Abstract
Pharmacogenetics is the study of genetic differences in an individual that leads to variability in drug response. Single-nucleotide polymorphisms (SNPs) prove to be important determinants in evaluating and predicting a patient’s response to certain medications and risk of adverse events. The Cytochrome P450 2D6 (CYP2D6) gene is particularly important in the metabolism of many clinically prescribed drugs. In this study, we designed a multiplexed panel to interrogate 8 clinically relevant SNPs of CYP2D6 (*2 at C2856G, *2 at G4181C, *3, *4, *5,*6, *7, and *8). We PCR-amplified a 4.7 kB segment of the CYP2D6 locus containing all the SNPs of interest using genomic DNA extracted from whole blood. Using single base extension and capillary electrophoresis separation, peaks corresponding to the SNPs resolve within a 25–60 bp window. We subsequently analyzed 25 samples using this protocol and compared results to traditional DNA sequencing using an ABI 3730. All samples were 100% concordant between the two methods. This assay can be performed with <24 h turnaround time and minimal hands-on effort. This multiplex SNP panel can be used for interrogation of 8 SNPs within the 2D6 gene, with application to identifying poor metabolizers of 2d6. Patients harbouring SNPs in 2D6 could be triaged to alternative therapies in an effort to maximize therapeutic efficacy and reduce adverse drug reactions.
Introduction
Pharmacogenetics refers to the study of genetic differences that result in interindividual variability of drug pharmacokinetics, that is, drug absorption, distribution, metabolism and excretion. It has been long observed that given the same drug at the same dosage, patients can exhibit substantial heterogeneity in drug response, efficacy and toxicity both on a population scale and among individuals [Citation1,Citation2]. Besides obvious factors such as age, gender and lifestyle, genotypic traits such as the presence of specific single-nucleotide polymorphisms (SNPs) prove to be important determinants in evaluating and predicting a patient's response to medications and risk of adverse events. Identifying and characterizing efficiently such genetic variants in a patient's genotypic profile can therefore allow for the individual tailoring of therapeutic intervention, drug selection and dose adaptation. The ultimate goal of personalized medicine is to optimize therapeutic response as well as drug safety, thus improving clinical outcome for each patient [Citation2,Citation3].
Understanding factors that contribute to interindividual variability in drug metabolism is central to explaining differences in pharmacologic responses and clinical outcomes among patients. Cytochrome P450 (CYP), a monooxygenase superfamily of over 60 drug-metabolizing enzymes, is responsible for mediating 70–80% of all phase I-dependent metabolism of commonly prescribed drugs as well as metabolism of countless xenobiotic chemicals in the liver [reviewed in Citation4].
Among the various isoforms, cytochrome P450 2D6 (CYP2D6) is a particularly important and extensively studied CYP responsible for the phase-I biotransformation of approximately 25% of all clinically prescribed medications, including many blockbuster drugs such as paroxetine, metoprolol and risperidone [Citation5]. With over 100 single-nucleotide polymorphisms (SNPs), 26 null alleles and other alleles with decreased activity identified to date [Citation6], its highly polymorphic nature has given rise to different metabolic phenotypes for 2D6 substrates: poor metabolizers (PM) with no functional alleles, intermediate metabolizers (IM) with one functional allele, extensive metabolizers (EM) with normal alleles and ultrarapid metabolizers (UM) with gene duplications. Among Caucasians, 7–10% are considered PMs of CYP2D6 substrates, where 99% of those PMs can be correlated with the presence of SNPs CYP2D6*3, *4, *5, *6, *7 and *8 in the gene.
In this study, we designed a multiplexed panel to interrogate 8 clinically relevant SNPs of CYP2D6 (*2 at C2856F, *2 at G4181C, *3, *4, *5, *6, *7, *8) in a high-throughput and time-efficient fashion. We amplified the CYP2D6 gene from genomic DNA extracted from whole blood, targeted SNP extension using specific oligonucleotide primers, and interrogated the genotypes of the SNPs of interest after capillary electrophoresis separation using the Genetic Expression Profiler (GeXP) technology. The development of such an assay has major clinical applicability. By readily and accurately identifying important SNPs of this important drug-metabolizing enzyme, our assay can provide physicians with valuable insights of a patient's ability to metabolize countless major classes of commonly prescribed medications. This assay thus can provide the information a physician needs to predict a patient's response to any drug metabolized by CYP2D6, thereby improving drug selection and dosing while decreasing risks of overdosing and other side effects.
Materials and methods
Genomic DNA extraction
Genomic DNA was extracted from whole blood using the Beckman SPRI-TE automated nucleic acid extraction platform using the gDNA kit (Beckman Coulter Inc, Fullerton, CA, USA). Briefly, 300 μL of whole blood were used to obtain 50 or 100 μL of genomic DNA (100 mg/L).
Long-range Polymerase Chain Reaction (PCR) of the 2D6 gene
Long-range PCR was performed using an Expand Long Template PCR system (Roche, Indianapolis, IN, USA). 0.5 μmol/L of the forward primer P100 and reverse primer P200 (see and [Citation7], 1 mol/L betaine (Sigma, St. Louis, MO, USA), 200 μmol/L dNTP Mix (Qiagen, Valencia, CA, USA) and 50 ng genomic DNA were added to 0.6 U of thermostable Taq DNA in a final volume of 20 μL. The thermocycling conditions for this PCR were as follow: initial denaturation at 94°C for 2 min, 35 cycles of 92°C for 15 s, 55°C for 30 s and 68°C for 10 min, and final extension at 68°C for 7 min. To verify the amplification of the desired 4.7kb CYP2D6 gene product, 5 μL of PCR product was electrophoresed on a pre-cast 1.2% agarose gel (Invitrogen, Carlsbad, CA, USA). In addition, 3 μL of PCR product was digested using BamHI restriction enzyme (NEB Inc., Ipswich, MA, USA) and the presence of 3 fragments was verified on 1.2% agarose gel.
Table I. Primer sequences used for this report.
Single-Nucleotide Polymorphism (SNP) extension reactions
50 μL of the PCR product was purified using a QIAquick® DNA purification kit (Qiagen, Valencia, CA, USA) and resuspended in 50 μL of elution buffer. The GenomeLab™ SNPStart Primer Extension (Beckman Coulter, Fullerton, CA, USA) kit was used for SNP interrogation. 1 μmol/L of the appropriate SNP interrogation primer () was added to 3 μL of purified PCR product, 4 μL of the MasterMix, 1.5 mol/L of betaine and brought to a final reaction volume of 11 μL. A positive control containing 4 μL of the MasterMix, 1 μL of Positive control and 5 μL of nuclease-free PCR grade water was also prepared. The thermocycling conditions for this SNP extension PCR were as follows: initial denaturation at 95°C for 1 min, followed by 30 cycles of 90°C for 10 s and 48°C for 20 s. In the case of SNP multiplex interrogation, thermocycling conditions were identical to singlet interrogation but with 40 cycles instead of 30. After the SNP reaction, excess dye-labeled terminators were degraded by Shrimp Alkaline Phosphatase (USB, Cleveland, OH, USA) at 37°C for 40 min.
Multiplexed SNP analysis using the GeXP analyzer
1 μL of each purified SNP reaction product was loaded to a mixture containing 38.5 μL of Sample Loading Solution from the SNPStart kit (Beckman Coulter Inc., Fullerton, CA, USA) and 0.5 μL of GenomeLab DNA Size Standard 80 (Beckman Coulter Inc., Fullerton, CA, USA) in a sample plate. Once overlaid with one drop of mineral oil in each well to prevent evaporation, the sample was loaded onto the Genetic Expression Profiler (GeXP) for capillary electrophoresis of the SNP reaction products. The experiment was run using the SNP-1 method program at a capillary temperature of 50°C, and the raw data was analyzed with the GenomeLab SNP Software.
Sequencing of CYP2D6
The 2d6 gene was sequenced using an ABI 3730 at Memorial Sloan-Kettering Cancer Center.
Results
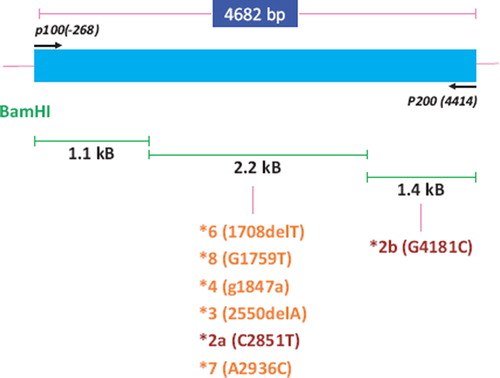
A schematic diagram of the genomic locus of cytochrome P4502d6 is shown in , with the location of the SNPs of interest also depicted. PCR amplification using genomic DNA extracted from blood generates a single product of ∼4.7 kB (data not shown). provides details for all primer sequences used. BamHI digest of this amplicon results in three bands of 1.1, 2.2, and 1.4 kB. As can be seen in the figure, nearly all SNPs of interest are located within the central fragment of 2.2 kB. One SNP is located in the distal fragment of 1.4 kB.
Figure 1. Schematic diagram of the genomic locus of cytochrome P4502D6 and the location of the various SNPs of interest.
Our assay entails the following major steps: genomic DNA extraction from whole blood, PCR amplification of the 2d6 locus followed by exonuclease-shrimp alkaline phosphatase (ExoSAP) treatment, single base extension analysis followed by shrimp alkaline phosphatase (SAP) treatment, and finally capillary electrophoresis separation. These steps, along with the approximate duration, are schematically diagrammed in . Details on the single base extension analysis can be found in . We used this assay to genotype 25 samples for each of the eight SNPs detected with our assay. We observed 100% concordance for a total of 200 SNPs when compared with sequencing using an ABI 3730.
Figure 2. Workflow for multiplexed SNP analysis using capillary electrophoresis (right) and length of time required at each step (left).
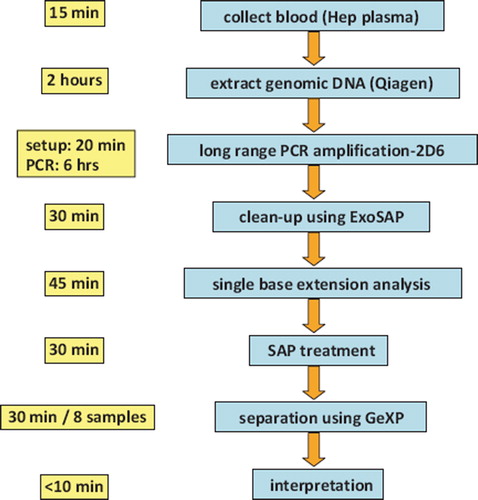
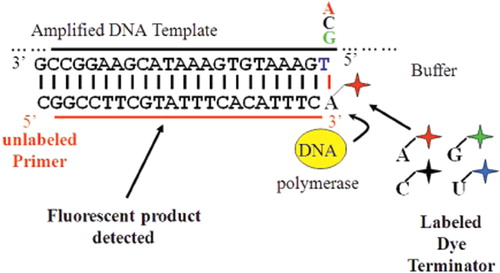
Figure 3. Schematic diagram of single base extension analysis. An oligonucleotide primer, sequence ending one nucleotide upstream of SNP of interest, is extended by one base with DNA polymerase. Each base if labelled with a different colour fluorophore allowing for interrogation of the SNP of interest at detection.
Discussion
The gene for cytochrome P4502D6 encodes the enzyme debrisoquine hydrolase, which is responsible for the metabolism of approximately 25% of all clinically prescribed drugs (4). With over 100 different alleles (6), this gene product is one of the most highly polymorphic in the human genome. These various alleles produce effects that can stratify individuals based on the enzymatic activity conferred on the 2d6 gene product into poor metabolizers, intermediate metabolizers, extensive metabolizers, and ultrarapid metabolizers.
We describe herein the design of a multiplex Cytochrome P4502D6 panel in order to interrogate several SNPs within this gene. The entire workflow, including PCR and SNP analysis, can be performed with total turnaround time <24 h (). Such a workflow can be generally applied for multiplexed SNP analysis and provides feasibility evidence for the design of such a panel.
We sequenced 25 random samples and demonstrate 100% correlation to our GeXP-based assay, demonstrating the feasibility of our assay for accurate genotyping analysis. Our panel, which includes 2D6 *2 (2 distinct loci), *3, *4, *6, *7, and *8, can identify individuals that are poor metabolizers and may exhibit a compromised ability to metabolize 2d6 substrates. In addition, the initial PCR amplification step can identify individuals that are *5, i.e. gene deletion of cytochrome P4502D6. Identification of such individuals would be useful so that these patients could be placed on alternative medications to enhance therapeutic benefit or to reduce adverse drug reactions, a costly problem and health burden both in the United States and worldwide ([Citation8] and references therein).
Future experiments may entail the addition of other SNPs to our multiplex panel. These SNPs could be either in the 2d6 gene or from other genes encoding drug metabolizing enzymes. The ability to rapidly make changes to the original multiplex panel exploits the flexibility of this assay design to add/remove SNPs of interest with ease.
In conclusion, we have designed a multiplex assay to assist in genotyping patients for 2d6 status. Identification of poor metabolizers of the 2d6 gene allows for triaging such individuals to alternative therapeutic regimens which are not metabolized through the 2d6 pathway. Such intervention will enhance therapeutic benefit and/or reduce the incidence of adverse drug reactions. The development of such assays, which can be used to genotype patients, in combination with biomarker assays that are used to stratify disease, will help to usher in the new era of personalized medicine.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Evans WE, McLeod HL. Pharmacogenomics–drug disposition, drug targets, and side effects. N Engl J Med. 2003 Feb 6;348(6):538–49.
- Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. Nature. 2004;429(6990): 464–8.
- Ely S. Personalized medicine: individualized care of cancer patients. Transl Res. 2009;154(6):303–8. Epub 2009 Sep 1.
- Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41(2):89–295.
- Owen RP, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2D6. Pharmacogenet Genomics. 2009 Jul;19(7):559–62.
- http://www.cypalleles.ki.se/cyp2d6.htm (Accessed 100510).
- Roberts RL, Kennedy MA. Rapid detection of common cytochrome P450 2D6 alleles in Caucasians. Clin Chim Acta. 2006;366(1–2):348–51. Epub 2005 Dec 20.
- Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286(18): 2270–9.