Abstract
The human genome contains multiple stretches of CGG trinucleotide repeats, which act as transcription- and translation-regulatory elements but at the same time form secondary structures that impede replication and give rise to sites of chromosome fragility. Proteins binding to such DNA elements may be involved in divergent cellular processes such as transcription, DNA damage, and epigenetic state of the chromatin. We review here the work done on CGG repeats and associated proteins with special focus on a factor called CGGBP1. CGGBP1 presents with an interesting example of factors that do not have any single dedicated function, but participate indispensably in multiple processes. Both experimental results and data from cancer genome sequencing have revealed that any alteration in CGGBP1 that compromises its function is not tolerated by normal or cancer cells alike. Based upon a large amount of published data, information from databases, and unpublished results, we decipher in this review how CGGBP1 is a classic example of the ‘one factor, divergent functions’ paradigm of cytoprotection. By taking cues from the studies on CGGBP1, more such factors can be discovered for a better understanding of the evolution of mechanisms of cellular survival.
Keywords:
Introduction
The enormous complexity of the human genome can only be understated. The functional annotation of the genomic regions coding for proteins and non-coding RNAs is rapidly progressing, whereas our knowledge about the functions and mechanisms of regulation of repetitive DNA has lagged behind. The relatively slow progress can mainly be ascribed to technical problems associated with the sequencing of repetitive DNA and to the relative lack of experimental protocols for functional studies on them.
The tandem repeats constitute a major part of our genomes (Citation1). The longest tandem repeats are located at the transcriptionally inactive telomeric and centromeric DNA, whereas the shorter repeats are scattered across the genome (Citation2) or present as extra-chromosomal circular DNA (Citation3). Depending on their length and location, they exert different effects on genome function and stability. The GC-rich tandem repeats, such as the CGG trinucleotide repeats, are special because they are highly rich in CpG sites that can be methylated (Citation4) and are more likely to occur in the gene-rich regions with high GC-content (Citation1). In fact, the CGG repeats in promoters and transcribed regions of some genes were identified as early as 1992 (Citation5). The CGG repeats pose unique challenges to our genome: they act as transcription- and translation-regulatory elements (Citation6), adopt secondary structures that hinder replication fork progression (Citation7), and give rise to sites of chromosome fragility (Citation8). CGG repeats are under constant CpT-to-TpG drift due to spontaneous deamination of methylated cytosines in the CpG context (Citation1). Given these properties of the CGG repeats, it is surprising that perfect and imperfect CGG repeats of varying lengths have accumulated in our genomes. Knowledge about how the cells deal with the CGG repeats, perhaps through a host of CGG-binding factors, can shed light on the general nature of the mechanisms the cells employ to keep the repetitive DNA under check and counteract their adverse effects.
CGG repeats and CGGBP1
The existence of a CGG repeat-binding protein was experimentally demonstrated in 1990 as a factor that specifically binds to the 5′UTR CGG repeat of the human BCR gene (Citation9). Subsequently, a CGG triplet repeat-binding protein was identified through its affinity in vitro for both double-stranded (ds) CGG repeats and single-stranded (ss) CCG oligonucleotides and termed CGG-BP1 (Citation10). Later, Deissler and co-workers found various factors from nuclear extracts of human, mouse, fish, and insect cells that form complexes with CGG oligonucleotides in vitro (Citation6). HeLa cell nuclear extracts were subsequently used to isolate one of these protein-DNA complexes, and after characterization using mass spectrometry a 20 kDa protein was identified (Citation11). In the absence of any knowledge about its biological functions, the protein was given the generic name CGGBP1 (Citation10).
In addition to CGGBP1, these studies revealed the binding of many other proteins, some with unknown functions, to the CGG repeats (Citation9). Identification of the CGG repeat-binding proteins has shed light on the mechanisms of regulation of CGG repeats. The proteins identified in these studies, other than CGGBP1, include XRCC5, XRCC6, WRN, CBF-A (HNRNPAB), heterogenous nuclear ribonucleoprotein-related telomere-binding proteins (UP1), and ZF5 (Citation9). CGG triplet repeats are inherently prone to single-strand hairpin structure formation which causes errors during DNA replication including DNA replication fork stalling (Citation12) and spontaneous expansion due to polymerase stuttering (Citation7). Interestingly, unlike CGGBP1, many of these proteins had other previously established functions, which include stabilization/destabilization of hairpin structures of the repeats, effects on replication and expansion through collaboration with DNA polymerase, and transcription regulation (Citation13).
Until recently, CGGBP1, unlike other CGG-binding proteins, was portrayed as a dedicated CGG repeat-binding protein with CGG repeat-associated transcription-regulatory functions only (Citation6). Some recent developments in our knowledge about CGGBP1 have, however, revealed that it also shares functionalities with other CGG-binding proteins. These functions include DNA damage/repair and telomere metabolism with indications of its involvement in mRNA metabolism as well. Currently we are only beginning to understand the seemingly complex functions of CGGBP1 as indicated by its conservation amongst mammals, ubiquitous expression pattern, and in vitro and in situ functional assays. The majority of direct functional studies on CGGBP1 were performed by Doerfler and colleagues (Citation6) and more recently by our group (Citation14). These findings have been supplemented and supported by information about the structure of CGGBP, its evolution, and various data from large-scale experiments not aiming to investigate CGGBP1 specifically. The collective information on CGGBP1 reveals its role in a vast repertoire of vital cellular functions. Here we review and analyse the information about CGGBP1 available in different databases and integrate it with published data as well as unpublished results on different aspects of CGGBP1, which encompass its evolution, expression pattern, and molecular and biological functions. It appears that CGGBP1 participates in growth signal-induced gene expression, silencing of interspersed repeats, CpG methylation, endogenous DNA damage, chromosomal segregation, and cytokinesis. Thus, CGGBP1 emerges as a central regulator of cell growth and proliferation with indispensable cytoprotective functions.
Structure and evolution of CGGBP1
CGGBP1 is a 167 amino acid long 20 kDa protein (Citation11) with a nuclear localization signal from amino acid (aa) 80-84 (Citation15), which includes a double lysine residue at position 81-82. A C2H2-type Zn finger domain is located between aa 43 and 67 as predicted by the amino acid sequence (RCSB Protein Data Bank). By mutational analysis the DNA-binding activity of CGGBP1 was traced to a small region between aa 67-71 and a large C-terminal region from aa 95-167 (Citation15); the C2H2 DNA-binding domain (DBD) overlaps with the former (), and the latter seems to be a modulator of the DNA-binding property. In vitro, incubation of crude nuclear extracts with CGG repeat oligonucleotides gives rise to multiple mobility-retarded bands in electrophoretic mobility shift assays (Citation10). It has been argued that some of these multiple bands could be due to binding of CGG repeats to other nuclear proteins such as MECP1 (Citation10). However, it has been proposed that CGGBP1 binds to target DNA in vitro as oligomers and this could also account for multiple mobility-shifted bands (Citation15).
Figure 1. Evolution and structure of CGGBP1. A: A schematic depicting the known and predicted domains and functional sites in human CGGBP1. The SH2 domain, the C2H2 domain, and the nuclear localization signal (NLS) are highlighted. The three tyrosine residues (positions 20, 150, 155) and one serine residue (position 164) are marked out. The cellular effects of phosphorylation of these amino acids have been studied. B: An I-TASSER structure prediction using CGGBP1 amino acid sequence predicts sequence-based structural similarities with proteins Hermes DNA transposase (2BW3; in the C-terminal half) and with ZNF346 from Xenopus laevis (1ZU1; all throughout the peptide sequence). The NLS and C2H2 DNA-binding domain (DBD) have been highlighted. C: The predicted 3-dimensional structure of CGGBP1 from different angles of view. The C- and N-termini are marked as ‘C’ and ‘N’, respectively. The two cysteine and histidine residues forming the C2H2 Zn finger domain are also identifiable through their side chains that converge at the zinc ion (shown).
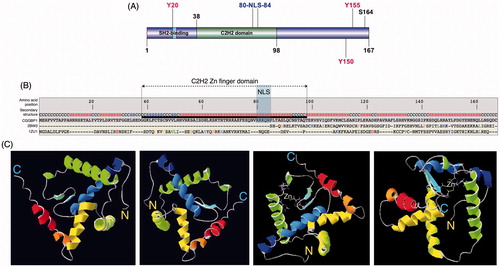
The 3D structure of CGGBP1 has not yet been solved. I-TASSER (Iterative Threading ASSEmbly Refinement) (Citation16) predicts a structure that justifies the amino acid sequence-based domain prediction. The template protein to which CGGBP1 structure shows highest similarity is the Hermes DNA transposase (Musca domestica), a member of the Hobo, Ac, Tam3 (hAT) family of DNA transposases, and a Zn-finger domain dsRNA-binding protein Znf346 from Xenopus laevis () (Singh and Westermark, unpublished observation). The predicted structure shows that the N-terminal half of CGGBP1 forms a C2H2 Zn finger domain comprising two major alpha helices and two beta sheets, followed by an approximately 10 aa long linker region from aa 83 to 92. The remaining C-terminal region is predicted to be organized into three alpha helices, which together constitute a dimerization domain () (Singh and Westermark, unpublished observation). Such a presence of two major domains of roughly equal size is supported by heteronuclear single quantum coherence NMR spectrum data for CGGBP1 (Singh, Berglund, Pedersen, Westermark, unpublished observations). While the N-terminal region of CGGBP1 has a DNA-binding function, the C-terminal half might enable formation of complexes between different CGGBP1 molecules, including its own oligomerization as has also been predicted earlier (Citation15).
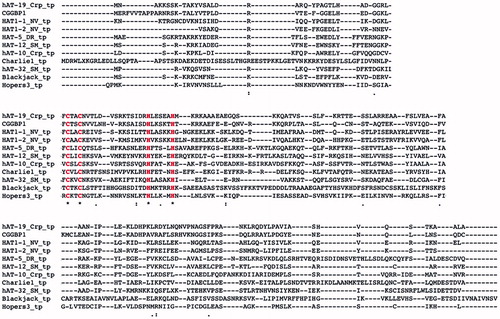
The C2H2 Zn finger DNA-binding domain of CGGBP1, formed by cysteine residues 43 and 46 and histidine residues 61 and 67, shows significant similarity to the BED Zn finger domain of the hAT family DNA transposases (Singh and Westermark, unpublished findings). Although the C2H2 domain of CGGBP1 is not predicted as a BED domain, it does have a conserved aromatic amino acid phenylalanine at positions 42 and 74 flanking the C2H2 domain, a feature of a BED domain (Singh and Westermark, unpublished findings). The Zn finger transcription factors related to the Ac subgroup of the hAT family of DNA transposases (to which the Hermes transposase belongs) are derived from one of the two proposed independent domestication events (Citation17). One of these gave rise to the transcription factor ZBED1, which is conserved in all jawed vertebrates except amphibians (Citation17). Interestingly, an NCBI Homologene search shows that CGGBP1 is also conserved between all amniotes and is absent from amphibians. A genomic analysis of the CGGBP1 locus also shows similarities between mammals and chicken, enough for a cross-hybridization in Southern blot analysis using a human CGGBP1 probe (Citation6). However, the same probe fails to hybridize with frog, fish, or drosophila DNA samples (Citation6). A guided structural similarity search using ZBED1 NMR structure as template indicates that ZBED1 and CGGBP1 have similar structures (Singh and Westermark, unpublished findings) and probably both originated from the Hermes transposase. An independent approach to identify Zn finger transcription factors originating from the hAT superfamily of DNA transposases in the human genome revealed CGGBP1 as the strongest candidate (Smit, unpublished findings). An amino acid sequence alignment shows a high sequence conservation between the DBD of the DNA transposase of hAT Charlie and CGGBP1 with the two cysteine and histidine residues of the C2H2 domain preserved () (Smit, unpublished findings). Overall the information supports the view that CGGBP1 has evolved from DNA transposons and could thus share its ability to bind a variety of DNA sequences as an oligomer. Interestingly, as is discussed in detail below, subsequent to its evolution from DNA transposons, CGGBP1 seems to have acquired transposon-regulatory functions.
Figure 2. Sequence similarity between DNA transposons and CGGBP1 indicates a common origin. A sequence alignment of CGGBP1 against Charlie group of hAT transposases suggests that CGGBP1 evolved from the DBD of these transposases. Interestingly, the two cysteine and histidine residues constituting the C2H2 domain are conserved across all the sequences analysed, suggesting an evolutionary pressure to preserve the DBD.
Genomic location and regulation of CGGBP1
The human CGGBP1 gene is located at cytogenetic band 3p11.1, the most proximal band to the centromere (NCBI Genome database) (Citation11). Four promoters (p1, p2, p3, and p5) have been identified using the capped analysis of gene expression. While p1, p3, and p5 are clustered within co-ordinates 88108083 and 88108203, p2 lies upstream (transcription from the reverse strand) between co-ordinates 88199008 and 88199035. Transcription from p2 yields an mRNA which is over 9 kb longer than the p1/p3/p5-derived transcripts, with a 5′UTR and first introns larger than those produced from downstream promoters (Citation18). All transcripts contain the same open reading frame and code for an identical 167 aa long protein. The existence of multiple transcripts was also shown through Northern hybridization (Citation19). However, the significantly longer 5′UTR is likely to predispose the p2 transcripts to additional post-transcriptional mechanisms of gene regulation, such as miRNA targeting and translational regulation by RNA-binding proteins. In addition, the p2 promoter may be controlled by cis elements other than the clustered p1, p3, and p5 promoters. Interestingly, transcription from the p1 and p2 promoters accounts for most of the CGGBP1 mRNA expression, with p1 as the strongest promoter (Citation20). The p2-driven CGGBP1 gene encompasses transcription start sites for ZNF654 and C3ORF38 in anti-sense direction (FANTOM database: http://fantom.gsc.riken.jp/zenbu/gLyphs/#config = b-IMGb5lG53ntH8qgNeChB;loc = hg19::chr3:88076621.88223495+) (Citation21). This raises a possibility of mutual post-transcriptional gene expression regulation between CGGBP1-p2, ZNF654, and C3ORF38 transcripts. As discussed later, an analysis of CAGE data at FANTOM database (Citation22) reveals enhanced p2 activity in cancer, unlike other CGGBP1 promoters.
Two enhancer elements associated with p1 and p2 promoters each (genomic co-ordinates 88079560 to 88079697 and 88208920 to 88209337 on chromosome 3) have been identified, both being permissive in nature (Citation20). The levels of CGGBP1 during development and differentiation might be regulated through these enhancers. Mouse Cggbp1, although >99% conserved with human CGGBP1 (Citation23), differs in the genomic organization. There is only one cluster of multiple promoters but no distant p2-like promoter (Citation22). Also, unlike in humans, the Zfp654 gene is non-overlapping with Cggbp1 with no possibilities of post-transcriptional gene regulation through anti-sense RNA. This points towards subtle differences between the mouse Cggbp1 and human CGGBP1 genes and precludes a simple extrapolation of findings about CGGBP1 regulation from one species to another.
The human CGGBP1 is ubiquitously expressed, generally at high levels. The ‘Bgee’ database (Citation24) describes CGGBP1 as expressed in 134 tissue types and 136 developmental stages, whereas ‘Genevisible’ (Citation25) describes it as expressed at medium-to-high level in 325 tissues. Mouse Cggbp1 is also expressed ubiquitously (Citation26). A direct evidence of developmentally regulated expression of Cggbp1 is the selective increase in Cggbp1 expression during ear development from otic vesicle to inner ear (Citation27).
More objective analysis of CGGBP1 expression by Northern hybridization was performed by Doerfler and colleagues (Citation19). The human CGGBP1 gene seems to contain several mini cistrons in the 5′UTR that probably do not code for peptides (Citation19). The 3′UTR has two prominent poly-adenylation sites that result in two transcripts of 1.2 kb and 4.3 kb (Citation19). Expression of both transcripts is detectable in various human and mouse tissues, with the 4.3 kb transcript being more strongly expressed (Citation19). The mouse and human (non-p2) CGGBP1 promoters are highly conserved, and both have a CGGx6 repeat upstream of a cluster of binding sites for CCAAT enhancer-binding protein and SP1 (Citation19). The CpG dinucleotides in the CGGBP1 promoter are heavily unmethylated and associated with high transcriptional activity of the promoter (Citation19). In vitro assays have shown that the CCAAT boxes and SP1-binding sites are required for driving luciferase expression using CGGBP1 promoters (Citation19). In vitro methylation of the promoter, however, silences the luciferase activity driven by CGGBP1 promoters (Citation19).
The presence of CGG repeats in CGGBP1’s own promoter suggests a feedback loop through which CGGBP1 can regulate its own expression. Since the CGG repeat is not required for expression, it may be working as a methylation-dependent silencing element for the CGGBP1 gene. Whether CGGBP1 binds to this repeat or not is not known.
Functions of CGG repeat-binding proteins: a precedent for functions of CGGBP1
Although CGG repeats are known in the 5′ ends of some genes, the spontaneously expandable CGG repeat in the 5′ end of the human FMR1 genes has been a major tool for studying the effects of CGG repeats on gene expression. Until recently, the functions of CGGBP1 other than regulation of transcription have been unknown. The strong focus on the FMR1-associated CGG repeat has been prejudicial to an unbiased approach to unveil novel functions of CGGBP1. To this end, a survey of the commonality in functions of other CGG-binding proteins can prospectively shed light on hitherto unknown functions of CGGBP1.
The Werner’s syndrome nuclease protein WRN is a DNA helicase and exonuclease that plays important roles in DNA repair and is recruited to CGG repeats. WRN is preferentially recruited to repetitive DNA sequences, such as telomeres, that can form secondary structures. It is required for telomeric integrity, and it prevents telomere fusions, genomic instability, and premature senescence (Citation28). CGG repeat-binding factors XRCC5 and XRCC6 are involved in DNA non-homologous end joining (NHEJ) and repair of double-strand breaks. The XRCC5/6 heterodimer is also required for efficient DNA recruitment of DNA protein kinase (DNA-PK), a key enzyme of the PI3 kinase-like kinase family that co-operates with Ku protein to detect double-strand breaks and phosphorylates H2AX at serine 139. It thus stabilizes dsDNA breaks and initiates repair through NHEJ. XRCC5/6 strikes the balance between NHEJ-mediated lengthening of telomeres and homologous recombination (HR)-mediated shortening of telomeres (Citation29). HNRNPAB, though identified as a DNA (CGG repeat)-binding protein, is an RNA-binding protein with mRNA-editing function through interactions with APOBEC1 (Citation30). It has been demonstrated to be involved in editing of apolipoprotein B mRNA. Members of the APOBEC family, which function through their cytidine deaminase activity on RNA as well as DNA, act as negative regulators of CpG methylation (Citation31). Another CGG-binding factor with mRNA-editing activity is UP1, a proteolytic fragment of HNRNPA1, which is a protein important for assembly of mRNA into hnRNP particles, nucleus-to-cytoplasm mRNA transport, and splice site selection (Citation32). ZBTB14 (ZF5) is a protein with multiple C2H2 Zn finger domains and has transcriptional silencing functions at promoters of growth-supporting genes such as MYC and thymidine kinase (Citation33). These proteins serve to destabilize the intra-strand tetrahelical structures at the CGG repeats and facilitate a smooth passage of DNA polymerase during replication, although the XRCC proteins might also stabilize secondary structures formed by the CGG repeats (Citation29). There is also evidence that the RNA binding of some of the CGG-binding factors may affect the mRNA translation and stability (Citation34). Overall the functions of these known CGG-binding proteins seem not to be restricted to CGG repeats but extend to telomere homeostasis, DNA damage/repair, mRNA stability, splicing, and translation.
Although the functions of CGG repeat-binding proteins can indicate novel functions of CGGBP1, there is a unique feature of CGGBP1. Of all the CGG repeat-binding factors described above, CGGBP1 was the one specifically identified as binding only to unmethylated, and not to methylated, CGG repeats (Citation15); it is interesting to note that CGG repeats constitute dense methyl-able CpG target sites. These findings were obtained by studying the transcription regulation of the FMR1 gene by CGGBP1 binding to a CGG repeat in its upstream region (Citation15). Interestingly, the CGG repeat-binding factors ZF5 and CGGBP1 co-operate to regulate FMR1 expression (Citation33).
Functions of CGGBP1
As the binding of CGG oligonucleotides to CGGBP1 was detected in nuclear extracts, the nuclear presence of CGGBP1 was expected (Citation10). The nuclear localization was confirmed by expressing transgenic CGGBP1 in human cells (Citation15). In addition, GFP-tagged CGGBP1 was shown to bind to the short arms of human acrocentric chromosomes (Citation15). These regions contain the rRNA gene clusters, which are rich in CGG repeats, thereby reaffirming the affinity of CGGBP1 to CGG-rich DNA in situ (Citation15). A closer examination of these findings reveals that significant above-background binding of GFP-tagged CGGBP1 occurs also on the GC-rich R-bands of all chromosomes. These sequences may be rich in small CGG repeats as well as other hitherto unidentified GC-rich CGGBP1-binding sequences. While the nuclear expression of CGGBP1 is extremely strong, different databases and published findings report extra-nuclear presence of CGGBP1 as well (Citation35). We have also observed endogenous as well as transgenic CGGBP1 in nuclei and in cytoplasm in interphase cells (Citation14). In mitotic cells, CGGBP1 localizes to the condensed chromatin during metaphase with predominant presence at the telomeric termini and centromeric regions of the chromosomes (Citation35). Further, it localizes to the spindle mid-zone during anaphase and eventually to midbodies during telophase (Citation35).
Transcription
The first series of functional studies on CGGBP1 was directed to find out its role in transcription regulation. These studies were devised to observe cis-regulatory effects on endogenous FMR1 promoter using CGGBP1 over-expression systems (Citation15). It has been shown that CGGBP1 binds to the CGG repeat in FMR1 5′UTR only when it is unmethylated, and represses FMR1 transcription (Citation15). Interestingly, an absence of CGGBP1 binding to this region is associated with CpG methylation and constitutive FMR1 gene silencing. This suggests that through binding to an unmethylated CGG repeat in FMR1 5′UTR, CGGBP1 prevents CpG methylation and shields the FMR1 gene from constitutive silencing (Citation6). Thus, CGGBP1 binding keeps FMR1 in a CpG methylation-free, transcriptionally repressed state and simultaneously shields it from of CpG methylation.
In the light of the findings that CGGBP1 represses RNA Pol II-mediated transcription by preventing constitutive gene silencing through CpG methylation-independent mechanisms, the role of histone modifications in transcription regulation by CGGBP1 becomes important. It is plausible that this might involve histone modifications at nucleosomal regions, as has been shown at CDKN1A and GAS1 promoters (Citation36), and a possible direct repressive effect at nucleosome-free sites. Such a CpG methylation-independent regulation of transcription is likely to be highly flexible and fits with the findings that CGGBP1-mediated gene repression is rapidly affected by external stimuli such as acute heat shock (Citation14).
We have reported that CGGBP1 is a heat shock-induced regulator of HSF1 expression. CGGBP1, along with its interacting partners NFIX and HMGN1, constitutes a complex that acts as a bidirectional regulator of HSF1 transcription such that the imperfect short CGG repeat in the HSF1 promoter is both required for driving basal levels of transcription as well as for repressing excessive levels of expression that are permitted only after heat shock induction (Citation14). The transcriptional regulation by CGGBP1 in response to heat shock is associated with enhanced nuclear presence, co-localization with DAPI-positive heterochromatin, and a change in solubility or antigenicity of CGGBP1 making it undetectable in the soluble fraction of cellular lysates (Citation14). This disappearance of CGGBP1 from the soluble fraction is associated with the disintegration of the Pol II-regulatory complex between NFIX, HMGN1, and CGGBP1 and renders transcription regulation by these factors ineffective at least at the HSF1 locus (Citation14).
In addition to heat shock-induced stress, external growth signals also modulate gene expression by CGGBP1. We have recently demonstrated that CGGBP1 regulates gene expression transcriptome-wide in response to serum (Citation37). Further, CGGBP1 participates in growth factor signal transduction downstream of EGF and PDGFB and undergoes tyrosine phosphorylation at Y20, which is required for its normal nuclear localization (Citation37). Strikingly, whereas CGGBP1-depletion in growth-stimulated cells leads to global changes in gene expression (with the change in expression not restricted to genes of specific functional categories or genes with common promoter sequence motifs), this effect is absent in quiescent cells. These findings, with support from additional experiments, have shown that CGGBP1 is also a trans-regulator of RNA Pol II-transcribed genes (Citation37).
The CGGBP1 gene itself seems to get turned on upon heat shock, leading to an acute induction of CGGBP1 transcript level (Citation14). Evidence for a role of CGGBP1 as a negative regulator of RNA Pol II also comes from direct experiments demonstrating transcriptional repression of CDKN1A and GAS1 genes and binding of CGGBP1 on their promoters (Citation36). Interestingly, the GAS1 gene is rich in interrupted short CGG repeats, whereas the CDKN1A promoter does seemingly not contain any CGG-rich region. The increase in expression of these genes is associated with a decrease in transcription-repressive histone modification H3K9-me3 in their promoter regions where CGGBP1 binds (Citation36). This further supports the view that transcriptional repression by CGGBP1 occurs through a histone modification mechanism, which is amenable to rapid changes, unlike gene silencing by CpG methylation.
While this direct evidence makes CGGBP1 a bona fide regulator of genes transcribed by RNA Pol II, there is evidence to suggest that genes transcribed by other RNA polymerases are also regulated by CGGBP1. For example, on metaphase-like chromosomal preparations, CGGBP1 exhibits a very strong binding to rRNA gene clusters in situ (Citation15). These rRNA gene clusters are located on small arms of acrocentric human chromosomes and are rich in CGG repeats (Citation15). In vitro binding assays also show that the CGG-rich genomic DNA from WT 28S RNA is shifted by incubation with CGGBP1 and super-shifted by an antibody against CGGBP1. A mutation that replaces CGG with AGG abrogates this binding (Citation15). In the absence of a functional study that measures the effect of CGGBP1 loss-of-function on rRNA levels, the evidence for CGGBP1-mediated regulation of RNA Pol I promoters remains strong but as yet incomplete.
Recently, strong evidence has emerged that RNA Pol III is regulated by CGGBP1. We analysed the global DNA-binding pattern of CGGBP1 in normal human fibroblasts and showed that RNA Pol III promoter sequences at Alu-SINEs and RNA Pol II promoter sequences at L1-LINE interspersed repeats are the primary binding sites for CGGBP1 (Citation37). The CGGBP1-bound Alus identified in that study were located hundreds of kilobases away from the nearest known genes. Moreover, CGGBP1 target sequences in this study were rich in repetitive DNA with the total repeat content more than 80% (Citation37). The most enriched binding sites included satellite DNA in addition to Alu-SINEs and L1-LINEs. It is possible that due to the nature of the techniques used in this study the simple tandem repeats were not detected because small sequence reads from ChIP sequencing experiments cannot be uniquely aligned to tandem repeats at any particular annotated genomic location. Identification of long stretches of tandem repeats such as peri-centromeric and telomeric DNA is also rendered difficult by the lack of unambiguous location-specified sequence data. While the net binding of CGGBP1 on L1-LINE elements increased slightly upon serum stimulation of quiescent cells, the increase in binding on Alu-SINEs was disproportionately high (Citation37), suggesting that under different circumstances of cellular growth stimulation CGGBP1 regulates RNA Pol III targets differently. Remarkably, despite strong sequence similarities between Pol III promoters at 7SL genes (the precursors of the Alu-SINEs) and Alu-SINEs, CGGBP1 exhibited binding discriminately to the latter. Binding of CGGBP1 was concentrated at the RNA Pol III promoter that exists downstream of the transcription start site. This region included the A-box and downstream sequence up to the B-box sequence and has been denoted the Alu enhancer element (ATE). Binding of CGGBP1 to ATE correlated inversely with the recruitment of RNA Pol III components to the promoters of Alu-SINEs both in vitro and in vivo. Thus, CGGBP1 acts as an inhibitor of transcription of Alu-SINEs by RNA Pol III (Citation37).
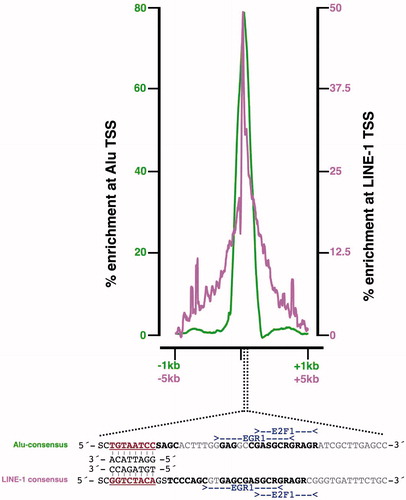
Curiously, the L1-LINE and Alu-SINE promoter elements are both devoid of CGG repeats or similar sequences to which CGGBP1 is shown and hence expected to bind. The peak of CGGBP1 binding in both cases is located 30–50 bases downstream of the transcription start site (TSS) at a region that bears striking functional similarity to Alu-SINEs and L1-LINEs () (Singh and Westermark, unpublished findings) (Citation37). A sequence alignment of these two regions with transcription factor (TF) sites mapped on them demonstrates the similarity of these regions and binding sites of TFs with which CGGBP1 can co-operate to regulate transcription (Singh and Westermark, unpublished findings). This study raises the possibility that CGGBP1 can potentially bind to sequences other than CGG repeats, although more experiments are needed to establish whether the binding between these DNA sequences and CGGBP1 is direct or indirect through interaction with other site-specific factors. Some of our preliminary results show that, in electrophoretic mobility shift assays, recombinant CGGBP1 can directly bind to these sequences.
Figure 3. Common sequence features in CGGBP1-binding sites in Alu-SINEs and L1-LINEs. Binding sites of CGGBP1 on Alu and L1 elements have sequence similarity. An alignment of Alu and L1 DNA sequences of regions at which CGGBP1-binding peaks shows degeneracy of sequence such that the transcription factor-binding sites for EGR1 and E2F1 (deduced using JASPAR and Transfac) are conserved (region in bold, marked with EGR1 and E2F1). An additional region of similarity (brown underlined bold) seems to be conserved and complementarily inverted between L1 and Alu elements. This region (5′-GGAYTACA-3′) is a part of the Alu transcription enhancer region and a major binding site for CGGBP1. (Citation37).
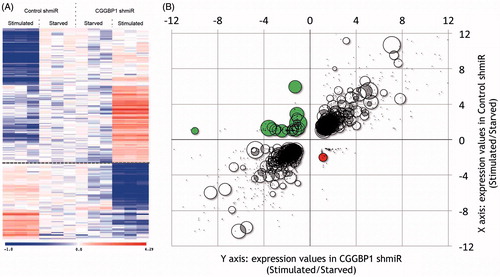
Cis-regulation of gene expression by CGGBP1 at promoters devoid of CGG repeats might occur through the presence of Alu or LINE elements. Indeed, a deeper analysis of recently published data proves this possibility. Genes that are down-regulated upon serum stimulation in the presence of CGGBP1 are rich in L1-LINEs in their 1kb proximal promoter, whereas promoters of genes that are up-regulated upon serum stimulation in the absence of CGGBP1 are poor in L1-LINE content () (Singh and Westermark, unpublished findings) (Citation37). This is striking because L1-LINEs are usually enriched in GC-poor regions and under-represented in GC-rich promoters (Citation38). A closer analysis reveals that these are truncated GC-rich fragments of L1 elements in the promoters of some genes that undergo expression changes differently in response to growth stimulation in a manner dependent on the levels of CGGBP1 (Singh and Westermark, unpublished findings). CGGBP1, in complex with NFIX, regulates HSF1 expression levels, thereby affecting the expression of genes induced by HSF1 (Citation14). CGGBP1 depletion alters the levels of HSF1 and secondarily affects the transcription of HSF1 target genes. Interestingly, such a cis-regulation of expression by CGGBP1–HSF1 axis could affect Alu transcription as well. This possibility is supported by the finding that, like CGGBP1, HSF1 also regulates Alu transcription (Citation39). HSF1 regulates Alu transcription in sense as well as antisense directions (Citation39), and it will be interesting to investigate if the Alu antisense (Citation39) and LINE-1 antisense (Citation40) transcription is also regulated by CGGBP1.
Figure 4. L1-LINEs function as CGGBP1-dependent cis-regulatory elements for growth-responsive genes. A: A subset of genes undergo expression changes upon growth stimulation (Stimulated) of quiescent cells (Starved) in a CGGBP1-dependent manner. The presence of CGGBP1 in normal levels, or its depletion, can dictate their induction of silencing upon growth stimulation (10% serum used in this case). B: Genes which are suppressed by serum stimulation in the presence of CGGBP1 are rich in L1 content in their 1kb proximal promoters unlike genes which are induced by serum stimulation in the presence of CGGBP1. The top left quadrant has 25% of all genes containing L1 elements recognized by Repeatmasker. The bottom right quadrant has only one gene containing L1. The areas of the circles represent the percentage of L1 content in the 1kb promoter region. The top right and bottom left quadrants with grey/black data points represent those genes which are unaffected by CGGBP1 levels and serum stimulation or starvation.
DNA methylation
CpG methylation is a major transcription-silencing mechanism (Citation4). The ability of CGGBP1 to bind only to unmethylated templates in vitro (Citation15) indicates that although CGGBP1 does not employ CpG methylation as a transcription-regulatory mechanism, it may itself play a role in maintaining or blocking CpG methylation at target sequences. We have measured global changes in CpG methylation by high throughput sequencing and observed that CGGBP1 deficiency leads to an increase in CpG methylation at already heavily methylated sequences (Citation43). More specifically, the increase in methylation was observed at repetitive sequences including at Alu and LINE1 repeats. Targeted evaluation of methylation at Alu and LINE1 elements genome-wide has shown that CpG methylation at these normally heavily methylated sequences is further augmented by CGGBP1 deficiency. Interestingly, while the change in LINE1 methylation is a unidirectional increase, on the Alu elements there is a bidirectional change with an increase at the majority of Alus but a decrease of methylation at a small subset of Alus (Citation43). It is an interesting coincidence that the Alu elements that are target sites for CGGBP1 binding are mainly of the young Alu family AluY, which is not constitutively inactivated by mutations and still retains the potential to be transcribed (Citation41). It is pertinent yet daunting to decipher what kinds of Alu elements are induced upon CGGBP1 deficiency and if that is accompanied by a loss of methylation. To interpret how CGGBP1 might regulate CpG methylation levels, several possibilities could be envisaged. A positive regulation on a minority of Alu elements could be caused by CGGBP1 binding and recruitment of heterochromatin-inducing factors such as SUV39H2 (a CGGBP1-interacting partner) (Citation42), which in turn can recruit DNA methyl transferases. Through mechanisms not yet clear, CGGBP1 discriminates between sequences at which CpG methylation has to be augmented and those at which CpG methylation has to be antagonized. Most likely, CGGBP1 binds to the latter sequences without recruiting positive regulators of CpG methylation such as SUV39H2. If CGGBP1 binding to these sequences creates a steric hindrance for the DNA methyl transferases, then the increase in CpG methylation brought about by CGGBP1 deficiency must involve de novo methylation, which can be caused by DNMT3B and DNMT3A, as well as by DNMT1 on heavily methylated templates (Citation43).
An alternative possibility is that CGGBP1 binding at heavily methylated repetitive elements potentiates the activity of factors that remove methylated cytosine residues by base oxidation followed by a base excision repair mechanism. A closer look at the gene expression changes brought about by CGGBP1 depletion lends support to the latter possibility (Citation37,Citation43). Analysis of the effect of CGGBP1 on expression change of genes known to participate in DNA methylation regulation shows that the genes that participate in base oxidation and removal of base excision repair are down-regulated whereas those acting to maintain methyltransferase activity are up-regulated (Citation37,Citation43). CGGBP1 thus seems to be required for expression of genes involved in DNA repair, and loss of CpG methylation is an associated consequence of this. Moreover, CpG methylation at retrotransposons silences them and reduces faulty recombinations, thereby minimizing their deleterious effects on the genome. Indeed, there is clear experimental evidence that CGGBP1 is a regulator of endogenous DNA damage (Citation44).
Genomic integrity
The human CGGBP1 has a C-terminal SQ motif that constitutes a strong phosphorylation site by PI3 kinase-like kinase family of enzymes that includes the DNA damage sensors ATM and ATR kinases (Citation45). An ATR substrate screen has also identified CGGBP1-S164 as a target (Citation46). Depletion of CGGBP1 leads to DNA damage identified as γH2AX-positive foci which is remarkably recapitulated by over-expression of a dominant negative S164A mutant form of CGGBP1 (Citation44). A large fraction but not all of the DNA damage foci induced by CGGBP1 dysfunction co-localizes with telomere-specific FISH probes (Citation44). S164 phosphorylation by ATR is required for proper binding of the telomere protector protein POT1 on telomeres. Lack of POT1 binding renders telomeres unstable and leads to their shortening (Citation44). The telomere fusions that take place in S164A over-expressing cells most likely occur through NHEJ repair of unprotected telomere ends. The endogenous DNA damage response that ensues activates ATM, ATR, and downstream checkpoint kinases and leads to premature cellular senescence (Citation44). CGGBP1, through maintaining telomeres in an ATR-induced phosphorylation-dependent manner, acts as a mediator of protective effects of ATR on telomeres (Citation44).
The pattern of telomeric damage caused by CGGBP1 dysfunction is reminiscent of what is observed upon functional deficiency of shelterin proteins, the guardians of telomeric integrity (Citation47). Telomeres are one of the largest fractions of simple tandem repeats that form hairpin and quadruplex structures. DNA damage on telomeres is easily visible and identifiable (Citation48). Other such sequences including the CGG repeats constitute smaller fractions of the genome and, unlike the telomeres, they are scattered. DNA damage at these shorter tandem repeats is difficult to detect. Proteins like WRN that are necessary for integrity of telomeres also bind to CGG repeats (Citation28). CGGBP1 also binds to telomeres in vivo (Citation44). These findings indicate some overlap in the mechanisms behind endogenous DNA damage at telomeres and at other tandem repeats like CGG repeats. In addition, dysfunction of proteins like WRN and CGGBP1 would also initiate repair at the stalled replication sites at CGG repeats, and perhaps at other simple interspersed repeats to which CGGBP1 binds. In addition to the centromeric and telomeric repeats which are maintained by special dedicated mechanisms, long simple tandem repeats that can compromise genomic stability, such as the CGG repeat fragile sites (Citation49), are relatively uncommon. Interspersed repeats are, however, widespread, and upon loss of CpG methylation they can undergo faulty recombinations resulting in chromosomal fusions and gross genomic instability (Citation50). Investigations into DNA damage elicited by CGGBP1 dysfunction have been limited to telomeric repeats. Studies of centromeric and interspersed repeats will shed more light on the various ways through which CGGBP1 regulates genomic stability.
An interesting aspect of telomeric damage caused by insufficient CGGBP1 S164 phosphorylation is that the ensuing telomere fusions disturb the faithful segregation of chromatin between dividing cells resulting in delayed cytokinetic abscission and lengthening of midbodies (Citation44). The reason behind the persistent presence of CGGBP1 on midbodies has remained elusive. An interesting explanation worth experimental evaluation is that cells might recruit CGGBP1 to midbodies to detect the presence of any unsegregated DNA in the cytokinetic bridge and to delay or abort mitosis as a response. A similar function of abscission checkpoint control has been attributed to AURKB (Citation51), an abscission checkpoint control protein with which CGGBP1 shows striking spatial and temporal co-localization (Citation35). AURKB serves to detect unsegregated chromatin in cytokinetic bridges and delays abscission to allow more time for resolution of the lagging chromatin, and, in the event of persisting chromatin-positive bridges, it leads to cleavage furrow regression and tetraploidization (Citation51). It is noteworthy that upon CGGBP1 depletion as well as lack of phosphorylation, normal human fibroblasts exhibit longer metaphase, pointing to a delayed metaphase-to-anaphase transition and delayed abscission as well as tetraploidization, indicating that the cell succumbs to the abscission checkpoint (Citation44).
Cell cycle
We have reported that CGGBP1 is required for cell cycle progression in normal as well as cancer cells (Citation36). Because DNA damage is the biggest elicitor of checkpoint response and cell cycle arrest (Citation52), it is pertinent to look at the cell cycle arrest caused by CGGBP1 loss-of-function as an effect of DNA damage. Nonetheless, cell cycle arrest caused by CGGBP1 dysfunction may also be initiated or at least potentiated by the unfolded protein response that ensues upon CGGBP1 depletion (Citation14). In totality, the changes in gene expression, CpG methylation, retrotransposon activation, telomere fusions, and UPR may all add up to signal a cell cycle arrest when CGGBP1 function is impaired. The dominance of either of these mechanisms may depend on the cell type as, unlike normal cells, most cancer cell types have inactivating mutations in or epigenetic silencing of checkpoint control genes (Citation53).
The cell cycle arrest caused by CGGBP1 deficiency in normal cells is an S-phase and G2/M phase arrest (Citation35), whereas in cancer cells it is a G1/G0 phase arrest (Citation36). The G1/G0 arrest in cancer cells is compatible with the expected cellular response to DNA damage during interphase. Even an early S-phase DNA damage response due to replisome stalling at long tandem repeats in the absence of CGGBP1 will lead to a sustained DNA damage-repair tug-of-war leading to an arrest. These early S-phase arrested cells will show up as a G1/G0 population in flow cytometry assays. Cancer cells have an unstable genome combined with loss of DNA damage-sensing genes such as p53 or Rb1. In combination with a continued stress of DNA replication, they may be more sensitive to CGGBP1 deficiency than normal cells and become arrested in G1. Normal cells, with all the repair mechanisms intact and smaller load of replication stress, would be able to repair the endogenous DNA damage elicited due to CGGBP1 deficiency and slowly progress through the S-phase to later stages of the cell cycle. According to this interpretation, as compared to cancer cells, normal cells are more likely to progress through the cell cycle in the absence of CGGBP1. Indeed, we have observed that under chronic depletion of CGGBP1, a small population of normal cells does recover and continue to divide, albeit slowly, with barely detectable amounts of CGGBP1 protein (Singh and Westermark, unpublished findings) (Citation37).
Some prominent stress and cell cycle-regulatory genes that are induced upon CGGBP1 depletion include DNA damage-induced gene DDIT3, ER stress regulator XBP1, heat shock chaperone HSP70 and 90, HSF1, NFIX, CDKN1A, GAS1, and Alu-SINEs (Citation14). This clearly indicates that although DNA damage response may initiate the checkpoint response in the absence of CGGBP1, protein unfolding and stress response also play an important role in the cell cycle arrest.
Recent findings reveal another mechanism through which CGGBP1 is indispensable for the cell cycle and justifies why cancer cells are likely to be more dependent on CGGBP1. CGGBP1 binds to Alu promoters and inhibits RNA Pol III recruitment on the A- and B-box sequences (Citation37). By inhibiting Alu RNA production, CGGBP1 ensures that RNA Pol II remains free from inhibition caused by Alu RNA. Indeed the net amount of mRNA yielded per unit amount of DNA is reduced upon CGGBP1 depletion. In parallel, cells manage to recruit RNA Pol III only to the growth-supporting 7SL and tRNA genes, which are required for cellular growth and cycling. This property of CGGBP1 discriminately to interfere with RNA Pol III recruitment only at Alu promoters but not at growth-supporting genes is dependent on Y20 phosphorylation and nuclear localization upon growth stimulation of cells. Given the large number of Alu elements in the human genome (>10% of the entire sequence), this mechanism suggests that CGGBP1 helps cells direct their resources for transcription of growth-supporting genes without performing wasteful indiscriminate transcription (Citation37). A schematic summary of the roles of CGGBP1 in signal transduction, subsequent nuclear localization, and participation in nuclear processes such as transcription, DNA damage, replication at repeats, and retrotransposon silencing is presented in .
Figure 5. A schematic view of the mechanisms through which CGGBP1 acts as a sensor of growth-induced stress and acts in the nuclei as a cytoprotector agent. The topmost box depicts the signalling pathways that extranuclear CGGBP1 is demonstrated or indicated to participate in. The CGGBP1 then localizes to the nuclei and executes a multitude of functions as depicted in the lower boxes to which the arrows lead. CGGBP1 participates in signal transduction and undergoes Y20 phosphorylation. The EGF and PDGF-induced phosphorylation of CGGBP1 has been proven in human cells, whereas other interactions with MAGI3, TYRO3, and MAP4K4 are deduced from protein–protein interaction studies in yeast. Anticlockwise from the top, CGGBP1 in nuclei binds to unmethylated DNA and promotes transcription of factors that promote cytosine demethylation. It also binds to transcription-regulatory regions of Alu-SINEs in growing cells thereby freeing RNA Pol III from unnecessary binding there. On the tRNA genes, CGGBP1 allows transcription under conditions of growth by not binding there. In quiescent cells, CGGBP1 does not bind to tRNA or Alu-SINEs. This aids in deployment of RNA Pol III at growth-promoting targets. Nuclear CGGBP1 binds to mitotic chromosomes, and its dysfunction causes telomeric damage resulting in chromosomal fusions and abscission failures. CGGBP1 presence at midbody seems to regulate abscission checkpoint and prevent tetraploidization. Whether CGGBP1 detects the presence of unsegregated DNA at mitotic bridges is an interesting matter of investigation. The nature of CGG repeats, CGGBP1-binding sites, suggests that CGGBP1 dysfunction might result in endogenous DNA damage by allowing formation of secondary and tertiary structures by CGG repeats, such as G4 quadruplexes, that can halt replication fork progression.
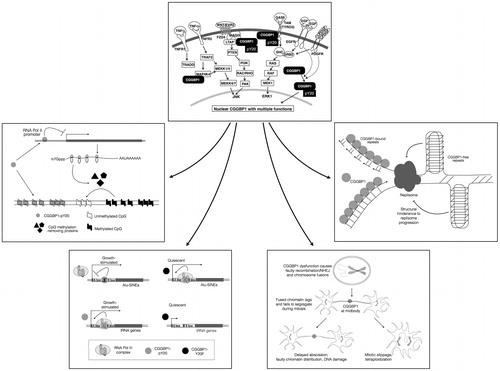
CGGBP1 in cancer
Given the precedents described above, it becomes important to ask how CGGBP1 functioning is associated with cancer. Different cancer genome-sequencing experiments have not identified CGGBP1 as a frequently mutated gene. Some studies suggest that expression levels of CGGBP1 might serve as a biomarker in some cancers (Citation54). Supporting this view, a survey of the NCBI Gene Expression Omnibus database shows that CGGBP1 is expressed in most cancer samples evaluated to date. These findings are supported by high levels of CGGBP1 expression detected by the Human Protein Atlas project (Citation55) and expression data available at the cBioportal/TCGA database (Citation56). A survey of the cancer genome atlas shows that CGGBP1 point mutations, deletions, and amplifications are all observed only with extremely low frequency in a variety of cancers.
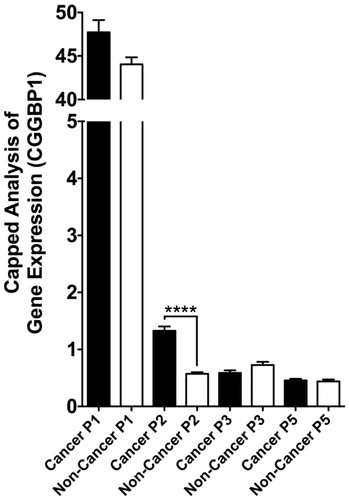
However, epigenetic mechanisms might operate to alter CGGBP1 levels in cancer. As mentioned above, the p2 promoter of CGGBP1 gains hyperactivity in cancer () (Citation22). Even if the net levels of CGGBP1 transcripts might remain unaltered, the protein levels might change and be under different sets of post-transcriptional gene-regulatory mechanisms. Additional mechanisms involving post-translational modifications of CGGBP1 such as tyrosine phosphorylation in response to growth signals, which make CGGBP1 more nuclear and growth compatible (Citation37), are likely to be hyperactive in cancer cells. Nonetheless, analysis of transcriptome-wide data from different cancer samples in the TCGA database provides significant insights.
Figure 6. Quantification of differential promoter usage at CGGBP1 locus in normal and cancer samples (cBioportal and TCGA databases). The quantitative CAGE data available for different transcript termini (5′ end) for each were assorted into ‘non-cancer’ and ‘cancer’ groups manually, and t test was performed to detect differential promoter usage in non-cancer versus cancer tissues/cells. While p1 is the most dominant promoter, p2 clearly has the most significant cancer-specific induction. The effects of longer 5′UTR associated with p2-specific transcription in regulation of CGGBP1 p2 transcript are currently unknown.
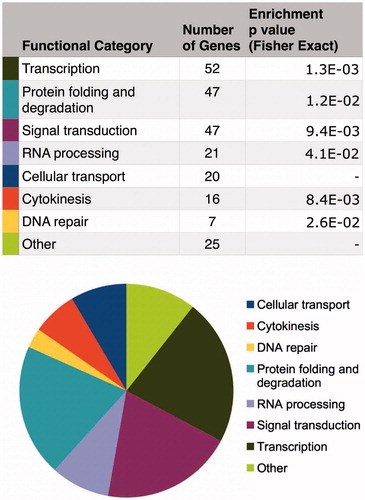
From such databases, identification of genes co-expressed with CGGBP1 (those genes the mRNA levels of which exhibit positive or negative correlation with that of CGGBP1) shows that indeed the genes involved in protein folding/degradation, DNA damage/repair, mRNA transport/splicing/stability, and cell division/cytokinesis are the prominent functional categories that co-vary with CGGBP1 (Singh and Westermark, unpublished findings). Interestingly, these are the same processes in which CGGBP1 or other CGG repeat-binding proteins have been shown to be involved (). This underscores the functional relevance of CGGBP1 in the regulation of cancer cells at all three levels: DNA, RNA, and protein, with consequences on the cell cycle. An analysis of the 1kb core promoter sequences of genes, whose expression levels either positively or negatively co-vary with that of CGGBP1, shows an unexpected presence of Alu. The promoters of positively co-varying genes (genes whose expression is higher when CGGBP1 levels are higher) contain less than expected Alu content (7%; expected approximately 10%), whereas the promoters of negatively co-varying genes (genes whose expression is lower when CGGBP1 levels are higher and vice versa) contain higher than expected Alu sequences(17%; expected approximately 10%). The Alu subsequence GGATTACA, which is located at the epicentre of the CGGBP1-binding region on Alus (Citation37), was identified as a common motif in promoters and present in nearly 50% of all the negatively co-varying genes. In contrast, the positively co-varying gene promoters did not have any common motif. These observations support the view that full-length or truncated Alu-SINEs that contain the ATE sequence (Citation37) act as negative CGGBP1-dependent cis-regulators of transcription. Thus it seems that transcription activation by CGGBP1 occurs through two different mechanisms: a trans-acting mechanism that involves CGGBP1-binding and repression of Alu elements located in gene-poor regions (Citation37), and a cis-acting mechanism that involves CGGBP1-binding to cis-regulatory Alu elements in promoters of target genes.
Figure 7. Direct and indirect gene expression regulation by CGGBP1 is directed at specific functional categories that justify known functions of CGGBP1 so far. The TCGA and cBIOPORTAL databases were mined to fish out genes that exhibit significant positive or inverse correlation with CGGBP1 expression in various cancers. These genes, defined as CGGBP1-co-varying genes, belong to specific functional categories that overlap with the functions where CGGBP1 has been shown to participate or has been implicated based on preliminary findings. The co-variance of these genes with CGGBP1 thus indicates that CGGBP1 acts as a common (direct or indirect) underlying regulator of their expression in cancer.
Functions of CGGBP1: indications from CGGBP1-interacting proteins
Knowledge about protein-interacting partners is extremely valuable in deciphering functions and regulation of any biological entity. The existing information about CGGBP1 clearly indicates that there is a lot about CGGBP1 that we do not know. Potentially, future work on CGGBP1 will not only increase our knowledge about the protein itself, but also reveal hitherto unknown cellular and molecular mechanisms. In order to begin to understand the diversity of CGGBP1-regulated processes, an important step is to unravel the spectrum of interacting proteins. Guided by studies from other groups and from our own unpublished work, we have identified a number of interacting partners of CGGBP1 () that can lead the way and help us formulate hypotheses about the diverse biological functions of CGGBP1.
Table I. A list of proteins that interact with CGGBP1. The unpublished findings (Singh and Westermark) are based on a yeast two-hybrid screen using full-length human CGGBP1 as bait against a human brain cDNA library.
Future directions
Future work on CGGBP1 can take multiple directions. Some questions are very obvious. For example, how does CGGBP1 regulate such a variety of functions? Is this a simple coincidental multiple usage of one factor in seemingly independent pathways, ranging from signal transduction to DNA damage/repair, transcription, CpG methylation regulation, protein integrity, and cytokinesis, or does CGGBP1 orchestrate these diverse functions? If the latter is the case, then how can a recently evolved protein, conserved strongly only amongst mammals (only poorly conserved with avians; NCBI Homologene), have found its way into otherwise well conserved cellular processes? Does CGGBP1 deficiency impair the cellular response to heat shock stress? How does CGGBP1 possibly act in a self-regulatory loop of transcriptional regulation? Does CGGBP1 indeed bind to peri-centromeric heterochromatin and regulate its integrity? Is centromeric CGGBP1 important for kinetochore-spindle attachment and chromosomal migration in anaphase? What directs CGGBP1 off-loading from chromatin onto spindle fibres and midbodies? What is the structure of CGGBP1, and how does it complex with DNA? The DNA-binding domain of CGGBP1 seems to have structural similarities with RNA-binding domains also. If so, does CGGBP1 bind to RNA? Does CGGBP1 modulate cancer incidence or progression?
Knowledge about alteration of levels and post-translational modifications of CGGBP1 in cancer will give valuable insights into how CGGBP1 regulates cellular transformation, cancer cell survival and ability of cancer cells to thrive under stress. With such a widespread functional footprint, studies on CGGBP1 will lead us to an improved holistic understanding of cellular health and disease that transcends different sub-disciplines of cell biology.
Funding
Our own work cited in the text was supported by grants from the Swedish Cancer Society and the Swedish Research Council. Since 28 May 2015, the contributions of Umashankar Singh to this review have been supported by the Indian Institute of Technology, Gandhinagar.
Declaration of interest
The authors report no conflicts of interest.
References
- Catasti P, Chen X, Mariappan SV, Bradbury EM, Gupta G. DNA repeats in the human genome. Genetica. 1999;106:15–36.
- Politz JC, Scalzo D, Groudine M. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu Rev Cell Dev Biol. 2013;29:241–70.
- Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. 2009;124:327–38.
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–6.
- Richards RI, Sutherland GR. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992;70:709–12.
- Deissler H, Behn-Krappa A, Doerfler W. Purification of nuclear proteins from human HeLa cells that bind specifically to the unstable tandem repeat (CGG)n in the human FMR1 gene. J Biol Chem. 1996;271:4327–34.
- Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci USA. 1994;91:4950–4.
- Ashley CT Jr, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–28.
- Zhu QS, Heisterkamp N, Groffen J. Unique organization of the human BCR gene promoter. Nucleic Acids Res. 1990;18:7119–25.
- Richards RI, Holman K, Yu S, Sutherland GR. Fragile X syndrome unstable element, p(CCG)n, and other simple tandem repeat sequences are binding sites for specific nuclear proteins. Hum Mol Genet. 1993;2:1429–35.
- Deissler H, Wilm M, Genç B, Schmitz B, Ternes T, Naumann F, et al. Rapid protein sequencing by tandem mass spectrometry and cDNA cloning of p20-CGGBP. A novel protein that binds to the unstable triplet repeat 5'-d(CGG)n-3' in the human FMR1 gene. J Biol Chem. 1997;272:16761–8.
- Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–8.
- Gaff C, du Sart D, Kalitsis P, Iannello R, Nagy A, Choo KH. A novel nuclear protein binds centromeric alpha satellite DNA. Hum Mol Genet. 1994;3:711–16.
- Singh U, Bongcam-Rudloff E, Westermark B. A DNA sequence directed mutual transcription regulation of HSF1 and NFIX involves novel heat sensitive protein interactions. PLoS One. 2009;4:e5050.
- Müller-Hartmann H, Deissler H, Naumann F, Schmitz B, Schröer J, Doerfler W. The human 20-kDa 5'-(CGG)(n)-3'-binding protein is targeted to the nucleus and affects the activity of the FMR1 promoter. J Biol Chem. 2000;275:6447–52.
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38.
- Hayward A, Ghazal A, Andersson G, Andersson L, Jern P. ZBED evolution: repeated utilization of DNA transposons as regulators of diverse host functions. PLoS One. 2013;8:e59940.
- Fantom Database. Available at: http://www.ensembl.org/Homo_sapiens/Gene/Summary?db = core;g = ENSG00000163320;r =3:88051944-88149885. Date accessed 11 June 2015.
- Naumann F, Remus R, Schmitz B, Doerfler W. Gene structure and expression of the 5'-(CGG)(n)-3'-binding protein (CGGBP1). Genomics. 2004;83:106–18.
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61.
- Fantom Database. Available at: http://fantom.gsc.riken.jp/5/sstar/EntrezGene:8545. Date accessed 11-June-2015.
- FANTOM Consortium, the RIKEN PMI, and CLST (DGT); Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, Haberle V, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70.
- NCBI HomoloGene Database. Available at: http://www.ncbi.nlm.nih.gov/homologene/?term = cggbp1. Date accessed 11-June-2015.
- Bastian F, Parmentier G, Roux J, et al. Bgee: integrating and comparing heterogeneous transcriptome data among species. In: Bairoch A, Cohen-Boulakia S, Froidevaux C, editors. Data Integration in the Life Sciences. Vol. 5109. Berlin/Heidelberg: Springer; 2008. p. 124–31.
- Faisal A, Peltonen J, Georgii E, Rung J, Kaski S. Toward computational cumulative biology by combining models of biological datasets. PLoS One. 2014;9:e113053.
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE; Mouse Genome Database Group. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–36.
- Ficker M, Powles N, Warr N, Pirvola U, Maconochie M. Analysis of genes from inner ear developmental-stage cDNA subtraction reveals molecular regionalization of the otic capsule. Dev Biol. 2004;268:7–23.
- Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–802.
- Uliel L, Weisman-Shomer P, Oren-Jazan H, Newcomb T, Loeb LA, Fry M. Human Ku antigen tightly binds and stabilizes a tetrahelical form of the fragile X syndrome d(CGG)n expanded sequence. J Biol Chem. 2000;275:33134–41.
- Lau PP, Zhu HJ, Nakamuta M, Chan L. Cloning of an Apobec-1-binding protein that also interacts with apolipoprotein B mRNA and evidence for its involvement in RNA editing. J Biol Chem. 1997;272:1452–55.
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77.
- Xu R-M, Jokhan L, Cheng X, Mayeda A, Krainer AR. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–70.
- Gulyĭ PV, Orlov SV, Dizhe EB, Kuteĭkin-Tepliakov KB, Ignatovich IA, Zhuk SV, et al. [The role of ZF5 and CGGBP-20 transcription factors in expression regulation of human FMR1 gene responsible for X-fragile syndrome]. Tsitologiia. 2009;51:1005–12.
- Iber H. Sequence specific binding of cytosolic proteins to a 12 nucleotide sequence in the 5' untranslated region of FMR1 mRNA. Biochim Biophys Acta. 1996;1309:167–73.
- Singh U, Westermark B. CGGBP1 is a nuclear and midbody protein regulating abscission. Exp Cell Res. 2011;317:143–50.
- Singh U, Roswall P, Uhrbom L, Westermark B. CGGBP1 regulates cell cycle in cancer cells. BMC Mol Biol. 2011;12:28.
- Agarwal P, Enroth S, Teichmann M, Wiklund HJ, Smit A, Westermark B, et al. Growth signals employ CGGBP1 to suppress transcription of Alu-SINEs. Cell Cycle. 2014 Nov 21. [Epub ahead of print].
- Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400.
- Pandey R, Mandal AK, Jha V, Mukerji M. Heat shock factor binding in Alu repeats expands its involvement in stress through an antisense mechanism. Genome Biol. 2011;12:R117.
- Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–85.
- Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988;85:4775–8.
- Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200.
- Agarwal P, Collier P, Fritz MH, Benes V, Wiklund HJ, Westermark B, et al. CGGBP1 mitigates cytosine methylation at repetitive DNA sequences. BMC Genomics. 2015;16:390.
- Singh U, Maturi V, Jones RE, Paulsson Y, Baird DM, Westermark B. CGGBP1 phosphorylation constitutes a telomere-protection signal. Cell Cycle. 2014;13:96–105.
- Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays. 2005;27:397–407.
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6.
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–7.
- Juranek SA, Paeschke K. Cell cycle regulation of G-quadruplex DNA structures at telomeres. Curr Pharm Des. 2012;18:1867–72.
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92.
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40.
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–84.
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–35.
- Fearon ER. Human cancer syndromes: clues to the origin and nature of cancer. Science. 1997;278:1043–50.
- Cody NAL, Shen Z, Ripeau J-S, Provencher DM, Mes-Masson A-M, Chevrette M, et al. Characterization of the 3p12.3-pcen region associated with tumor suppression in a novel ovarian cancer cell line model genetically modified by chromosome 3 fragment transfer. Mol Carcinog. 2009;48:1077–92.
- The Human Protein Atlas database. Available at: http://www.proteinatlas.org/search/cggbp1. Date accessed 11-June-2015.
- The cBioPortal for Cancer Genomics. Available at: http://www.cbioportal.org/. Date accessed 11-June-2015.
