Abstract
Aim: Biomarker testing detects actionable driver mutations to inform first-line treatment in advanced non-small-cell lung cancer (aNSCLC) and metastatic colorectal cancer (mCRC). This study evaluated biomarker testing in a nationwide database (NAT) versus the OneOncology (OneOnc) community network. Patients & methods: Patients with aNSCLC or mCRC with ≥1 biomarker test in a de-identified electronic health record–derived database were evaluated. OneOnc oncologists were surveyed. Results: Biomarker testing rates were high and comparable between OneOnc and NAT; next-generation sequencing (NGS) rates were higher at OneOnc. Patients with NGS versus other biomarker testing were more likely to receive targeted treatment. Operational challenges and insufficient tissue were barriers to NGS testing. Conclusion: Community cancer centers delivered personalized healthcare through biomarker testing.
Plain language summary
Improving biomarker testing in advanced non-small-cell lung cancer and metastatic colorectal cancer: experience from a large community oncology network in the USA What is this article about? Cancer therapies often work better in certain subgroups of patients. Tumors may have characteristics that can predict which therapies may be more likely to work. These cancer biomarkers may be identified by special testing, such as next-generation sequencing (NGS). If a biomarker is detected, the patient can potentially be treated with medicine that targets that biomarker. This study looked at biomarker testing of lung and colon cancers in two community cancer practices (OneOncology [OneOnc] and nationwide database [NAT]). What were the results? The biomarker testing rates were high (≥81%) and similar between OneOnc and NAT. NGS testing rates were higher at OneOnc than at NAT (58 vs 49% for non-small-cell lung cancer, 55 vs 42% for metastatic colorectal cancer [mCRC]), suggesting the success of OneOnc’s networkwide educational, pathway and operational programs. NGS testing was lower in community practices due to operational challenges and insufficient tissue collection. Patients who had NGS versus other biomarker testing were more likely to receive treatment specifically for that biomarker. However, some patients started treatment before their biomarker results were reported, usually because of their disease and a long wait time for biomarker test results. What do the results of the study mean?Community cancer centers can treat patients with targeted medicine based on biomarker testing results. There are opportunities to increase the number of patients getting NGS testing, shorten turnaround times and reduce the number of patients who start treatment before getting their biomarker test results.
Tweetable abstract
Community cancer centers deliver biomarker testing to inform 1L targeted treatment for the actionable oncogene mutations detected in aNSCLC and mCRC. Higher NGS testing rates at OneOnc versus Flatiron Health reflect the success of OneOnc’s educational and operational initiatives.
Graphical abstract
Biomarker testing is critical in identifying actionable oncogenic mutations and selecting appropriate biomarker-driven targeted therapies that are approved by the US FDA [Citation1–3]. This is especially relevant for the treatment of advanced non-small-cell lung cancer (aNSCLC) and metastatic colorectal cancer (mCRC), for which there are several actionable driver mutations with established targeted therapies as first-line (1L) treatments [Citation4,Citation5]. Fueled by technological advances in the field of next-generation sequencing (NGS) that have driven faster and more cost-effective solutions, the biomarker landscape has evolved in recent years from single-gene testing to comprehensive genomic profiling to whole exome/genome sequencing [Citation6,Citation7]. These advances aim to enable timely and precise identification of actionable mutations to aid in treatment decisions to deliver on personalized healthcare (PHC) [Citation8]. Several policies have been implemented to foster the use of NGS testing in routine clinical care, including the Centers for Medicare and Medicaid Services national coverage determination for initial and further NGS testing in patients with solid tumors and a companion diagnostic claim in 2018 [Citation9] and in patients with clinical risk factors of germline breast or ovarian cancer in 2020 [Citation10].
Approximately 85% of cancer care in the USA is delivered at community centers [Citation11]. OneOncology (OneOnc) [Citation12] is a network of community oncology practices that was formed in 2018 with the goal of providing state-of-the-art cancer care to patients close to home. The practices provide clinical research services, use a single electronic health record (EHR) system with embedded care pathways determined by physician leaders, and share operational procedures. One major clinical initiative is the promotion of precision oncology to deliver PHC. To achieve this goal, educational programs for physicians and support staff were developed and disseminated across the practices. Systems were built into the EHR to support appropriate biomarker testing for different diseases and disease stages. The results of test monitoring and feedback were shared with individual physicians and practices to promote continuous quality improvement.
This study aims to leverage EHR data to quantify the recent adoption and utility of biomarker testing to direct treatment for patients with aNSCLC or mCRC in a predominately community care setting; to share experiences with programs to foster PHC among the OneOnc community network; and to discuss opportunities to optimize PHC.
Patients & methods
We conducted a mixed methods study. Quantitative insights were generated from a retrospective cohort analysis of the Flatiron Health database. The Flatiron Health database is a longitudinal database, comprising de-identified patient-level structured and unstructured data, curated via technology-enabled abstraction [Citation13,Citation14]. During the study period, the de-identified data originated from approximately 280 USA cancer clinics (∼800 sites of care). Institutional review board approval of the study protocol for data collection from the real-world cohort was obtained prior to study conduct and included a waiver of informed consent. Studies were conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, Council for International Organizations of Medical Sciences, Belmont Report, US Common Rule). Qualitative insights were obtained from a survey distributed via SurveyMonkey to 200 oncologists practicing in the OneOnc network in April 2022, followed by two email reminders before survey closure.
Study Cohorts
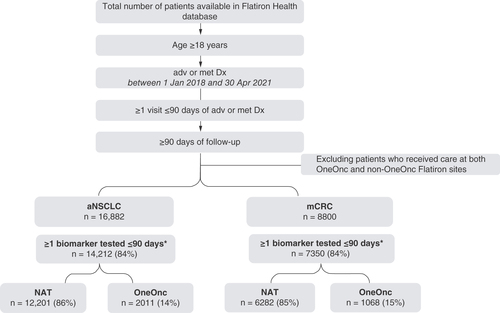
Patients in the Flatiron Health database who (i) were aged ≥18 years; (ii) had a primary diagnosis of stage IIIB or IV aNSCLC (International Classification of Diseases, Tenth Revision [ICD-10] C34x or C39.9) or stage IV mCRC (ICD-10 C18x, C19x, C20x, or C21x); (iii) were diagnosed between 1 January 2018 and 30 April 2021; (iv) had ≥1 visit ≤90 days after advanced or metastatic diagnosis and (v) had ≥90 days of follow-up until the study end date of 31 July 2021 were included in this study. Patients receiving care at both OneOnc and Flatiron Health Nationwide (NAT, excluding OneOnc) were excluded. The patients who met the study criteria were stratified into two cohorts: (i) the OneOnc cohort, which comprised a sample of patients from selected OneOnc sites (West Cancer Center & Research Institute, Tennessee Oncology, Eastern Connecticut Hematology and Oncology [ECHO], New York Cancer and Blood Specialists [NYCBS], Los Angeles Cancer Network [LACN] and Astera Cancer Care); and (ii) the NAT cohort.
Variables
Biomarker test types were categorized as (i) NGS tests and (ii) non-NGS or other biomarker tests. The NGS tests were found by searching for keywords related to testing technology platform (e.g., Illumina, HiSeq), NGS testing providers (e.g., FoundationOneCDx, Caris Life Science) and physician notes calling out the term “NGS”. The non-NGS tests were primarily defined as tests performed by non-NGS techniques, including, but not limited to, fluorescence in situ hybridization, immunohistochemistry and polymerase chain reaction. At the time of data abstraction, a selected list of actionable biomarkers recommended by the National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology (NCCN Guidelines®) [Citation4,Citation5] were included: ALK, BRAF, EGFR, PD-L1, KRAS and ROS-1 for aNSCLC; and BRAF, HER2, deficient DNA mismatch repair/microsatellite instability (dMMR/MSI), NRAS and KRAS for mCRC.
The survey solicited OneOnc oncologists’ input on the following: (i) factors that impact their decision to order NGS tests across tumor types in clinical practice; (ii) barriers that prevent completion of NGS testing for patients; and (iii) reasons why some patients received treatment before test results were available (Supplementary Table 1).
Outcomes
Adoption of biomarker testing was evaluated from initial diagnosis to ≤90 days after advanced/metastatic diagnosis, including rates of use of ≥1 biomarker test and NGS testing by year of advanced/metastatic diagnosis; the rate of testing uptake by calendar year over time; and turnaround time, defined as the time from advanced/metastatic diagnosis to the first test result.
The utility of biomarker testing was evaluated from initial diagnosis to ≤90 days after advanced/metastatic diagnosis to inform first-line biomarker-driven treatment in aNSCLC with ≥1 actionable oncogene mutation (ALK, BRAF, EGFR, ROS-1); aNSCLC with ≥1 oncogene and/or PD-L1 positivity (staining ≥1%); mCRC with BRAF v600 mutation and/or MMR/MSI-H; and mCRC with RAS mutation, including KRAS and NRAS. Since the reason for change in treatment was not included in the Flatiron Heath database, a proxy of switching treatment within 49 days (i.e., 7 weeks) from treatment initiation was used as a priori evidence of a biomarker-driven decision, based on the fact that for a typical practice, the minimum amount of time from start of treatment to imaging-derived change would be approximately 49 days.
A list of prespecified options was available for selection in the survey (Supplementary Table 1).
Statistical Analysis
Descriptive analysis of biomarker testing patterns, patient characteristics and survey results was conducted. The χ2 test was used to compare proportions in the OneOnc and NAT cohorts. Linear probability regression models were used to evaluate testing rates over time, investigating OneOnc and NAT separately by regressing the binary testing variable on time (days) and determining differences in rates between OneOnc and NAT by interacting time and a binary variable for OneOnc or NAT. The statistical significance threshold was set a priori using two-sided tests at p < 0.05. All analyses were conducted using R version 4.1.2 [Citation15].
Results
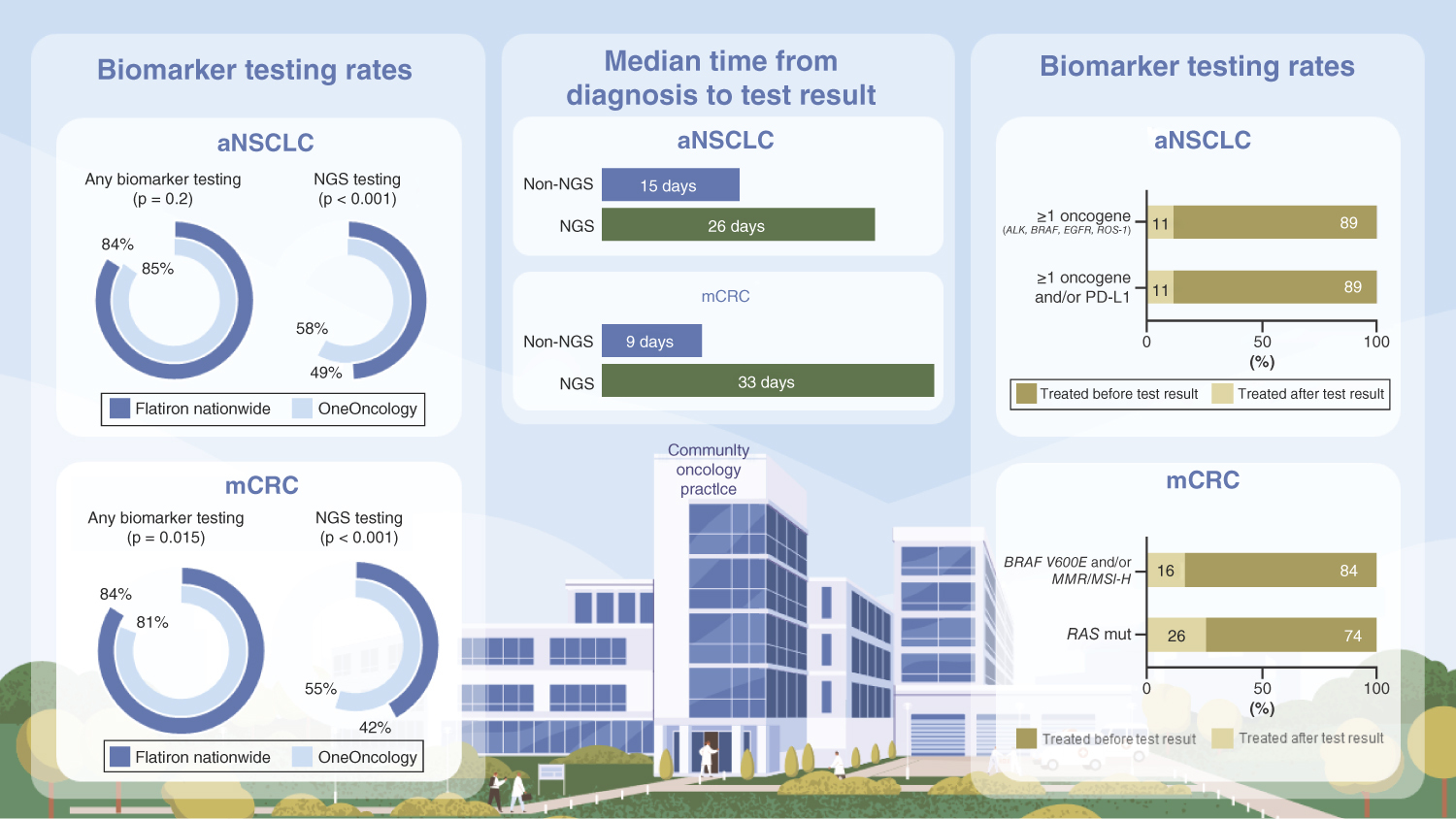
Of the Flatiron Health patients who met the inclusion criteria, 14,212 of 16,882 (84%) with aNSCLC and 7350 of 8800 (84%) with mCRC received ≥1 biomarker test ().
*Any biomarker testing up to 90 days after advanced or metastatic diagnosis. To minimize selection bias, the database follows a selection process: broad cohort selection based on ICD codes for the disease of interest (ICD-9 162.x or ICD-10 C34x or C39.9 for aNSCLC; ICD-9 153.x or 154.x or ICD-10 C18x, C19x, C20x or C21x for mCRC), followed by a proportion of patients probabilistically sampled and confirmed via manual review on their selection criteria to be included in the final cohort for analysis in the database.
adv: Advanced; aNSCLC: Advanced non-small-cell lung cancer; Dx: Diagnosis; mCRC: Metastatic colorectal cancer; met: Metastatic; NAT: Flatiron Health Nationwide, excluding OneOnc; OneOnc: OneOncology.
Statistically significant differences were observed in age, sex, race/ethnicity, insurance, smoking history and histology between cohorts without versus with ≥1 biomarker test in aNSCLC at both NAT and OneOnc. Statistically significant differences were observed in race/ethnicity between cohorts without versus with ≥1 biomarker test in mCRC at both NAT and OneOnc ().
Table 1. Patient demographics and characteristics: patients without versus with ≥1 biomarker test from initial diagnosis to ≤90 days after advanced or metastatic diagnosis in Flatiron Health Nationwide, excluding OneOncology and OneOncology cohorts.
Patients who underwent ≥1 biomarker test between initial diagnosis and ≤90 days after advanced/metastatic diagnosis had similar age, sex and insurance type (the majority were commercially insured) in both the OneOnc and NAT cohorts (). Statistically significant differences in race/ethnicity and practice type were observed between the cohorts: OneOnc had more White patients than NAT (aNSCLC, 74 vs 63%; mCRC, 69 vs 56%). Most NAT cohort patients were treated at community practice sites (range, 88–91%) rather than academic centers, whereas all patients in the OneOnc cohort received care at community practice sites ().
Table 2. Patient demographics and characteristics: patients with ≥1 biomarker test from initial diagnosis to ≤90 days after advanced or metastatic diagnosis in Flatiron Health Nationwide, excluding OneOncology and OneOncology cohorts.
Adoption of biomarker testing
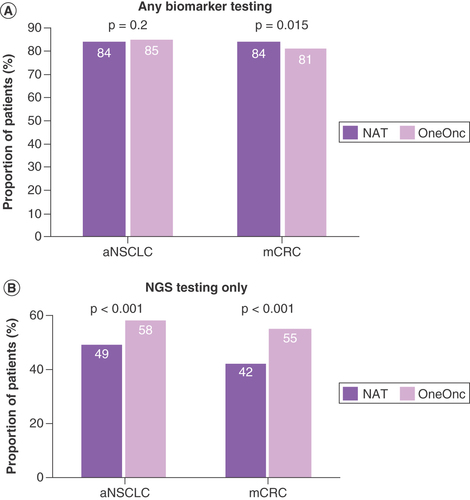
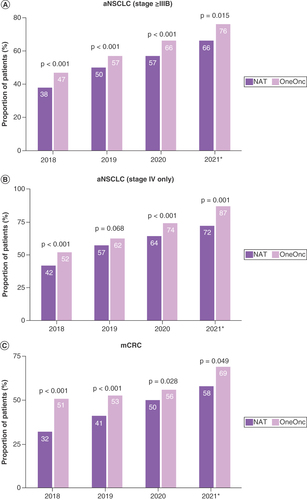
Biomarker testing rates were relatively high in both OneOnc and NAT patients with aNSCLC and mCRC and were relatively comparable between NAT and OneOnc (A). Rates of any biomarker testing increased over time in both cohorts (A & C).
(A) Any biomarker testing, including NGS and other biomarker tests. (B) NGS testing only.
aNSCLC: Advanced non-small-cell lung cancer; mCRC: Metastatic colorectal cancer; NAT: Flatiron Health Nationwide, excluding OneOnc; NGS: Next-generation sequencing; OneOnc: OneOncology.
(A) aNSCLC (stage ≥IIIB). (B) Stage IV aNSCLC only. (C) mCRC.
*Until April 2021.
aNSCLC: Advanced non-small-cell lung cancer; mCRC: Metastatic colorectal cancer; NAT: Flatiron Health Nationwide; OneOnc: OneOncology.
Overall, the proportion of patients who received NGS tests was higher in OneOnc versus NAT (aNSCLC, 58 vs 49% [p < 0.001]; mCRC, 55 vs 42% [p < 0.001]; B). While testing rates increased over time, the rate of NGS testing uptake was similar for patients with aNSCLC (p > 0.05) and faster in NAT than in OneOnc for patients with mCRC (p < 0.05; A & C).
(A) aNSCLC. (B) Stage IV only aNSCLC. (C) mCRC.
*Until April 2021.
aNSCLC: Advanced non-small-cell lung cancer; mCRC: Metastatic colorectal cancer; NAT: Flatiron Health Nationwide; NGS: Next-generation sequencing; OneOnc: OneOncology.
The majority of the NGS testing in aNSCLC was tissue (80% at OneOnc vs 82% NAT), followed by liquid (19% OneOnc vs 17% NAT) and both (0.5% OneOnc vs 1% NAT). Tissue biopsy was also the most common type of sample for NGS testing in mCRC (97% at OneOnc and NAT), followed by liquid (3% OneOnc vs 3% NAT; Supplementary Table 1).
The higher NGS testing rate was associated with a significantly greater proportion of OneOnc than NAT patients who had all 1L-actionable mutations tested (aNSCLC, 64 vs 57%; mCRC, 56 vs 48%; p < 0.001; ). Overall, patients who received NGS test results before initiating 1L treatment, regardless of their mutation status, had a greater likelihood of enrolling in a clinical trial or receiving targeted treatment ().
Table 3. Biomarker testing rate from initial diagnosis to ≤90 days after advanced or metastatic diagnosis from 1 January 2018 to 30 April 2021 in Flatiron Health Nationwide and OneOncology cohorts.
Table 4. First-line treatment pattern after test results (next-generation sequencingTable Footnote† vs other biomarker testTable Footnote†), regardless of mutation status.
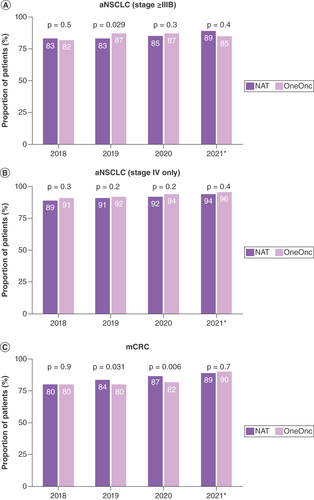
After the 2018 revision of the American Joint Committee on Cancer system, some patients with aNSCLC in the Flatiron Health database had their disease reclassified from stage IIIA to stage IIIB [Citation16]. Among patients with aNSCLC in this study, 13% (2131 of 16,882) were newly diagnosed with stage IIIB. Sensitivity analysis revealed that the testing rate in patients with stage IV aNSCLC was comparable to the overall rate in patients with stage IIIB, IIIC and IV disease, which indicates that the impact of the American Joint Committee on Cancer revision was minimal (A & B & A & B).
31 of 200 oncologists at OneOnc (16%) responded to the online survey. Common factors impacting oncologists’ decision to order NGS tests included diagnosis of advanced cancer (94%), a later line of advanced cancer (93%), NCCN Guidelines® and American Society of Clinical Oncology guideline recommendations (90%), availability of a liquid biopsy (81%) and availability of a FDA-approved NGS test (70%). The most common barriers to NGS testing were insufficient tissue for testing (87%) and operational challenges within the hospital or pathology department (81%; Supplementary Table 2).
Clinical utility of biomarker testing
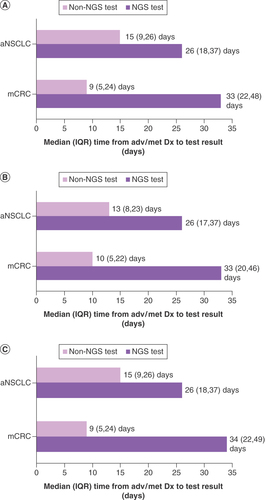
From the OneOnc oncologist survey, the two most common reasons that led to treatment initiation before receiving biomarker test results were the patient’s clinical condition necessitating urgent treatment (97%) and the average turnaround time for test results being too long (87%; Supplementary Table 2). In the Flatiron Health analysis, we observed patients who underwent ≥1 biomarker test, regardless of whether testing identified ≥1 actionable mutation; the median turnaround time from advanced/metastatic diagnosis to availability of the first test result was longer for NGS than for other biomarker testing (aNSCLC, 26 vs 15 days; mCRC, 33 vs 9 days) The median turnaround times were relatively comparable between OneOnc and NAT ().
(A) Overall. (B) OneOnc only. (C) NAT only.
aNSCLC: Advanced non-small-cell lung cancer; Dx: Diagnosis; IQR: Interquartile range; mCRC: Metastatic colorectal cancer; NGS: Next-generation sequencing.
Due to the limited sample size of patients with ≥1 actionable oncogene mutation in the 1L setting, we combined the OneOnc and NAT cohorts to evaluate the utility of biomarker testing in informing biomarker-driven treatment decisions. Most patients identified with ≥1 actionable oncogene mutation in the 1L setting initiated treatment after biomarker test results were available (range, 74–89%; ). However, not all patients who initiated 1L treatment after receiving biomarker test results received targeted therapy. Among patients with aNSCLC with ≥1 actionable oncogene mutation (n = 1014), 73% initiated targeted therapy for the studied mutations. Of patients with aNSCLC with ≥1 actionable oncogene mutation and/or PD-L1 positivity (n = 7226), 29% received cancer immunotherapy (CIT) + chemotherapy (chemo) ± other, 27% CIT only and 21% targeted therapy ± chemo ± other. Most patients with mCRC received chemo + bevacizumab (41% of those with BRAF V600E mutation and/or dMMR/MSI-H [n = 567]; 62% of those with RAS mutation [n = 1543]; ).
Actionable oncogene mutations studied were ALK, BRAF, EGFR and ROS-1.
aNSCLC: Advanced non-small-cell lung cancer; mCRC: Metastatic colorectal cancer; MMR: Mismatch repair; MSI-H: Microsatellite instability-high; mut: Mutant; NAT: Flatiron Health Nationwide; OneOnc: OneOncology; wt: Wild-type.
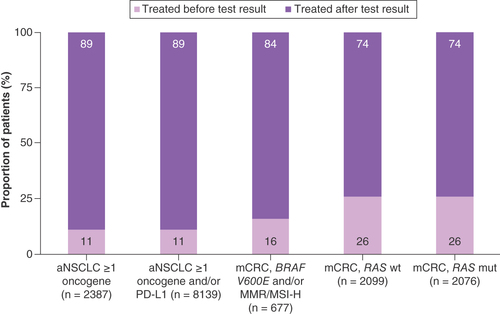
Table 5. Treatment patterns after availability of biomarker test results: patients with advanced non-small-cell lung cancer or metastatic colorectal cancer identified with ≥1 actionable oncogene mutation in the first-line treatment setting.
For patients with ≥1 actionable oncogene mutation who initiated 1L treatment before biomarker test results were available, the most common treatment was CIT ± chemo and/or targeted treatment among patients with aNSCLC (40% of patients with ≥1 actionable oncogene mutation [n = 262] vs 45% with ≥1 studied actionable oncogene mutation and/or PD-L1 positivity [n = 913]). Chemo + bevacizumab was most common among patients with mCRC (62% with BRAF V600E and/or MMR/MSI-H [n = 110]; 62% with RAS mutation [n = 533]; ).
Table 6. Treatment patterns before and after availability of biomarker test results among patients with advanced non-small-cell lung cancer or metastatic colorectal cancer identified with ≥1 actionable oncogene mutation in the first-line treatment setting who received treatment before their test results.
After test results became available, some patients with ≥1 actionable oncogene mutation changed treatments. The median time from test result to a change in treatment or loss to follow-up was 111 days in patients with aNSCLC (n = 919) and 192 days in patients with mCRC (n = 667). In addition, not all patients switched to targeted treatment within 49 days of treatment initiation. Among patients with aNSCLC with ≥1 actionable oncogene mutation, 18% (48 of 262) switched treatment within 49 days of treatment initiation; most (~75%) of these patients switched to targeted treatment. Almost 11% (102 of 913) of patients with aNSCLC with ≥1 actionable oncogene mutation and/or PD-L1 positivity switched treatments: 35% to targeted treatment and 30% switched to CIT + chemo. Similarly, 7% of patients (8 of 110) with mCRC with BRAF V600E and/or dMMR/MSI-H switched treatments; the majority (6 of 8; 75%) subsequently received CIT. Of the patients with mCRC with RAS mutation, 3% (14 of 533) switched; of the six patients initially treated with cetuximab or panitumumab, fewer than half switched to chemo ± bevacizumab ().
Discussion
The biomarker testing rates observed in OneOnc (100% community) and NAT (~80% community, ~20% academic) indicate that the majority of community oncologists recognize the importance of biomarker testing at the time of advanced/metastatic diagnosis, per American Society of Clinical Oncology 2022 guideline recommendations [Citation17] and consistent with findings from another large US community network [Citation18].
The statistical differences in race/ethnicity between cohorts without versus with ≥1 biomarker test, as well as with versus without NGS testing, align with recent literature reporting Black patients with metastatic NSCLC were significantly less likely to undergo NGS testing compared with white patients [Citation19]. Racial/ethnic inequity on NGS testing was also more pronounced in non-Latinx Black and Latinx patients with metastatic NSCLC in the post national coverage determination era [Citation20]. Future studies are warranted to better understand drivers to address racial/ethnic inequity in biomarker testing.
The high biomarker testing rates observed in OneOnc and NAT demonstrate that community practices are routinely doing biomarker testing. The percent of NGS testing is increasing over time, although there is still room for improvement to achieve as high as the ≥81% rate observed in any biomarker testing and ultimately universal testing for all patients. Overall, NGS testing rates were higher among OneOnc compared with NAT patients, which may be attributed to OneOnc’s networkwide precision medicine clinical initiatives dedicated to driving broad and uniform use of guideline-adherent NGS testing with ongoing monitoring and provider feedback. Another differentiating factor for OneOnc is the development of weekly molecular tumor boards that review every NGS report from the participating network sites, with nonbinding consensus recommendations for treatment from the tumor board network experts communicated back to the provider for relevant cases. A monthly educational tumor board has also been created as part of a broader education effort to increase the understanding and utility of both common and rarer genomic alterations that may be actionable [Citation12]. Supporting this interpretation, most OneOnc survey respondents agreed or highly agreed that the availability of molecular tumor boards impacted the clinician’s decision to perform NGS. The integration of clinical predictive factors, histology and biomarkers assists physicians in identifying specific pathways and clinics in prioritizing the most appropriate and preferred treatment regimens, as biomarker results are directly reported into searchable data fields.
One important barrier to ordering NGS identified by OneOnc survey respondents was the operational challenges of working with pathology departments that are tasked with delivering tissue specimens for NGS testing. OneOnc has piloted some operational enhancements related to PHC to include simultaneous submission of liquid and tissue biopsies to the NGS laboratory and immediate performance of liquid biopsies, followed reflexively by tissue testing if liquid biopsy results prove uninformative. In addition, informatics improvements to the EHR are being made to allow NGS ordering and delivery of results directly into searchable fields. Finally, pathologists must make additional effort to retrieve blocks, analyze specimens for the greatest amount of tumor tissue and package and ship samples to the third-party NGS provider. Since pathologists are often geographically and organizationally removed from community oncology practices, communication and procedural structure may be less than optimal, causing delays in mailing samples to referral labs and increasing turnaround time.
The value of NGS to inform biomarker-driven therapy was underscored by our findings that patients with aNSCLC or mCRC who underwent NGS testing versus other biomarker testing had a greater likelihood of receiving targeted treatment or being considered for a clinical trial; however, the proportion of patients enrolled in a clinical trial is small overall at ≤3%.
While most patients with ≥1 studied actionable oncogene mutation received 1L treatment only after molecular testing, not all patients received biomarker-specified targeted treatments. Similarly, only a minor subset of patients with ≥1 oncogene who received treatment before receiving test results switched treatment within 49 days of treatment initiation, based on the driver mutations identified (aNSCLC, ≤18%; mCRC, ≤7%). These findings highlight opportunities to make treatment decisions based on actionable biomarkers identified. Among patients with aNSCLC who received treatment before biomarker results were available, CIT ± chemo was the most common treatment. This treatment practice does not adhere to NCCN Guidelines [Citation4], which recommend that clinicians consider holding immunotherapy for one cycle until results of molecular testing are available. This recommendation is further supported by a recent study of routine clinical practice, in which survival was significantly compromised in patients with aNSCLC treated with 1L CIT ± chemo who subsequently proved to carry an oncogenic driver mutation (hazard ratio [HR], 2.35; p < 0.0001) [Citation21]. Of the patients with aNSCLC who initiated targeted treatment before test results were available, the majority (83% [44 of 53]) received EGFR-targeted treatment, which may reflect potential treatment decisions based on predictive factors (e.g., age, Asian race, female sex and nonsmoker). It is critical to leverage biomarkers to inform treatment decisions as recent research has demonstrated that companion diagnostic testing coupled with versus without biomarker-driven treatment resulted in a greater survival benefit in patients with 1L-actionable mutations and stage IV NSCLC (adjusted HR, 0.72) [Citation22].
The long median time from test result to initiation of treatment switch (≥111 days) reflects potential treatment changes occurring at disease progression and/or due to biomarker test results. However, the specific reasons for the change in treatment were not available in the Flatiron database. We developed a conservative proxy of treatment switch due to biomarker test results by restricting the period to ≤49 days from the start of treatment initiation, which accounted for standard-practice evaluation of aNSCLC or mCRC progression and/or response to initial treatment. These findings underline another area for improvement in providing timely, biomarker-driven, targeted treatment for optimal treatment outcomes. To this point, a recent study observed that a >3-week delay in the initiation of ALK inhibitor treatment in patients with aNSCLC and an ALK mutation was associated with twice the risk of death [Citation23].
There are several limitations to our study findings. The Flatiron Health database is an open network; patients may have received undocumented care elsewhere, resulting in under-representation or missing data. Due to the lack of information on testing order date, a proxy of turnaround time from advanced/metastatic diagnosis to first test result was estimated. Missing data also resulted in under-representation of non-Hispanic Black patients (e.g., ~8.3% of patients with aNSCLC in this study were Black vs 12% in the Surveillance, Epidemiology and End Results Program database [Citation13], 35% in the West Cancer Center & Research Institute, 20% in Tennessee Oncology, 8% in Astera, 11% in NYCBS, 3% in ECHO and 1% in LACN) [Citation12]. In addition, the Flatiron OneOnc cohort is a sample of data from six selected sites and does not reflect data from all practices in the OneOnc network. Insights on the higher NGS testing rates at OneOnc were contextualized with input from OneOnc authors and survey results, which reflect the experiences of OneOnc oncologists only; therefore, findings may not be generalizable to other clinical practices in the USA. Future studies are warranted to evaluate the effectiveness of PHC implementation programs in routine clinical settings. In addition, the survey selections were prespecified and may not have captured all possible factors impacting oncologists’ decisions.
Conclusion
Our study highlights the ability of community oncology centers to perform biomarker testing (including NGS) in multiple solid tumors to inform personalized treatment decisions. Adoption of NGS testing was higher at OneOnc centers than at other nationwide oncology centers, highlighting the importance of educational and operational initiatives focusing on improving clinical care and, importantly, providing molecular tumor boards to support community oncologists. Opportunities remain to optimize PHC, including improving the uptake of NGS, shortening biomarker test turnaround times and reducing the proportion of patients receiving treatment before receiving biomarker test results.
This study demonstrated community oncology centers’ ability to deliver personalized healthcare to patients with advanced non-small-cell lung cancer (aNSCLC) and metastatic colorectal cancer (mCRC) through biomarker testing, including NGS testing.
During the period from 1 January 2018 to 30 April 2021, the any-biomarker-testing rate was high (≥81%) and comparable between the OneOnc community network and other Flatiron nationwide sites (NAT).
NGS testing rates were higher at OneOnc versus NAT (aNSCLC: 58 vs 49%; p < 0.001; mCRC: 55 vs 42%; p < 0.001), which reflects the success of OneOnc networkwide educational, pathway and operational initiatives.
Patients who underwent NGS testing had a greater likelihood of receiving targeted treatment than patients who received other biomarker tests.
Some patients with ≥1 actionable mutation (aNSCLC, ≤11%; mCRC, ≤26%) received treatments before biomarker test results were available.
Although most patients with ≥1 actionable mutation received 1L targeted treatment after testing, not all received targeted treatment.
Based on a OneOnc oncologist survey, operational challenges and insufficient tissue collection were common barriers to completion of NGS testing in routine practice. In addition, patient clinical condition and long turnaround time were common factors attributed to treating patients before biomarker test results were available.
Opportunities remain to optimize personalized healthcare by increasing NGS uptake, shortening turnaround times and reducing the number of patients who receive treatment before receiving biomarker test results.
Author contributions
All authors contributed to, reviewed and approved the final draft of the paper. All authors had full access to all data in the study and had final responsibility for the decision to submit for publication. Concept and design: all authors. Acquisition, analysis or interpretation of data: all authors. Drafting of manuscript: all authors. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: all authors. Obtained funding: F. Hoffmann-La Roche Ltd/Genentech, Inc. Administrative, technical or material support: Ma, E. Supervision: Schwartzberg, L.; Ma, E.
Financial & competing interests disclosure
This study was sponsored by F. Hoffmann-La Roche Ltd/Genentech, Inc. This study was funded by F. Hoffmann-La Roche Ltd/Genentech, Inc., which contributed to the study design, data interpretation and writing of this report. L Schwartzberg: has consulted for Pfizer, Helsinn, Spectrum, AstraZeneca, Genentech, Myriad, Bayer, Napo, Lilly, BeyondSpring, AbbVie, Coherus, Illumina, Sanofi, Mirati and GlaxoSmithKline; and has served on speakers bureaus for Puma, Coherus, Merck and Seagen. D Daniel: has received research funding from AstraZeneca, Genentech, Guardant Health, Janssen, Bristol Myers Squibb, G1 Therapeutics, Merck, Novartis, AbbVie, ARMO, Immunomedics, Lilly, Merus NV, Daiichi Sankyo, F. Hoffmann-La Roche AG, Celgene and Amgen. D Vaena: has consulted for Bristol Myers Squibb, Bayer, Genomic Health, Natera, Seagen, Exelixis, EMD Serono, Immunomedics and Eisai; received honoraria from HMP; received research funding from Bristol Myers Squibb, Novartis, AstraZeneca, Peloton, BioClin, Aeglea, Nektar, Calithera, Tizona, Compugen, TG, Merck, OBI, Incyte, Seagen and Roche/Genentech. D Slater: nothing to disclose. H Staszewski: nothing to disclose. B Fang: nothing to disclose. L Seneviratne: served on the speaker bureau for Novartis and received honoraria from Gilead and AstraZeneca. E Yu: employee of Genentech, Inc., and owns stock in Roche. R Price: employee of Genentech, Inc., and owns stock in Roche. T Szado: employee of Genentech, Inc., and owns stock in Roche. CS Meyer: at the time of the study was an employee of Genentech, Inc., and owned stock in Roche. A Shah: at the time of the study was an employee of Genentech, Inc.; received travel, accommodations, expenses from Genentech, Inc.; and owned stock in Roche. E Ma: employee of Genentech, Inc. and owns stock in Roche. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
The authors thank Denise Kenski of Health Interactions, Inc., and the Global Outcomes Group for providing editorial assistance, which was funded by Genentech, Inc., in accordance with Good Publication Practice (GPP 2022) guidelines.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Data sharing statement
The data that support the findings of this study have been originated by Flatiron Health, Inc. These deidentified data may be made available on request and are subject to a license agreement with Flatiron Health; interested researchers should contact [email protected] to determine licensing terms.
Supplemental Table 1. NGS Testing by Sample Type
Download MS Word (21.1 KB)Acknowledgments
Analytical output quality control and support were provided by Danny Sheinson and Ronrong Wang at Genentech, Inc., and Baiyu Yang at Roche Diagnostics. Survey distribution and analytical support were provided by Aaron Chrisman at OneOncology, Inc.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/fon-2022-1216
Additional information
Funding
References
- Aisner DL , RielyGJ. Non–small cell lung cancer: recommendations for biomarker testing and treatment. J Natl Compr Canc Netw.19(5.5), 610–613 (2021).
- Malone ER , OlivaM, SabatiniPJB, StockleyTL, SiuLL. Molecular profiling for precision cancer therapies. Genome Med.12(1), 8 (2020).
- Lindeman NI , CaglePT, AisnerDLet al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med.142(3), 321–346 (2018).
- Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for non-small-cell lung cancer. V.2.2023. © National Comprehensive Cancer Network, Inc (2023). All rights reserved. Accessed 12April2023. To view the most recent and complete version of the guideline, go online toNCCN.org
- Referenced with permission from the NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Colon Cancer V.1.2023. © National Comprehensive Cancer Network, Inc (2023). All rights reserved. Accessed 12April2023. To view the most recent and complete version of the guideline, go online toNCCN.org
- Freedman AN , KlabundeCN, WiantKet al. Use of Next-Generation Sequencing Tests to Guide Cancer Treatment: Results From a Nationally Representative Survey of Oncologists in the United States. JCO Precis Oncol.2, 1–13 (2018).
- Cobain EF , WuYM, VatsPet al. Assessment of Clinical Benefit of Integrative Genomic Profiling in Advanced Solid Tumors. JAMA Oncol.7(4), 525–533 (2021).
- Miller TE , YangM, BajorDet al. Clinical utility of reflex testing using focused next-generation sequencing for management of patients with advanced lung adenocarcinoma. J. Clin. Pathol.71(12), 1108–1115 (2018).
- Centers for Medicare & Medicaid Services . National Coverage Determination (NCD): Next Generation Sequencing (NGS). https://www.cms.gov/medicare-coverage-database/view/ncd.aspx?NCDId=372 (Accessed 12April2023).
- Centers for Medicare & Medicaid Services . National coverage analysis (NCA): next-generation sequencing for Medicare beneficiaries with advanced cancer. (2020). https://www.cms.gov/medicare-coverage-database/view/ncacal-tracking-sheet.aspx?NCAId=296
- Unger JM , VaidyaR, HershmanDL, MinasianLM, FleuryME. Systematic Review and Meta-Analysis of the Magnitude of Structural, Clinical, and Physician and Patient Barriers to Cancer Clinical Trial Participation. J. Natl Cancer Inst.111(3), 245–255 (2019).
- Driving the Future of Cancer Care. OneOncology (2023). https://www.oneoncology.com/assets/pdf/COA/OO_AnnualReport2021_Digital_FINAL3.pdf (Accessed 12April2023).
- Ma X , LongL, MoonS, AdamsonBJS, BaxiSS. Comparison of Population Characteristics in Real-World Clinical Oncology Databases in the US: Flatiron Health, SEER, and NPCR. medRxiv.doi: 10.1101/2020.03.16.20037143 (2020).
- Birnbaum B , NussbaumN, Seidl-RathkopfKet al. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. arXiv.doi: 10.48550/arXiv.2001.09765 (2020).
- R: A language and environment for statistical computing. R Foundation for Statistical Computing (2013).
- Lim W , RidgeCA, NicholsonAG, MirsadraeeS. The 8th lung cancer TNM classification and clinical staging system: review of the changes and clinical implications. Quant Imaging Med Surg.8(7), 709–718 (2018).
- Chakravarty D , JohnsonA, SklarJet al. Somatic Genomic Testing in Patients With Metastatic or Advanced Cancer: ASCO Provisional Clinical Opinion. J. Clin. Oncol.40(11), 1231–1258 (2022).
- Robert NJ , EspiritoJL, ChenLet al. Biomarker testing and tissue journey among patients with metastatic non-small-cell lung cancer receiving first-line therapy in The US Oncology Network. Lung Cancer.166, 197–204 (2022).
- Bruno DS , HessLM, LiX, SuEW, PatelM. Disparities in Biomarker Testing and Clinical Trial Enrollment Among Patients With Lung, Breast, or Colorectal Cancers in the United States. JCO Precis Oncol.6, e2100427 (2022).
- Sheinson DM , WongWB, MeyerCSet al. Trends in Use of Next-Generation Sequencing in Patients With Solid Tumors by Race and Ethnicity After Implementation of the Medicare National Coverage Determination. JAMA Netw Open.4(12), e2138219 (2021).
- Smith RE , JohnsonML, GordanLNet al. Evaluation of outcomes in patients (pts) with stage 4 non-small-cell lung cancer (NSCLC 4) harboring actionable oncogenic drivers (AOD) when treated prior to report of mutation without tyrosine kinase inhibitors (TKI): An Integra Connect Database (ICD) retrospective observational study. J. Clin. Oncol.60(Suppl. 16), abstr 1530 (2022).
- John A , ShahRA, WongWB, SchneiderCE, AlexanderM. Value of Precision Medicine in Advanced non-small-cell Lung Cancer: Real-World Outcomes Associated with the Use of Companion Diagnostics. Oncologist.25(11), e1743–e1752 (2020).
- Sheinson D , WongWB, WuN, MansfieldAS. Impact of delaying initiation of anaplastic lymphoma kinase inhibitor treatment on survival in patients with advanced non-small-cell lung cancer. Lung Cancer.143, 86–92 (2020).
