNAs have been cited as potential biomarkers in many cancer types [Citation60–62]. Cheng et al. [Citation60] showed that the expression of piR-651 in gastric, colon, lung and breast cancer tissues was higher than that in paired noncancerous tissues. The upregulated expression of piR-651 was confirmed in several cancer cell lines including gastric, lung, mesothelium, breast, liver and cervical cancer cell lines. Moreover, the growth of gastric cancer cells was inhibited by a piR-651 inhibitor and arrested at the G2/M phase. Therefore, their findings suggest that piR-651 might be involved in the development of gastric cancer and other cancers and is a potential marker for cancer diagnosis.
In the same year, Cui et al. [Citation61] stated that piRNAs may be valuable biomarkers for detecting circulating gastric cancer cells. In their study, they observed that the levels of piR-651 and piR-823 in peripheral blood from patients with gastric cancer were significantly lower than those in healthy controls. Moreover, they demonstrated that piR-823 levels were positively associated with tumor-node-metastasis stage and distant metastasis.
Accordingly, Cheng et al. [Citation62] found that the expression level of piR-823 in gastric cancer tissues was significantly lower than that in noncancerous tissues. They also observed that after increasing the level of piR-823 in gastric cancer cells, cell growth was inhibited, suggesting that piR-823 is a putative biomarker for gastric cancer diagnosis.
Due to the possible role of piRNAs in various carcinogenesis steps, there are new promising opportunities in this field. Among these, an exciting possibility consists in application of piRNA investigations to provide early diagnosis, or even prevention of cancer development. This hypothesis is supported by preliminary analyses of piRNA profiles in samples adjacent to tumor tissue, showing a profile of expressed piRNAs that is more similar to cancer tissue pattern of expression than normal mucosa. In this way, piRNAs might be considered possible biomarkers of the cancer field effect.
According to the field effect theory, tissues near to a cancer already exhibit molecular abnormalities compared with normal tissues, although these tissues are not cancer according to classical pathological rules [Citation63]. Mainly, these molecular abnormalities might develop before the establishment of the complete set of cancer characteristics. The potential clinical benefit of identifying cancer risk marks in tissues that are considered as normal by conventional pathology seems to be very attractive.
Another possible application of piRNAs as cancer biomarkers results from the previously discussed potential tissue signatures based on the piRNA expression patterns. These tissue signatures might define prognostic marks, risk factors for specific types of metastasis or clinical features. Nevertheless, the promise of using piRNAs as biomarkers in clinical practice related to cancer depends on the confirmation of this hypothetical potential in large sample clinical investigations.
PIWI proteins in cancer
Given that many researchers acknowledge that PIWI proteins are expressed mainly in cancer tissues and stem cells and germ cells in adulthood [Citation49], these proteins could constitute an excellent target for cancer therapy compared with conventional treatments because most noncancer cells would not be affected. Side effects for nonreproductive patients should be minimal, considering that stem cells are not easily injured due to their numerous mechanisms of self-preservation [Citation59,Citation64–66]. Even possible undesirable effects regarding reproductive capability and stem cell populations could be reduced by applying preservation strategies for both cell types, such as freezing reproductive cells before treatment or performing bone marrow transplantation.
In humans, there are four expressed PIWI proteins: PIWIL1/HIWI, PIWIL2/HILI, PIWIL3 and PIWIL4/HIWI2 [Citation67]. Some of these proteins are related to specific types of cancer and seem to influence tumor aggressiveness and long-term survival [Citation68].
Several studies have demonstrated the potential of PIWIs as cancer diagnostic and prognostic biomarkers. It was recently demonstrated that HIWI plays an oncogenic role in hepatocellular carcinoma because the downregulation of this protein in hepatocellular carcinoma cell lines reduced the proliferation and migration capacities of these cells [Citation69].
In gastric cancer, the expression of PIWI increases progressively from preneoplastic lesions to advanced cancer, and silencing these proteins reduces the proliferation of gastric cancer cell lines [Citation70]. Additionally, investigations into the expression patterns of these proteins in gastric cancer tissues revealed an important prognostic factor because high expression levels seem to be strongly related to poor 5-year survival of gastric cancer patients [Citation68].
Recently, Yao et al. [Citation71] demonstrated that PIWIL2 facilitates the binding of NME2 to the promoter region of the MYC gene. PIWIL2 silencing significantly reduced tumor cell proliferation and colony formation in vitro and inhibited tumor growth in vivo [Citation72]. These observations could shed light on the mechanism of MYC overexpression, a common finding in diverse human cancers that is known to lead to cell proliferation, invasion and metastasis [Citation73–75]. This highlights the potential importance of PIWI proteins in carcinogenesis.
piRNAs plus PIWI interactions in cancer & potential clinical implications
In cancer research, many papers report increased expression of PIWI proteins in the cytoplasm of cancer cells compared with preneoplastic and normal cells [Citation70,Citation76–78]. Other investigators mention the expression patterns of specific piRNAs as possible biomarkers in some cancer types [Citation79,Citation80,Citation81,Citation82,Citation83]. Again, the fragmented data require integrative interpretation.
Most researchers do not investigate the role of piRNAs in conjunction with PIWI protein findings or with other noncoding RNA expression profiles. It is not clear whether PIWI protein expression in the cytoplasm constitutes an independent mechanism of proliferation and invasiveness that is not correlated with piRNA expression. Alternatively, PIWI proteins that remain in the cytoplasm should be taken to the nucleus by piRNAs to perform epigenetic control of homeostasis, and in this case, the lack of guiding piRNAs could favor PIWI accumulation in the cytoplasm of cancer cells. Despite the increasing knowledge regarding the role of piRNA/PIWI in cancer, it remains unknown whether PIWI protein expression drives tumorigenesis or constitute secondary events.
The role of piRNAs and PIWI proteins in cancer remits to the cancer stem cell hypothesis. Currently, the theory of cancer stem cells is gaining increasing importance for many cancer types [Citation82]. Accordingly, instead of beginning in a differentiated cell exposed to genetic and/or epigenetic alterations, the cancer origin relies on stem cell deregulation. Because piRNAs were discovered to play a role in stem cell regulation and PIWI proteins are expressed in normal stem cells and in cancer cells, a link between cancer stem cells, piRNA and PIWI proteins became an immediate possibility.
Because cancer stem cells seem to be implicated in cancer recurrence and metastasis potential, many attempts at treating cancer stem cells have been employed and have failed [Citation80]. Cancer stem cells remain a desired target for therapy; nevertheless, the current approach is ineffective. This opens new possibilities for the exploration of the potential of piRNA regulation of stem cells and the pattern of PIWI expression either in normal stem cells or in cancer stem cells as future innovative approaches.
New therapeutic approaches using piRNAs & PIWI proteins
Using synthetic piRNAs as bullets to block the synthesis of cancer related proteins by binding to mRNAs, as have been attempted with miRNAs, is an appealing application and might have the advantage of not requiring processing by enzymes such as Dicer, which is required by miRNAs. Additional speculative advantages of piRNAs over miRNAs include the possibility of targets with better specificity because each miRNA regulates several mRNAs and there is the potential to access undesirable lncRNAs with possible implications in the cancer process.
Even without a complete and clear understanding of the functions and interactions of piRNAs and PIWI proteins, many speculative approaches have been proposed in this field. Among these possibilities for intervention, blocking the proliferative effect of PIWI proteins should be achieved, at least in theory, in transcriptional, post-transcriptional and post-translational levels.
PIWI antibodies could have a clinical impact on cancer proliferation and could be involved as a post-translational approach in combinatory therapies for diverse cancers. As previously discussed, the specificity of the target might increase the response without increasing toxicity.
miRNA and piRNA sequences should be chosen for post-transcriptional silencing because, at least for miRNAs, the putative regulators of mRNA PIWI expression are already known [Citation84]. This approach is strengthened by the observation of reduced expression of one of these miRNAs (hsa-miR-483) in gastric cancer compared with nontumor gastric mucosa [Citation85].
At a glance, blocking the production of a harmful component seems to be better than antagonizing the undesired effects of an already functional molecule. In this way, transcriptional silencing is appealing. Regarding miRNAs, the synthetic sequences of piRNAs should be produced in vitro. Unlike miRNAs, and even unlike piRNA post-transcriptional mRNA inhibition, specific synthetic piRNAs designed to couple to PIWI proteins and exert genomic silencing on PIWI genes at a transcriptional level is a possible strategy. PIWI proteins, which are piRNA partners, are problematic, and the solution lies in their ‘interacting elements’ ().
This strategy is similar to the ‘ping-pong’ mechanism of piRNA biogenesis [Citation47,Citation52], although in a reverse manner. Instead of providing additional piRNAs to result in self-enhancement, it should result in blocking the production of the PIWI protein.
These therapeutic interventions could also be applied in combination, including with more conventional treatments; for example, PIWI antibodies could be used to deliver drugs to cancer cells, which is a delivery strategy that is already applied using other antibodies, thus decreasing the side effects of conventional cytotoxic drugs and possibly improving the response to therapy.
Conclusion & future perspectives
Due to both transcriptional and post-transcriptional regulatory functions, which are certainly not restricted to silencing transposable elements, piRNAs seem to provide new insights into cancer epigenetics and present great potential for future interventions in the course of diseases, including cancer.
piRNAs appear to be potential players in an epigenetic network, involving other noncoding RNAs, DNA methylation patterns and chromatin modifications that function together and in a redundant manner to effect epigenetic control of gene expression. The understanding of these complex interactions will provide many novel interventions either in biological and medical discovery or in clinical practice, improving the understanding and management of many diseases, including cancer.
Nevertheless, despite these preliminary ideas, integrative approaches for understanding and exploiting piRNA capacities are urgently needed. Public data banks are an effective tool for this task, but they are not sufficient. Scientists must share their data and collaborate with each other to develop new and robust knowledge to allow clinical applications in the medical and biological fields and, in particular, in cancer management.
Epigenetics as reversible controllers of gene expression patterns
Epigenetics determine whether a specific gene is expressed without modifying the genomic code.
Epigenetic marks of each gene are mitotically transmitted through generations, ensuring the expression profiles of each tissue to be maintained.
Epigenetics marks are transgenerationally transmitted, which allows these marks to be hereditable in a highly precise manner.
Cancer & epigenetics
Epigenetics influence prognosis and therapeutic outcomes and may represent a therapeutic option for many cancer types.
Strategies using noncoding RNAs as cancer biomarkers have been applied in the clinical field and appeared to have great potential for becoming part of routine clinical practice.
Noncoding RNAs
Regulatory ncRNAs can be further classified as long ncRNAs or small ncRNAs.
Diverse miRNAs have been shown to play roles in tumor occurrence and aggressiveness and to have potential for incorporation in clinical practice.
Recently, piRNA was integrated into the complex epigenetic machinery with an emphasis on cancer.
piRNAs
piRNAs interact with PIWI proteins during early embryogenesis in germ cells and stem cells to silence transposable elements in the genome at the transcriptional level.
piRNAs can share post-transcriptional activities by interacting with other Argonaut proteins and silencing mRNAs at the cytoplasmic level.
piRNAs plus PIWI proteins exhibit transcriptional activity, blocking mRNA synthesis.
Profile of miRNAs & piRNAs as tissue signatures
piRNAs might also participate in the epigenetic network, and similar to miRNAs, the piRNA profiles may also provide tissue signatures.
In cancer, a shift to global hypomethylation and focal hypermethylation could imply in a modification of the set of necessary piRNAs for employing an epigenetic control.
Preliminary data from our team demonstrated that the diversity of expressed piRNAs increases from normal gastric mucosa to tumor samples.
piRNAs as biomarkers in cancer
piRNAs have been cited as biomarkers in many cancer types.
An exciting possibility consists in application of piRNAs investigations to provide early diagnosis, or even prevention of cancer occurrence.
Another possible application of piRNAs as cancer biomarkers results from the previously discussed capacity of providing tissues signatures by piRNA expression patterns.
PIWI proteins in cancer
PIWI proteins are expressed mainly in cancer tissues, stem cells and germ cells in adulthood.
Several studies showed the potential of PIWI as cancer diagnostic and prognostic biomarkers.
piRNAs plus PIWI interactions in cancer & potential clinical implications
Most researchers do not investigate the role of piRNAs in conjunction with PIWI protein findings or with other noncoding RNA expression profiles.
The role of piRNAs and PIWI proteins in cancer remits to the cancer stem cell hypothesis.
piRNA regulation of stem cells and the pattern of PIWI expression either in normal stem cells or in cancer stems cells represent future innovative approaches.
New therapeutic approaches using piRNAs & PIWI proteins
The understanding of piRNA interactions will provide several novel interventions either in biological and medical discovery or in clinical practice, thus improving the understanding and management of cancer.
Acknowledgements
The authors would like to thank the reviewers for their helpful and insightful comments.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- Boumber Y , IssaJP . Epigenetics in cancer, what’s the future?Oncology (Williston Park)25 ( 3 ), 220 – 226 , 228 ( 2011 ).
- Tsai HC , BaylinSB . Cancer epigenetics, linking basic biology to clinical medicine . Cell Res.21 ( 3 ), 502 – 517 ( 2011 ).
- You JS , JonesPA . Cancer genetics and epigenetics, two sides of the same coin?Cancer Cell22 ( 1 ), 9 – 20 ( 2012 ).
- Sharma A , HeuckCJ , FazzariMJet al. DNA methylation alterations in multiple myeloma as a model for epigenetic changes in cancer . Wiley Interdiscip. Rev. Syst. Biol. Med.2 ( 6 ), 654 – 669 ( 2010 ).
- Jin B , LiY , RobertsonKD . DNA methylation, superior or subordinate in the epigenetic hierarchy?Genes Cancer2 ( 6 ), 607 – 617 ( 2011 ).
- Ong CT , CorcesVG . Enhancer function, new insights into the regulation of tissue-specific gene expression . Nat. Rev. Genet.12 ( 4 ), 283 – 293 ( 2011 ).
- Sharma S , KellyTK , JonesPA . Epigenetics in cancer . Carcinogenesis31 ( 1 ), 27 – 36 ( 2010 ).
- Feng S , JacobsenSE , ReikW . Epigenetic reprogramming in plant and animal development . Science330 , 622 – 627 ( 2010 ).
- Dawson MA , KouzaridesT . Cancer epigenetics, from mechanism to therapy . Cell150 ( 1 ), 12 – 27 ( 2010 ).
- Delcuve GP , RastegarM , DavieJR . Epigenetic control . J. Cell. Physiol.219 ( 2 ), 243 – 250 ( 2009 ).
- Choi JD , LeeJ . Interplay between Epigenetics and Genetics in cancer . Genomics Inform.11 ( 4 ), 164 – 173 ( 2013 ).
- Bird A . Perceptions of epigenetics . Nature447 ( 7143 ), 396 – 398 ( 2007 ).
- Sabin LR , DelásMJ , HannonGJ . Dogma derailed, the many influences of RNA on the genome . Mol. Cell49 ( 5 ), 783 – 794 ( 2013 ).
- Bernstein BE , MeissnerA , LanderES . The mammalian epigenome . Cell128 ( 4 ), 669 – 681 ( 2007 ).
- Baylin SB , JonesPA . A decade of exploring the cancer epigenome - biological and translational implications . Nat. Rev. Cancer11 ( 10 ), 726 – 734 ( 2011 ).
- Allis CD , MuirTW . Spreading chromatin into chemical biology . Chembiochem12 ( 2 ), 264 – 279 ( 2011 ).
- Yang G , LuX , YuanL . LncRNA, A link between RNA and cancer . Biochim. Biophys. Acta1839 ( 11 ), 1097 – 1109 ( 2014 ).
- Virani S , ColacinoJA , KimJH , RozekLS . Cancer epigenetics, a brief review . ILAR J.53 ( 3–4 ), 359 – 369 ( 2012 ).
- Chik F , SzyfM , RabbaniSA . Role of epigenetics in cancer initiation and progression . Adv. Exp. Med. Biol.720 , 91 – 104 ( 2011 ).
- Gigek CO , ChenES , CalcagnoDQ , WisnieskiF , BurbanoRR , SmithMA . Epigenetic mechanisms in gastric cancer . Epigenomics4 ( 3 ), 279 – 294 ( 2012 ).
- Hanahan D , WeinbergRA . Hallmarks of cancer, the next generation . Cell144 ( 5 ), 646 – 674 ( 2011 ).
- Hinoue T , WeisenbergerDJ , LangeCPet al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer . Genome Res.22 ( 2 ), 271 – 282 ( 2012 ).
- Azad N , ZahnowCA , RudinCM , BaylinSB . The future of epigenetic therapy in solid tumours – lessons from the past . Nat. Rev. Clin. Oncol.10 ( 5 ), 256 – 266 ( 2013 ).
- Jones PA , TaylorSM . Cellular differentiation, cytidine analogs and DNA methylation . Cell20 , 85 – 93 ( 1980 ).
- Turcan S , FabiusAW , BorodovskyAet al. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT InhibitorDecitabine . Oncotarget4 ( 10 ), 1729 – 1736 ( 2013 ).
- Borodovsky A , SalmasiV , TurcanSet al. 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft . Oncotarget4 ( 10 ), 1737 – 1747 ( 2013 ).
- Fan HX , TangH . Complex interactions between microRNAs and hepatitis B/C viruses . World J. Gastroenterol.20 ( 37 ), 13477 – 13492 ( 2014 ).
- Pauli A , RinnJL , SchierAF . Non-coding RNAs as regulators of embryogenesis . Nat. Rev. Genet.12 ( 2 ), 136 – 149 ( 2011 ).
- Iyengar BR , ChoudharyA , SarangdharMA , VenkateshKV , GadgilCJ , PillaiB . Non-coding RNA interact to regulate neuronal development and function . Front. Cell. Neurosci.8 , 47 ( 2014 ).
- Place RF , NoonanEJ . Non-coding RNAs turn up the heat, an emerging layer of novel regulators in the mammalian heat shock response . Cell Stress Chaperones19 ( 2 ), 159 – 172 ( 2014 ).
- Volders PJ , VerheggenK , MenschaertGet al. An update on LNCipedia, a database for annotated human lncRNA sequences . Nucleic Acids Res.43 , D174 – D180 ( 2014 ).
- Felekkis K , TouvanaE , StefanouC , DeltasC . MicroRNAs, a newly described class of encoded molecules that play a role in health and disease . Hippokratia14 ( 4 ), 236 – 240 ( 2010 ).
- Mochizuki K , GorovskyMA . Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement . Genes Dev.18 , 2068 – 2073 ( 2004 ).
- Yao MC , ChaoJL . RNA-guided DNA deletion in Tetrahymena, an RNAi-based mechanism for programmed genome rearrangements . Annu. Rev. Genet.39 , 537 – 559 ( 2005 ).
- Kapranov P , ChengJ , DikeSet al. RNA maps reveal new RNA classes and a possible function for pervasive transcription . Science316 , 1484 – 1488 ( 2007 ).
- Taft RJ , GlazovEA , CloonanNet al. Tiny RNAs associated with transcription start sites in animals . Nat. Genet.41 , 572 – 578 ( 2009 ).
- Wei W , BaZ , GaoMet al. A role for small RNAs in DNA double-strand break repair . Cell149 ( 1 ), 101 – 12 ( 2012 ).
- Phuah NH , NagoorNH . Regulation of microRNAs by natural agents, new strategies in cancer therapies . Biomed. Res. Int. 2014 , 804510 ( 2014 ).
- Ha T . MicroRNAs in human diseases, from cancer to cardiovascular disease . Immune Netw.11 ( 3 ), 135 – 154 ( 2011 ).
- Paulmurugan R . MicroRNAs – a new generation molecular targets for treating cellular diseases . Theranostics3 ( 12 ), 927 – 929 ( 2013 ).
- Watanabe T , LinH . Post-transcriptional regulation of gene expression by Piwi proteins and piRNAs . Mol. Cell56 ( 1 ), 18 – 27 ( 2014 ).
- Weick EM , MiskaEA . piRNAs, from biogenesis to function . Development141 ( 18 ), 3458 – 3471 ( 2014 ).
- Zheng X , ZhuJ , KapoorA , ZhuJK . Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing . EMBO J.26 , 1691 – 1701 ( 2007 ).
- Matzke M , KannoT , DaxingerL , HuettelB , MatzkeAJ . RNAmediated chromatin-based silencing in plants . Curr. Opin. Cell Biol.21 , 367 – 376 ( 2009 ).
- Matzke MA , MosherRA . RNA-directed DNA methylation, an epigenetic pathway of increasing complexity . Nat. Rev. Genet.15 ( 6 ), 394 – 408 ( 2014 ).
- Wu L , ZhouH , ZhangQet al. DNA methylation mediated by a microRNA pathway . Mol. Cell38 ( 3 ), 465 – 475 ( 2010 ).
- Ross RJ , WeinerMM , LinH . PIWI proteins and PIWI-interacting RNAs in the soma . Nature505 ( 7483 ), 353 – 359 ( 2014 ).
- Thomson T , LinH . The biogenesis and function of PIWI proteins and piRNAs: progress and prospect . Annu. Rev. Cell. Dev. Biol.25 , 355 – 376 ( 2009 ).
- Han BW , ZamorePD . piRNAS . Curr. Biol.24 ( 16 ), 730 – 733 ( 2014 ).
- Luteijn MJ , KettingRF . PIWI-interacting RNAs: from generation to transgenerational epigenetics . Nat. Rev. Genet.14 ( 8 ), 523 – 534 ( 2013 ).
- Siomi MC , SatoK , PezicDet al. PIWI-interacting small RNAs: the vanguard of genome defence . Nat. Rev. Mol. Cell Biol.12 ( 4 ), 246 – 258 ( 2011 ).
- Rajasethupathy P , AntonovI , SheridanRet al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity . Cell149 ( 3 ), 693 – 707 ( 2012 ).
- Wei C , SalichosL , WittgroveCM , RokasA , PattonJG . Transcriptome-wide analysis of small RNA expression in early zebrafish development . RNA18 ( 5 ), 915 – 929 ( 2012 ).
- Guo Z , MakiM , DingRet al. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues . Sci. Rep.4 , 5150 ( 2014 ).
- Esteller M . Cancer epigenomics, DNA methylomes and histone-modification maps . Nat. Rev. Genet.8 ( 4 ), 286 – 298 ( 2007 ).
- Siddiqi S , MatushanskyIJ . Piwis and piwi-interacting RNAs in the epigenetics of cancer . Cell. Biochem.113 ( 2 ), 373 – 380 ( 2012 ).
- Zhang G , PradhanS . Mammalian epigenetic mechanisms . IUBMB Life66 ( 4 ), 240 – 256 ( 2014 ).
- Moreira FC , AssumpçãoM , HamoyIGet al. MiRNA expression profile for the human gastric antrum region using ultra-deep sequencing . PLoS ONE9 ( 3 ), e92300 ( 2014 ).
- Kvinlaug BT , HuntlyBJ . Targeting cancer stem cells . Expert Opin. Ther. Targets11 ( 7 ), 915 – 927 ( 2007 ).
- Cheng J , GuoJM , XiaoBXet al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells . Clin. Chim. Acta412 ( 17–18 ), 1621 – 1625 ( 2011 ).
- Cui L , LouY , ZhangXet al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as marks . Clin. Biochem.44 ( 13 ), 1050 – 1057 ( 2011 ).
- Cheng J , DengH , XiaoBet al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells . Cancer Lett.315 ( 1 ), 12 – 17 ( 2012 ).
- Slaughter DP , SouthwickHW , SmejkalW . Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin . Cancer6 , 963 – 968 ( 1953 ).
- Li L , BhatiaR . Stem cell quiescence . Clin. Cancer Res.17 ( 15 ), 4936 – 4941 ( 2011 ).
- Wollenberg B . Implication of stem cells in the biology and therapy of head and neck cancer . GMS Curr. Top. Otorhinolaryngol. Head Neck Surg.10 , Doc01 , doi:10.3205/cto000074 ( 2011 ) ( Epub ahead of print ).
- Shukla S , MeeranSM . Epigenetics of cancer stem cells, pathways and therapeutics . Biochim. Biophys. Acta1840 ( 12 ), 3494 – 3502 ( 2014 ).
- Sasaki T , ShiohamaA , MinoshimaS , ShimizuN . Identification of eight members of the Argonaute family in the human genome . Genomics82 ( 3 ), 323 – 330 ( 2003 ).
- Wang Y , LiuY , ShenXet al. The PIWI protein acts as a predictive marker for human gastric cancer . Int. J. Clin. Exp. Pathol.25 ( 4 ), 315 – 325 ( 2012 ).
- Xie Y , YangY , JiD , ZhangD , YaoX , ZhangX . Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells . Mol. Med. Rep.11 ( 2 ), 1455 – 1461 ( 2015 ).
- Liu X , SunY , GuoJet al. Expression of Hiwi gene in human gastric cancer was associated with proliferation of cancer cells . Int. J. Cancer118 , 1922 – 1929 ( 2006 ).
- Yao Y , LiC , ZhouXet al. PIWIL2 induces c-Myc expression by interacting with NME2 and regulates c-Myc-mediated tumor cell proliferation . Oncotarget5 ( 18 ), 8466 – 8477 ( 2014 ).
- Taubert H , GreitherT , KaushalDet al. Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma . Oncogene26 , 1098 – 1100 ( 2007 ).
- Calcagno DQ , LealMF , AssumpcaoPP , SmithMA , BurbanoRR . MYC and gastric adenocarcinoma carcinogenesis . World J. Gastroenterol.14 ( 39 ), 5962 – 5968 ( 2008 ).
- Calcagno DQ , FreitasVM , LealMFet al. MYC, FBXW7 and TP53 copy number variation and expression in gastric cancer . BMC Gastroenterol.13 , 141 ( 2013 ).
- de Souza CR , LealMF , CalcagnoDQet al. MYC deregulation in gastric cancer and its clinicopathological implications . PLoS ONE8 ( 5 ), e64420 ( 2013 ).
- Qiao D , ZeemanAM , DengW , LooijengaLH , LinH . Molecular characterization of Hiwi, a human member of the PIWI gene family whose overexpression is correlated to seminomas . Oncogene21 , 3988 – 3999 ( 2002 ).
- Lee JH , SchütteD , WulfGet al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway . Hum. Mol. Genet.15 , 201 – 211 ( 2006 ).
- Zhao YM , ZhouJM , WangLRet al. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection . Cancer118 , 2708 – 2717 ( 2012 ).
- Lim SL , RicciardelliC , OehlerMK , TanIM , RussellD , GrütznerF . Overexpression of piRNA pathway genes in epithelial ovarian cancer . PLoS ONE9 ( 6 ), e99687 ( 2014 ).
- Cheng J , GuoJM , XiaoBXet al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells . Clin. Chim. Acta412 ( 17–18 ), 1621 – 1615 ( 2011 ).
- Huang G , HuH , XueXet al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer . Clin. Transl. Oncol.15 ( 7 ), 563 – 568 ( 2013 ).
- Sebastian C . Tracking down the origin of cancer, metabolic reprogramming as a driver of stemness and tumorigenesis . Crit. Rev. Oncog.19 ( 5 ), 363 – 382 ( 2014 ).
- Kasai T , ChenL , MizutaniAet al. Cancer stem cells converted from pluripotent stem cells and the cancerous niche . J. Stem Cells Regen. Med.10 ( 1 ), 2 – 7 ( 2014 ).
- TargetScanHuman . Prediction of microRNA targets . www.targetscan.org .
- Assumpção MB , MoreiraFC , HamoyIGet al. High-throughput miRNA sequencing reveals a field effect in gastric cancer and suggests an epigenetic network mechanism . Bioinform. Biol. Insights ( 2015 ). ( In Press ).
