Abstract
Aim: The need for pharmacogenomic education is becoming more and more urgent. Our aim was to evaluate the progress in pharmacogenomics education since then, and to put forward further recommendations. Methods: A survey was sent to 248 schools of medicine, pharmacy, nursing and health professions around the world. Results: The majority of the study programs (87%) include pharmacogenomics education, which is generally taught as part of the pharmacology curriculum. On average, educators and teachers have selected appropriate and highly relevant pharmacogenomics biomarkers to include in their teaching programs. Conclusions: Based on the results, we can conclude that the state of pharmacogenomics education at the surveyed universities has improved substantially since 2005.
Pharmacogenomics (PGx) aims to determine the influence of genetic and genomic variations on drug responses, in other words, drug efficacy and toxicity, and it represents an essential step toward personalization of medicine [Citation1]. Since the completion of the Human Genome Project in 2003, the rate of discovery of new PGx biomarkers has dramatically increased, and one of the healthcare challenges over the last decade has been the implementation of these genomic biomarkers in clinical practice. Despite the initial enthusiasm with the advent of PGx, its incorporation into the clinic has been frustratingly slow, and so far it has not met the expectations of the academic community, physicians and healthcare professionals [Citation2,Citation3]. Many barriers that hinder this process have been identified, such as the lack of reimbursement of costs of PGx testing [Citation4–8], and the lack of knowledge of healthcare professionals in the interpretation of PGx tests and the consequent treatment decisions [Citation9]. Also, in many cases, the impact of the results of PGx testing is limited compared with other sources of variability in patient responses to a drug (i.e., nutrition, other drugs, drug interactions, alcohol consumption, microbiota, gene–gene interactions, etc) [Citation10,Citation11]. Several studies have shown that medical professionals agree on the importance of PGx in clinical practice, but that they are reluctant to use PGx due to inadequate knowledge of the interpretation of PGx tests [Citation12–23].
A first attempt for a call for action for PGx education was made in 2005, with the publication of Gurwitz et al. [Citation24]. In this article, which was addressed to medical, pharmaceutical and health school Deans of Education, the authors recommended inclusion of at least 4 h and ideally 8 h, of PGx teaching into the basic medical doctor (MD) education. For study programs in pharmaceutical, life sciences and public health schools they suggested more extensive teaching, without specifying the number of hours recommended. To the best of our knowledge, no further PGx education recommendations that are addressed to Deans of Education have been published. Therefore, our current survey used the above recommendations as a reference point for a global assessment of the current level of PGx education.
The aim of this study was to determine the current status of PGx education in medical, pharmaceutical and health study programs. We evaluated the progress in PGx education over the years since the 2005 recommendations were published, and on the basis of the survey findings, we make further recommendations.
Methods
Study design
A total of 248 schools of medicine, pharmacy, nursing or health professions were selected from six world continental regions (Europe, Asia, Africa, North America, South America, and Australia and Oceania). As this study received no direct funding, it was not possible to include all of the programs worldwide, and therefore only selected schools were invited. The selection was performed as outlined in the following section. Once an institution had been selected, contact persons were identified (e.g., Dean, Dean of Education, Chair of Department) and they were sent e-mails requesting them to complete an on-line questionnaire on PGx education in their institution (Supplementary Materials 1).
The outcome variables initially analyzed were: PGx subject type; plans for future PGx education implementation; number of teaching hours for PGx; changes since implementation; use of PGx biomarkers in education; PGx integration with other fields in the educational process; initial year of PGx implementation; PGx educational tools; and PGx education in PhD programs. Next, the following comparisons of the outcome variables were analyzed: between programs, as schools of medicine, pharmacy, nursing, or health professions (including, but not limited to, laboratory medicine, biochemistry, toxicology and others); between continents, as Europe, Asia, Africa, North America, South America, and Australia and Oceania; and between member countries in the Organization for Economic Co-operation and Development (OECD) and non-OECD countries. In addition, the use of PGx biomarkers in different study programs was compared with the number of PubMed publications and the number of drugs with prescribing guidelines published by the US FDA for a specific PGx biomarker.
University selection criteria
The participating universities were selected from the Quacquarelli Symonds (QS) World University Rankings (www.topuniversities.com/). The ranking lists by subject for Medicine and for Pharmacy and Pharmacology were used, with subsequent filtering for continents and locations (i.e., countries within a specific continent). Separate lists were made for each of these six continental regions for schools of medicine, pharmacy, nursing or health professions. The top-ranked university from each country that was included in the MD or Pharmacy top world ranking was added to the final list. In the case of North America, where there are only three countries, more than one top university per country was included. Thus, for North America, ten universities were selected: four each from the USA and Canada, and two from Mexico. For the continents with <10 universities selected (i.e., Africa, South America, Australia and Oceania), additional universities were selected from the Webometrics Ranking of World Universities (www.webometrics.info/en). The ranking lists by region for Africa, South America and Australia and Oceania were applied for the selection of additional universities from countries that were not on the QS World University Rankings list (as one university per country), and that have one of the programs listed above, according to their Webometrics ranking. As the eastern European countries were under-represented on the European regional QS World University Rankings list, an additional ten universities were selected from the Emerging Europe and Central Asia QS Ranking.
To increase response rates and limit the selection bias of top-ranked universities, additional universities were selected independent of their ranking, according to membership of the European Society of Pharmacogenomics and Personalized Therapy (ESPT; i.e., universities whose ESPT members were part of ESPT research groups, boards and national societies). Of note, only 50.8% of the invited ESPT universities came from the top 500 universities on the QS World University Rankings list.
Questionnaire on pharmacogenomics education in medical doctor, pharmacy & health science programs
The questionnaire was an updated and extended version of the questionnaire used by Gurwitz et al. in 2005 [Citation24]. It consisted of 12 questions (as mostly multiple choice) pertaining to PGx education, plus comments and suggestions sections (Supplementary Materials 1). All of the selected participants were invited by e-mail to participate in the on-line questionnaire, with its completion from September 8, 2016, to February 21, 2017. The on-line survey was created using 1 ka (www.1ka.si/d/en), an open-source application for creating, conducting and analyzing online surveys. Following the initial invitation, three reminders were sent to each participant at 2-week intervals.
PubMed records
The number of PubMed-listed publications for specific PGx biomarkers was obtained from PubMed (www.ncbi.nlm.nih.gov/pubmed/) on January 18, 2019, using the advanced search mode with the terms ‘pharmacogenetics’ or ‘pharmacogenomics’ together with the names of the relevant human genes from the HUGO Gene Nomenclature Committee.
The number of drugs with FDA drug labels for specific PGx biomarkers was obtained from The Pharmacogenomics Knowledgebase (PharmGKB; www.pharmgkb.org/; last accessed, January 18, 2019) using the names of the genes from the HUGO Gene Nomenclature Committee as the search term. The prescribing information was found under the tab Drug labels.
Statistical analysis
Descriptive statistics were used to present the global results. For comparisons between two groups, Fisher exact (for categorical data) and Mann–Whitney U (for non-Gaussian continuous or ordinal data) tests were used. For comparisons between three or more groups, Fisher exact (for categorical data) and Kruskal–Wallis (for non-Gaussian continuous or ordinal data) tests were used. For testing correlations between the use of PGx biomarkers and the number of PubMed publications or the number of drugs with prescribing guidelines for a specific PGx biomarker, nonparametric Spearman correlations were used. All of these tests were carried out using the SPSS statistical program (version 24; IBM, NY, USA). For all of the statistical tests, differences with p values ≤ 0.05 were considered significant.
Results & discussion
Description of the study cohort
Of the 248 invited institutions, 69 responded, which provided an overall response rate of 27.8%. There were substantial differences in the response rates between the different world regions, with the highest in European countries (34.7%), and the lowest in South America (9.0%). Detailed response rate data and cohort characteristics are listed in and , respectively. The questionnaire, as well as the emails to contact persons (and reminders), were written in English; moreover, ESPT members were included in the survey (see Methods). These factors may in part explain the higher response rate from Europe.
Table 1. Response rates by location of institution (overall and by continent).
Table 2. Description of the study cohorts.
Prevalence, form & future implementation of pharmacogenomics education
Among the study programs included, 13.4% reported no PGx education (A).
(A) By subject. Other subjects with pharmacogenomics topics not shown include: pharmacokinetics (three responders), oncology and psychiatry (two responders each), and drug development, molecular biology, biopharmacy, homeostasis and organ-system study modules (one responder each). (B) Plans for the future implementation of pharmacogenomics.
PGx: Pharmacogenomics.
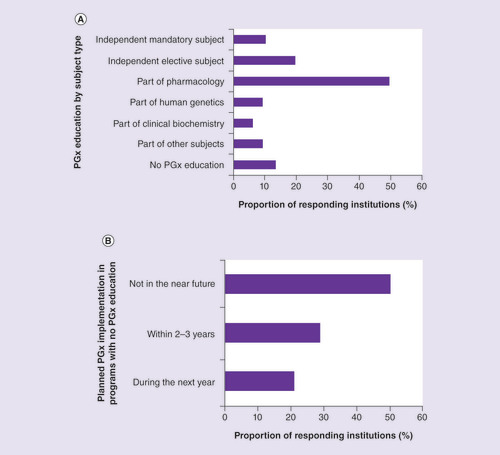
In about half of the programs that included PGx education, PGx was taught as part of the pharmacology curricula, followed by its teaching as independent elective (19.6%) and mandatory (10.3%) subjects (A). This finding is in line with the 2005 recommendation for medical, pharmacy and health schools to incorporate PGx as an integral part of their core pharmacology curricula [Citation24]. Significantly fewer programs in graduate schools reported PGx education as a part of pharmacology (15.4%) compared with MD (59.1%), pharmacy (50.0%) and nursing (66.7%) programs (Fisher’s exact test; p = 0.049).
The frequencies of study programs that included PGx in their pharmacology curricula were also compared between continents: PGx was reported as a part of the pharmacology curricula by 55.3% of the programs from Europe, 53.3% from Asia, 85.7% from Africa, 12.5% from North America and 36.4% from Australia and Oceania (Fisher exact p = 0.075). The distribution of the programs with PGx as an independent mandatory or elective subject did not differ between the subgroups. Among the responders, 9.3% indicated that PGx was included in other subjects not specifically listed in A. The subgroup analysis showed that PGx as a part of alternative subjects was reported by 18.4% of the responders from OECD countries, and by none from non-OECD countries.
These data are in line with majority of other published studies on PGx education in medical and pharmaceutical programs. Studies by Green et al. and Higgs et al. reported that in North America and Britain, 82 and 84%, respectively, of participating medical schools had PGx incorporated into their programs [Citation25,Citation26]. Similar to the present data, PGx was seldom taught as a standalone course, and it was most often part of the pharmacology course [Citation25,Citation26]. In the more recent study by Murphy et al., among responders from US schools and colleges of pharmacy, 89% reported PGx as included in their PharmD curriculum, and 21% where it was a standalone course, with others including it as a part of other mandatory or elective courses [Citation27]. A study by Pisanu et al. investigated the state of PGx education in southeastern Europe (i.e., Greece, Italy, Cyprus, Malta, Turkey, Albania, Bulgaria, Serbia, Montenegro, Bosnia and Herzegovina, Croatia). They reported that PGx teaching varied considerably between countries, and even between universities within the same country. In certain countries (i.e., Cyprus, Malta) PGx was not included in the curricula of any of their participating universities, while in others (i.e., Greece, Italy), it was taught in all of the programs studied, often as an independent course. In other southeast European universities included in their study, the incorporation of PGx into the health care curricula was variable, and taught mostly as a part of other courses that were offered, rather than as a standalone course [Citation20].
Number of teaching hours for pharmacogenomics, & changes since the implementation of pharmacogenomics education
Most programs offer 1–2 h of PGx teaching, followed by 3–4 h and >10 h in equal proportions (A). 63% of the programs offer at least the minimum recommended number of hours, as recommended by the 2005 ISP recommendations (i.e., 3–4 h) [Citation24], where more pharmacy (75.9%) and graduate school (80.0%) programs reached this 3-4 h minimum than for MD (48.5%) and nursing (0.0%) programs (B).
(A) Number of hours of pharmacogenomics education. (B) Distribution of minimal recommended number of hours (3–4 h) across schools. More pharmacy and graduate schools than MD programs offer the minimal recommended number of pharmacogenomics hours (Fisher’s exact test; p = 0.024).(C) Changes since implementation.
†Minimal recommended number of teaching hours.
‡Median (min-max) number of teaching hours, 21.5 h (12–50 h) per program.
§Median increase in hours (min–max): 5 h (1–40 h);
#Median increase in hours in a single case.
MD: Medical doctor; PGx: Pharmacogenomics.
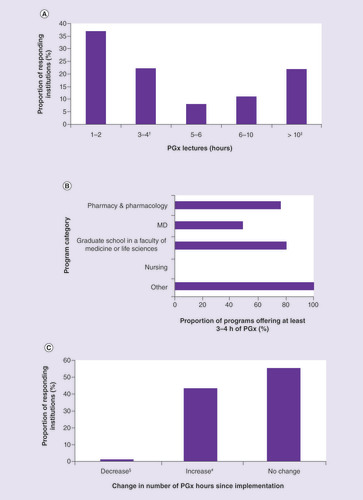
Other studies have also reported low numbers of hours of PGx coursework in medical school curricula, with only 21% of British medical schools [Citation26] and 28% of medical schools in the USA and Canada [Citation25] offering ≥4 h of PGx teaching. In the same two studies, the majority of the schools considered the offered amount of PGx teaching to be sufficient, and only 29% of North American medical schools planned to increase the number of hours for PGx [Citation25,Citation26]. Our survey shows that in the USA, pharmacy schools have integrated PGx into their curricula to a greater extent compared with medical schools. In line with this, Murphy et al. [Citation27]. reported that more than half of their participating institutions reported having ≥10 h of PGx coursework included in their pharmacy programs, and 46% of colleges planned to increase the number of hours of coursework dedicated to PGx instruction.
Use of pharmacogenomics biomarkers in education
The genomic markers used in PGx education and their frequency of use as reported by the survey responders, as well as their frequency of usage are illustrated in A. The most frequently taught PGx biomarkers were the three CYP genes (CYP2D6, CYP2C19, CYP2C9), which were all used at frequencies > 80%; this is in line with the 2005 ISP recommendations [Citation24]. These were followed by VKORC1, TPMT, UGT1A1 and the ABC transporter family genes, with frequencies of use from 50 to 70% (A). The use of biomarkers correlated significantly with their level of evidence according to PharmGKB, with those with higher levels of evidence used more frequently (Kruskal–Wallis p = 0.001) (A). Of note, the PharmGKB clinical annotation levels of evidence at least partly correspond to the Clinical Pharmacogenetics Implementation Consortium levels (CPIC A, B, C, D). CPIC assigns these CPIC levels to genes/ drugs with PharmGKB clinical cnnotation cevels of cvidence of 1A, 1B, 2A and 2B, or a PharmGKB PGx level for FDA-approved drug labels of ‘actionable PGx,’ ‘genetic testing recommended,’ or ‘genetic testing required,’ or based on nomination to CPIC for consideration [Citation28].
(A) Proportions of study programs teaching about each of the listed pharmacogenomics (PGx) biomarkers. The PGx biomarker PharmGKB levels of evidence are indicated as:
†1A; ‡1B; §2A; #2B; ¶3.
The 1A level of evidence corresponds to Clinical Pharmacogenetics Implementation Consortium A, except in the case of the ABC transporters, which are assigned a Clinical Pharmacogenetics Implementation Consortium A/B level of evidence. High level of evidence 1A markers were used significantly more frequently than lower level evidence markers (Kruskal–Wallis, p = 0.001). (B) Numbers of PGx biomarkers used per program, giving an overall median (min–max) of 7 (1–11). This median is significantly higher in Europe, Asia and North America compared with Africa and Australia and Oceania (Kruskal–Wallis, p = 0.033). (C, D) Relationships between PGx marker use in education and PubMed publications (C) and US FDA drug labels (D). Significant correlations are seen for number of PubMed publications per marker (C; Spearman’s rho, 0.643, p = 0.001) and number of FDA drug labels per PGx marker (D; Spearman’s rho, 0.635, p = 0.001).
PGx: Pharmacogenomics.
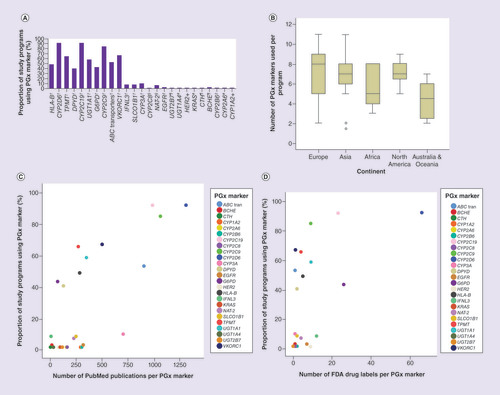
Overall, the median number of PGx biomarkers used per study program was seven (minimum–maximum, 1–11). Study programs from Europe included the highest median number of PGx biomarkers, followed by North America, Asia, Africa and Australia and Oceania (Kruskal–Wallis p = 0.033; B).
Next, we investigated the correlation between PGx biomarker use and the number of PubMed publications relating to the specific marker (C), and the number of drugs with an FDA drug label for a specific biomarker (D). As shown in A, the markers with CPIC A level of evidence (which corresponds to PharmGKB 1A) were used more frequently than those markers with lower levels of evidence. These results indicate that on average, educators and teachers select both appropriate and highly relevant PGx biomarkers to include in their teaching programs.
Pharmacogenomics integration with other fields of educational practice
In the present survey, 40% of responders reported integration of PGx with other -omics (A). We can therefore conclude that a minority of programs integrates PGx with other scientific fields, and thus in most study programs personalized medicine teaching is limited only to the genomic level.
(A) Integration of PGx education with other ‘-omics’ (OMICs), where the majority show little integration. The most commonly are epigenomics and bioinformatics. (B) Use of companion diagnostics in PGx education, where 74% of the study programs were positive. (C) Use of case studies in PGx education, where 71% of the study programs were positive.
PGx: Pharmacogenomics.
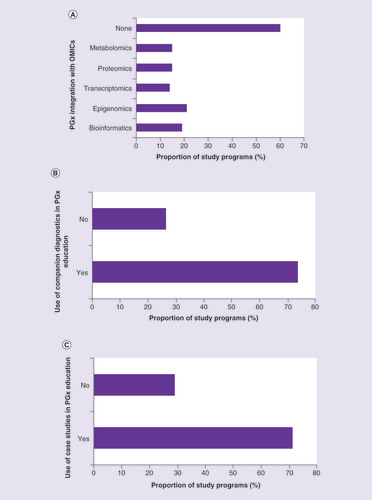
The situation is more encouraging considering the use of companion diagnostics and case studies in PGx education (B & C). This is in line with the 2005 ISP recommendations to include case studies in PGx teaching materials [Citation24].
The implementation of PGx education
On average, most programs started with PGx courses in 2007 (95% confidence interval, 2006–2009; min–max, 1995–2017), and programs from OECD countries started significantly earlier than those from non-OECD countries (mean, 2006 [min–max, 1995–2017] vs 2009 [min–max, 2000–2014]; t-test p = 0.035). Obviously, knowledge about the human genome sequence is crucial for the development of PGx markers, and indeed, 82.1% of the programs started to implement PGx topics after the completion of the Human Genome Project. Considering the clinical training associated with the study programs, the establishment of PharmGKB and CPIC might have been even more important time points for evaluation. PharmGKB was launched in 2000, and CPIC was established in 2009. Only 6.0% of the study programs surveyed started implementation of PGx topics prior to the release of PharmGKB, while 56.7% were already teaching PGx at the time of the establishment of CPIC.
PGx educational tools
The most frequently used educational tools for teaching PGx are original research papers (70.8%), followed by internet data bases (48.6%) and textbooks (41.7%) (A).
(A) Type of teaching sources used. (B) Most frequently reported internet databases used. (C) Most frequently reported textbooks used.(D) Use of original scientific papers, which was most frequent in pharmacy and least frequent in nursing study programs (Fisher’s exact test; p = 0.043).
†PhD theses, simulators and mobile apps.
‡Each reported once (see Supplementary Table 1).
§Each reported once (see Supplementary Table 2).
EMA: European Medical Agency; MD: Medical doctor; PGx: Pharmacogenomics.
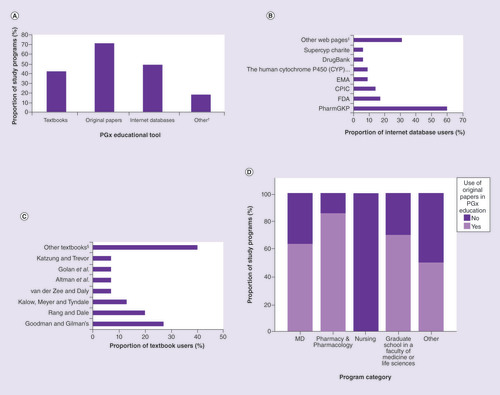
This situation has not changed much since the beginnings of PGx education, except for the wider availability of PGx textbooks [Citation29]. Our findings are expected, given that research papers are available on-line and provide up-to-date information, compared with textbooks. Moreover, according to a Directive of the European Commission of Open-Access Publishing, increasing numbers of journals offer open access, which is likely to lead to even higher use of original research papers in higher education. The use of original research papers for PGx education that was evident from our global survey is again in line with the 2005 recommendations [Citation24].
Internet databases were also very popular for PGx education, and equally so among all of the subgroups of responders. This is probably because they typically have open access, while many original research papers are still kept behind pay walls and are not accessible to all universities. This use of web resources in PGx education is also in line with the 2005 recommendations [Citation24]. By far the most frequently used website for PGx education was PharmGKB [Citation30], which was reported by 60.0% of all of the internet database users (B; Supplementary Table 1).
The majority of the textbooks used by our survey responders were in the field of pharmacology and toxicology, and in one instance biochemistry (Supplementary Table 2).
Pharmacogenomics education in PhD programs
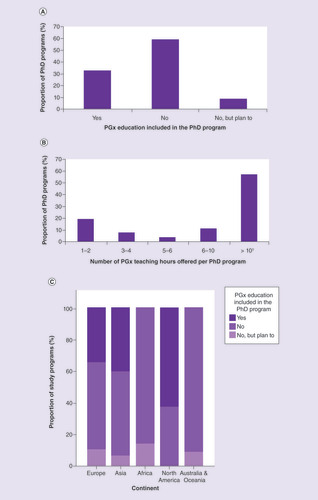
Finally, the responders were asked whether they offer PGx education to their PhD students; these findings are presented in . Of 30 PhD programs that offered PGx education, 43.3% were MD, 30.0% pharmacy, 23.3% graduate schools in faculties of medicine or life sciences, and 3.3% laboratory medicine. These data are similar to those of Murphy et al. [Citation27], who reported that 39% of their participating pharmacy schools and colleges teach PGx at the PhD level, and 10% offer PhD degrees focused on PGx. In a study by Pisanu et al. in southeastern Europe, PGx was taught in 23% of postgraduate programs (PhD, MSc, medical specialties studies), in 12.5% as a standalone course and in 87.5% as a part of other modules or courses [Citation20].
(A) PGx education included in PhD program. (B) Number of PGx teaching hours per PhD program. (C) PGx education in PhD programs by continent. None of the responders from Africa and Australia and Oceania include PGx topics in their PhD programs, with highest inclusion seen for North America (Fisher’s exact test; p = 0.127).
†Median (min–max) number of PGx teaching hours: 27.5 h (15–100 h).
PGx: Pharmacogenomics.
Limitations of this study
The major limitation of this study is that it had a relatively low response rate, which might have led to inaccurate representation of the global picture. However, the low response rate does not automatically imply low validity of the study, it simply indicates potentially greater bias [Citation31]. For example, evaluations of national surveys by Holbrook et al. [Citation32] with response rates ranging from 5 to 54% have concluded that studies with a much lower response rates were often only marginally less accurate than those with much higher response rates. More recently, Mealing et al. [Citation33]. considered relative risk estimates from two studies drawn from the same population in an Australian cohort study, and showed that despite the very different response rates (18 vs 60%) and different data collection methods (postal vs phone collection), the estimates of exposure and outcome relationships were particularly consistent where the same questions were asked. The most important indicator of the validity of a study are the differences between responders and nonresponders [Citation31]. Unfortunately, in this case, these differences, and consequently the study validity, could not be assessed due to the nature of the study design.
Another concern is the low absolute number of responding institutions per continent, and in particular for South America, where only two universities responded to the survey. Thus, the data obtained for South American schools are less likely to be representative, and therefore they were not included in the statistical analysis.
Surveying primarily top-ranked universities might introduce additional bias. Finally, the additional selection of universities from the ranks of the ESPT members (10 universities) might have led to the over-representation of European countries.
Conclusion
The majority of the recommendations that arose from the 2005 call for action [Citation24] that were assessed in the present survey are shown to have been addressed (). In short, the majority of the surveyed study programs include PGx education, which is mostly taught as part of a pharmacology curriculum. However, a minority of programs integrate PGx with other scientific fields, and thus in most study programs, personalized medicine teaching is limited only to the genomic level. Further recommendations based on our survey findings are given in Box 1. Future studies in the field of PGx education should also address PGx as part of the continuing education of healthcare professionals, and in addition, they must promote the dissemination of PGx knowledge in lay terms, for the general public. While these topics were not covered by our current survey, they will also be crucial for the success of implementing PGx tests for wider clinical use.
Table 3. Recommendations checklist from 2005, with the results realization and the future recommendations from the present study.
Overall, the state of pharmacogenomics (PGx) education has substantially improved since the publication of the first consensus paper [Citation24]. However, after reviewing all of the goals that were set, we find that there is still room for improvement ():
Although the majority of medical doctors (MD), pharmacy, nursing and public health school study programs offer PGx education in some form, it is mostly as a part of pharmacology and other topics. It would be unrealistic to recommend the implementation of PGx education as either an elective or a mandatory topic in most MD and nursing school programs. However, in pharmacy schools, an effort should be made to include PGx education as a stand-alone topic.
Most study programs offer the minimum recommended number of hours of PGx eduaction. However, < 50% of MD and nursing programs responding to our survey reached this goal. Thus, further efforts are needed to increase the number of PGx hours, with particular focus on MD and nursing programs.
The most encouraging finding of this survey is that educators have included relevant and clinically promising PGx markers in their teaching curricula. However, while the use of the CYP gene family markers is high, some 1A level of evidence markers (including some already widely applied in clinical practice) are being neglected. Thus, teachers should strive to increase the number of PGx markers taught, with a focus on those with high levels of evidence.
In addition to recommendations pertaining to the 2005 consensus paper [Citation24], we have identified several other points that are in need of improvement:
Programs with no PGx curricula were more common in non-Organization for Economic Co-operation and Development (OECD) countries, compared with OECD countries. We recommend the inclusion of PGx education in E-learning programs for low-income countries [Citation30] .
The number of PGx markers taught per program was significantly lower for Africa and Australia and Oceania, compared with other continents. Therefore, we recommend that educators strive to increase the number of PGx markers in their curricula to at least eight per program (i.e., median for Europe).
A minority of programs have integrated PGx with other scientific fields, and thus the teaching is limited to only the genomic level. We recommend the integration of PGx curricula with other ‘-omics’ (especially with epigenomics and metabolomics), and most importantly with bioinformatics (i.e., use of bioinformatics tools).
Most of the universities surveyed do not offer PGx education for PhD students, and in particular, none of the responders from Africa and Australia and Oceania. Conversely, >50% of the responders from North America implement PGx education in their biomedical PhD program. We strongly recommend the implementation of PGx education in particular for biomedical PhD programs globally, as students in these programs are the most likely to be employed by biomedical research institutions, private sector pharmaceutical and diagnostic companies, and the rapidly evolving E-health sector.
The majority of the study programs (87%) include pharmacogenomics (PGx) education, which is mostly taught as part of a pharmacology curriculum.
Sixty-three percent of programs offer at least the minimum recommended number of hours of PGx teaching (i.e., 3–4 h), as recommended by the 2005 consensus article.
The median number of PGx biomarkers used per study program was seven (1–11).
The most frequently taught PGx biomarkers were three CYP genes (CYP2D6, CYP2C19, CYP2C9), all of which were used at frequencies >80%, which is in line with the 2005 ISP recommendations.
The use of biomarkers in PGx education positively correlated with their level of evidence according to PharmGKB, the number of PubMed publications and the US FDA drug labels.
A minority of programs integrate PGx into other scientific fields, and thus in most study programs personalized medicine teaching is limited to only the genomic level.
The most frequently used educational tools for teaching PGx are original research papers (71%), followed by internet data bases (49%) and textbooks (41%).
Only 30% of study programs offer PGx as a part of the PhD curriculum.
Based on these results, we can conclude that the state of PGx education has substantially improved since 2005, although there remains room for improvement.
Supplemental information 1
Download Zip (382.1 KB)Supplemental information 2
Download PDF (172.8 KB)Supplemental information 3
Download PDF (201.2 KB)Supplemental document 4
Download MS Word (20.6 KB)Supplemental document 5
Download MS Word (21.8 KB)Acknowledgments
This study is dedicated to the memory of the founder and first President of the European Society of Pharmacogenomics and Personalized Therapy (ESPT), professor Gerard Siest (1936–2016), an educator and persistent supporter of PGx education.
Authors thank CP Berrie for the English language editing of the paper.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/pgs-2019-0009
Financial & competing interests disclosure
M Schwab was supported by the European Commission Horizon 2020 UPGx grant 668353, the ICEPHA Graduate School Tuebingen-Stuttgart (Germany) and the Robert Bosch Stiftung (Stuttgart, Germany). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Additional information
Funding
References
- Padmanabhan S . Handbook of Pharmacogenomics and Stratified Medicines.Academic Press, Cambridge, MA, USA (2014).
- Poste G . Bring on the biomarkers. Nature469(7329), 156 (2011).
- Luzum J , PakyzR , ElseyAet al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin.Pharmacol. Ther.102(3), 502–510 (2017).
- Manolio TA , ChisholmRL , OzenbergerBet al. Implementing genomic medicine in the clinic: the future is here. Genet. Med.15(4), 258 (2013).
- Shuldiner A , RellingM , PetersonJet al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin. Pharmacol. Ther.94(2), 207 (2013).
- Gurwitz D , ZikaE , HopkinsMM , GaisserS , IbarretaD. Pharmacogenetics in Europe: barriers and opportunities. Public Health Genomics12(3), 134–141 (2009).
- Lunshof J , GurwitzD. Pharmacogenomic testing: knowing more, doing better. Clin. Pharmacol. Ther.91(3), 387–389 (2012).
- Frueh F , GurwitzD. From pharmacogenetics to personalized medicine: a vital need for educating health professionals and the community. Pharmacogenomics5(5), 571–579 (2004).
- Sperber NR , CarpenterJS , CavallariLHet al. Challenges and strategies for implementing genomic services in diverse settings: experiences from the Implementing GeNomics In pracTicE (IGNITE) network. BMC Med. Genomics10(1), 35 (2017).
- Turner RM , ParkBK , PirmohamedM. Parsing interindividual drug variability: an emerging role for systems pharmacology. Wiley Interdiscip. Rev.7(4), 221–241 (2015).
- Wilkinson EM , IlhanZE , Herbst-KralovetzMM. Microbiota-drug interactions: Impact on metabolism and efficacy of therapeutics. Maturitas112, 53–63 (2018).
- Dodson C . Attitudes of oncology nurses concerning pharmacogenomics. Personal. Med.12(6), 559–562 (2015).
- Frick A , BentonC , SuzukiOet al. Implementing clinical pharmacogenomics in the classroom: student pharmacist impressions of an educational intervention including personal genotyping. Pharmacy (Basel)6(4), pii:E115 (2018).
- Haga SB , BurkeW , GinsburgGS , MillsR , AgansR. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin. Genet.82(4), 388 (2012).
- Haga SB , O’DanielJM , TindallGM , MillsR , LipkusIM , AgansR. Survey of genetic counselors and clinical geneticists’ use and attitudes towards pharmacogenetic testing. Clin. Genet.82(2), 115 (2012).
- Just K , SteffensM , SwenJ , PatrinosG , GuchelaarH , StinglJ. Medical education in pharmacogenomics – results from a survey on pharmacogenetic knowledge in healthcare professionals within the European pharmacogenomics clinical implementation project Ubiquitous Pharmacogenomics (U-PGx). Eur. J. Clin. Pharmacol.73(10), 1247–1252 (2017).
- Mahmutovic L , AkcesmeB , DurakovicCet al. Perceptions of students in health and molecular life sciences regarding pharmacogenomics and personalized medicine. Human Genomics12(1), 50 (2018).
- Mai Y , MitropoulouC , PapadopoulouXEet al. Critical appraisal of the views of healthcare professionals with respect to pharmacogenomics and personalized medicine in Greece. Personal. Med.11(1), 15–26 (2014).
- Marcinak R , ParisM , KinneySRM. Pharmacogenomics education improves pharmacy student perceptions of their abilities and roles in its use. Am. J. Pharm. Ed.82(9), 6424 (2018).
- Pisanu C , TsermpiniE , MavroidiE , KatsilaT , PatrinosG , SquassinaA. Assessment of the pharmacogenomics educational environment in southeast Europe. Public Health Genomics17(5–6), 272 (2014).
- Stanek E , SandersC , TaberKet al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin. Pharmacol. Ther.91(3), 450 (2012).
- Taber KaJ , DickinsonBD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmacogenomics Pers. Med.7, 145 (2014).
- Malentacchi F , ManciniI , BrandslundIet al. Is laboratory medicine ready for the era of personalized medicine? A survey addressed to laboratory directors of hospitals/ academic schools of medicine in Europe. Clin. Chem. Lab. Med.53(7), 981–988 (2015).
- Gurwitz D , LunshofJ , DedoussisGet al. Pharmacogenomics education: International Society of Pharmacogenomics recommendations for medical, pharmaceutical, and health schools deans of education. Pharmacogenomics J.5(4), 221 (2005).
- Green J , O’BrienT , ChiappinelliV , HarralsonA. Pharmacogenomics instruction in US and Canadian medical schools: implications for personalized medicine. Pharmacogenomics11(9), 1331 (2010).
- Higgs JE , AndrewsJ , GurwitzD , PayneK , NewmanW. Pharmacogenetics education in British medical schools. Genomic Med.2(3–4), 101 (2008).
- Murphy JE , GreenJS , AdamsLA , SquireRB , KuoGM , McKayA. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am. J. Pharm. Ed.74(1), 7 (2010).
- Caudle KE , KleinTE , HoffmanJMet al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab.15(2), 209–217 (2014).
- Gurwitz D , WeizmanA , RehaviM. Education: teaching pharmacogenomics to prepare future physicians and researchers for personalized medicine. Trends Pharmacol. Sci.24(3), 122–125 (2003).
- Owen RP , KleinTE , AltmanRB. The education potential of the pharmacogenetics and pharmacogenomics knowledge base (PharmGKB). Clin. Pharmacol. Ther.82(4), 472–475 (2007).
- Morton SM , BandaraDK , RobinsonEM , CarrPE. In the 21st Century, what is an acceptable response rate?Aust. N. Z. J. Public Health63(2), 106–108 (2012).
- Holbrook A , KrosnickJ , PfentA. The causes and consequences of response rates in surveys by the news media and government contractor survey research firms. In: Advances in Telephone Survey MethodologyLepkowskiJM, TuckerNC, BrickJM, DeLeeuw ED, JapecL, LavrakasPJ ( Eds). Wiley, New York, NY, USA (2007).
- Mealing NM , BanksE , JormLR , SteelDG , ClementsMS , RogersKD. Investigation of relative risk estimates from studies of the same population with contrasting response rates and designs. BMC Med. Res. Methodol.10, 26 (2010).
