Abstract
Patients with chronic kidney disease have an increased risk of cardiovascular disease and mortality. Since DNA methylation is an important mechanism modulating the gene expression associated with aging, inflammation, and atherosclerosis, the objective of this study was to determine the possible effect of the uremic milieu on global DNA methylation and DNA methyltransferase (DNMT) expression in uremic status by comparing chronic hemodialysis (HD) patients with the normal population. Twenty normal subjects and twenty chronic dialysis patients with similar ages, sex, and body mass indexes (BMIs) were included. We evaluated the clinical characteristics; the levels of homocysteine, total indoxyl sulfate (IS), and total p-cresol sulfate (PCS); and the DNMT messenger RNA expression and global DNA methylation in the peripheral blood leukocytes. The chronic HD patients had significantly higher blood urea nitrogen (BUN), creatinine (Cr), uric acid, Ca, P, intact parathyroid harmone (iPTH), cholesterol, high-sensitivity C-reactive protein (hs-CRP), total indoxyl sulphate (IS) and p-cresol sulphate (PCS), and homocysteine than the normal subjects. The expression of DNMT 1 and 3a did not differ significantly between these two study groups. The chronic HD patients had significantly decreased DNMT 3b expression in the leukocytes. There were no significant correlations between the global DNA methylation and the levels of IS, PCS, and homocysteine. We concluded that chronic HD patients may have lower DNMT 3b expression than normal subjects. However, the status of global DNA methylation may not change significantly in uremic patients when compared with the normal population.
INTRODUCTION
The prevalence of chronic kidney disease (CKD) has increased during the last decade.Citation1 Uremic patients suffer from increased oxidative stress, chronic inflammation, and malnutrition.Citation2 Clinically, CKD patients have a higher risk of cardiovascular disease (CVD) and mortality than the normal population.Citation3
The environment could change the phenotype via modifications of the epigenome.Citation4,5 Due to the fact that environmental factors are critical for the normal functioning of the genome, the associations between the nonphysiological uremic environment and the epigenotype should be important in patients with CKD. However, the question of whether the metabolic alterations and coexisting inflammation in CKD could change the phenotype of CKD patients via epigenetic changes remains to be answered. DNA methylation is an important mechanism modulating the gene expression associated with aging, inflammation, and atherosclerosis.Citation6 In addition, DNA methyltransferases (DNMTs) are the key enzymes for the regulation of DNA methylation.Citation7 Accumulated evidences reveal that DNA methylation regulated by DNMTs is associated with disease development and progression such as malignancies and autoimmune disease.Citation8,9 However, the impact of the uremia milieu on DNMT expression is currently unknown.
Hyperhomocysteinemia is commonly seen in uremic patients and can be associated with global DNA hypomethylation.Citation10–12 Previous studies have shown that epigenetic alterations were associated with inflammation and cardiovascular disease in patients with CKD.Citation13,14 However, the contents of uremic toxins are more complex than hyperhomocysteinemia. Clinical data about the difference in DNA methylation status between normal and uremic subjects have been limited. Protein-bound uremic toxins such as indoxyl sulfate (IS) and p-cresol sulfate (PCS) are difficult to remove by dialysis. The putative association between the protein-bound uremic toxins other than homocysteine and aberrant DNA methylation is still unclear.
The objective of this study was to ascertain the possible effect of the uremic milieu on global DNA methylation and DMNT expression in uremic status by comparing chronic hemodialysis (HD) patients with the normal population.
METHODS
Study Subjects
We conducted a case–control study that included 20 chronic HD patients. The average duration on dialysis was 6.4 ± 3.9 months. For comparison, an age-, gender-, and body mass index (BMI)-matched control group of 20 normal renal function patients was found in the same community. Patients with diabetes mellitus, hypertension, malignancy, chronic hepatitis, autoimmune disease, and recent acute infection were excluded. Each chronic HD patient received standard three times per week HD. We analyzed the demographic data and measured the blood urea nitrogen (BUN), creatinine (Cr), calcium (Ca), phosphate (P), intact parathyroid hormone (iPTH), total cholesterol, hemoglobin (Hb), high-sensitivity C-reactive protein (hs-CRP), uric acid, and albumin. The serum samples were collected after overnight fasting. For the HD patients, the blood samples were collected before dialysis. The concentration of homocysteine was measured by the ELISA method (IBL International GmbH, Hamburg, Germany) according to the instruction manual. This study design was approved by the Institutional Review Board of Chang Gung Memorial Hospital: 97-2159A3.
Indoxyl Sulfate and p-Cresol Sulfate Measurements
The method for the IS and PCS measurement was described in our previous report.Citation15 Briefly, serum samples were deproteinized by the addition of three parts methanol to one part serum for determination of the total IS and total PCS. All analyses were performed on a Waters Acquity ultra performance liquid chromatography (UPLC) system (Milford, MA, USA). Standard IS was pursued (Sigma, St. Louis, MO, USA), and standard PCS was provided by the Kureha Corporation (Tokyo, Japan). The IS and PCS were detected at 280 and 260 nm. The buffer flow was 0.4 mL/min using 10 mM NH4H2PO4 (pH = 4.0) (A) and 100% acetonitrile (B) with a gradient from 82.5% A/17.5% B to 55% A/45% B over 9 min. The limits of detection of this assay were 0.225 mg/L for the IS and 1 mg/L for the PCS. Calibration curves were constructed by plotting the peak areas versus the concentrations of each analysate. Quantitative results were obtained and calculated as concentrations (mg/L).
DNA and RNA Extraction from Leukocytes
Whole blood was collected in disodium EDTA containers and stored at −20°C. After thawing, the leukocytes were isolated with erythrocyte lysing buffer (0.155 M, NH4Cl; 10 mM, KHCO3; 0.1 mM, Na2 EDTA; pH, 7.4) and SE buffer (75 mM, NaCl; 25 mM, Na2 EDTA; pH, 8.0) containing 100 mg/mL of Proteinase K and 1% of sodium dodecyl sulfate. The chromosome DNA and total RNA were extracted with commercial kits (QIAamp DNA Blood Mini Kit and RNeasy Kit, Qiagen) according to the manufacturer’s instructions.
Quantitative Real-Time PCR
Five micrograms of total RNA were then reverse-transcribed by reverse transcriptase (Bio-Rad) with random primers. Real-time PCR was performed in 25 μL SYBR Green PCR Master Mix (Applied Biosystems) containing 0.6 mol/L primers and 1 μg cDNA using an iQ5 real-time PCR detection system (Bio-Rad). Forward and reverse primer sequences were as follows: β-actin, 5′-TCACCCACACTGTGC CCATCTACGA-3′ and 5′-CAGCGGAACCGCTCATTGCCAATGG-3′; DNMT1, 5′-CGGTTCTTCCTCCTGGAGAATGTCA-3′ and 5′-CACTGATAGCCCATGCG GACCA-3′; DNMT3A, 5′-CAATGACCTCTCCATCGTCAAC-3′ and 5′-CATGCA GGAGGCGGTAGAA-3′, and DNMT3B: 5′-CCATGAAGGTTGGCGACAA-3′ and 5′-TGGCATCAATCATCACTGGATT-3′.Citation16 The thermocycling parameters were: 3 min at 94°C for denaturation and 40 cycles consisting of 45 s at 94°C, 30 s at 55°C, and 45 s at 72°C. Each PCR was performed in triplicate, and the mean CT value was used for statistical analysis. Messenger RNA (mRNA) expression was standardized to the β-actin expression levels, followed by normalization to the control group.
Global Methylation Assay
Purified genomic DNA of leukocytes (100 ng) was used for the measurement of global DNA methylation status by MethylampTM Global DNA Methylation Quantification Ultra Kit (Epigentek Group, Inc., Farmingdale, NY, USA).Citation17 The assay was performed according to the product instructions. The absorbance of each reaction at 450 nm was read on a microplate reader. Each sample was measured in triplicate. The standard curve was constructed according to the manufacturer’s instructions and the slope of the standard curve was calculated using linear regression analysis. The percentage of methylation was calculated by the following formula: methylation (%) = optical density (sample − blank) × slope/100.
Statistical Analysis
The data were expressed as means ± SD. Differences in parameters between groups were compared using two-tailed unpaired Student’s t-tests or χ2 tests with the Fisher’s exact test. The correlation between two variables was analyzed using a two-tailed Pearson correlation. A value of p < 0.05 was considered statistically significant.
RESULTS
Patient Characteristics and Laboratory Results
Twenty normal controls and twenty chronic HD patients were included in the study. The general and clinical characteristics of the study subjects are listed in . The average ages of the normal subjects and chronic HD patients were not significantly different (61.4 ± 2.8 vs. 62.2 ± 2.2 years). The gender distribution and BMI did not differ significantly between these two study groups. The serum BUN and Cr levels of the chronic HD patients were significantly higher than the normal subjects (BUN: 96.6 ± 20.2 vs. 10.2 ± 1.8 mg/dL, p < 0.05; Cr: 9.6 ± 3.8 vs. 0.76 ± 0.13 mg/dL, p < 0.05). The chronic HD patients had significantly higher uric acid, Ca, P, iPTH, cholesterol, and hs-CRP levels when compared with the normal subjects. The serum albumin and Hb levels of the chronic HD patients were lower than those of the normal subjects.
Table 1. General and clinical characteristics of the study subjects.
IS, PCS, and Homocysteine Levels
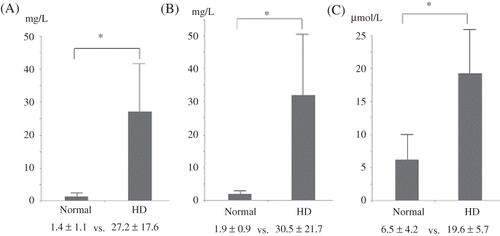
The serum levels of the uremic toxins are plotted in . Protein-bound uremic toxins, IS and PCS, were measured. The total IS and PCS concentrations of the chronic HD patients were significantly higher than those of the normal subjects (IS: 27.2 ± 17.6 vs. 1.4 ± 1.1 mg/L, p < 0.05; PCS: 30.5 ± 21.7 vs. 1.9 ± 0.9 mg/L, p < 0.05). The chronic HD patients also had significantly higher homocysteine levels than the normal subjects (19.6 ± 5.7 vs. 6.5 ± 4.2 μmol/L, p < 0.05).
Expressions of DNMTs
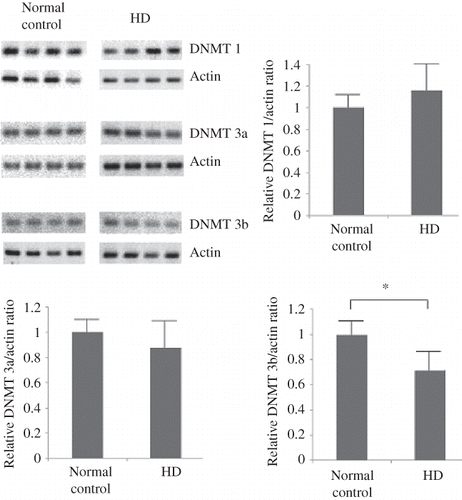
The results of the quantitative PCR analysis for DNMT 1, 3a, and 3b mRNA expression in the leukocytes are shown in . The mRNA expression of DNMT 1 and 3a was not significantly different between the normal subjects and chronic HD patients. The DNMT 3b mRNA expression significantly decreased in the chronic HD patients when compared with the normal subjects.
Figure 2. Quantitative PCR analysis for DNMT 1, 3a, and 3b mRNA expression in the leukocytes of the study subjects. The mRNA expression of DNMT 1 and 3a was not significantly different between the normal subjects (n = 20) and the chronic HD patients (n = 20). The DMNT 3b mRNA expression decreased significantly in the chronic HD patients. *p < 0.05.
Global DNA Methylation
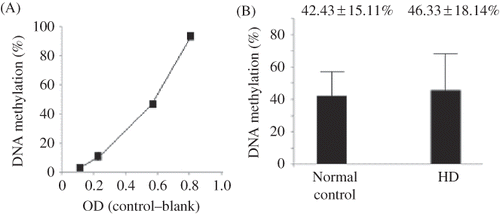
The percentages of global DNA methylation are plotted in . The slope of the standard curve was 144.10 ± 23.17 (standardized coefficients = 0.975, p < 0.05). The percentages of global DNA methylation of the normal subjects and HD patients were 42.43 ± 15.11% and 46.33 ± 18.14%, respectively. The global DNA methylation amounts were not significantly different between the normal subjects and the HD patients (p > 0.05).
Figure 3. Global DNA methylation analysis of the study subjects. (A) Standard curve for DNA methylation analysis, and (B) the percentage of global DNA methylation for the normal subjects (n = 20) were 42.43 ± 15.11% and 46.33 ± 18.14%, respectively. The global DNA methylation of the normal subjects and HD patients was not significantly different. p > 0.05.
Correlation between the Global DNA Methylation and the Uremic Toxins
We also analyzed the correlation between the global DNA methylation amount and the levels of BUN, total IS, PCS, and homocysteine, respectively, in the study subjects (n = 40). The results are listed in . No significant correlation between the global DNA methylation and the BUN was found. The correlation between the global DNA methylation and the levels of total IS, PCS, and homocysteine was also not significant.
Table 2. Results of correlation analysis between the global DNA methylation and the levels of BUN, total IS, PCS, and homocysteine (n = 40).
DISCUSSION
Hyperhomocysteinemia, uremic toxins accumulation, and chronic inflammation have been reported to lead to aberrant DNA methylation globally and in specific genes.Citation10,11,14,18 Phosphatemia, common in CKD patients, has also been reported to be related to SM22α promoter methylation in vitro.Citation19 However, clinical data about the impact of uremic milieus on global DNA methylation have been rare. A previous study of stage 2–4 CKD patients revealed that global DNA methylation was not associated with renal function.Citation14 Another clinical study with advanced stage CKD patients and normal subjects also showed that no significant correlations were observed between global DNA methylation and estimated GFR.Citation14 Our study showed a similar result in that the global DNA methylation did not differ significantly between the normal and chronic HD subjects. Aging of cells affects DNA methylation.Citation6 Uremic patients have shorter blood cell survivalCitation20; therefore, more younger cells are in the blood samples. The differences in the age of blood cells between HD and control groups may counterbalance other effects on the DNA methylation. The contents of the uremic milieu are complex.Citation2 The effects of the uremic milieu on aberrant global DNA methylation may be complex and context-sensitive. The impact of the uremic milieu on the DNA methylation should be the sum of the effects of uremic toxins. As demonstrated in this study, chronic dialysis patients had increased protein-bound uremic toxins such as IS and PCS. However, current studies about the putative effects of these uremic toxins on the DNA methylation regulation are limited.
DNMT 1 is the most abundant DNMT, and it is considered to be the key maintenance methyltransferase in mammals.Citation7,21 The results of this study showed that the DNMT 1 expression in the leukocytes was not different for the normal subjects and chronic HD patients. DNMT 3a and DNMT 3b were cooperative factors with DNMT 1 during DNA methylation.Citation7 Our data revealed that the expression DNMT 3b was decreased in chronic dialysis patients. However, the decreased DNMT 3b expression in chronic dialysis patients did not change the DNA methylation status significantly when compared with the normal subjects in this study. The related changes of DNMT expression and activity in the uremia state require further studies for clarification.
Our study was limited by its small population, which precluded a more reliable statistical analysis of the data. Considering the negative results of the association between the uremic state and global DNA methylation in this and previous studies, global DNA methylation might not be a suitable epigenetic biomarker to define the impact of uremia on epigenetic regulation. Studies with site-specific DNA methylation analysis are warranted.
In conclusion, chronic HD patients may have lower DNMT 3b expression than normal subjects. The status of global DNA methylation may not change significantly in uremic patients when compared with the normal population.
ACKNOWLEDGMENTS
We thank the staff of Keelung Chang Gung Memorial Hospital Research Center for their assistance with this investigation. We also thank the Kureha Corporation (Tokyo, Japan) for providing the PCS for our UPLC analysis.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
This work was funded by a grant from the CMRPG280091.
REFERENCES
- Hwang SJ, Tsai JC, Chen HC. Epidemiology, impact and preventive care of chronic kidney disease in Taiwan. Nephrology. 2010;15(Suppl 2):3–9.
- Yavuz A, Tetta C, Ersoy FF, . Uremic toxins: A new focus on an old subject. Semin Dial. 2005;18:203–211.
- Ayodele OE, Alebiosu CO. Burden of chronic kidney disease: An international perspective. Adv Chronic Kidney Dis. 2010; 17:215–224.
- Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141.
- van Vliet J, Oates NA, Whitelaw E. Epigenetic mechanisms in the context of complex diseases. Cell Mol Life Sci. 2007; 64:1531–1538.
- Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Aging Res Rev. 2009;8:268–276.
- Turek-Plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647.
- Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–2085.
- Pan Y, Sawalha AH. Epigenetic regulation and the pathogenesis of systemic lupus erythematosus. Transl Res. 2009;153:4–10.
- Ingrosso D, Cimmino A, Perna AF, . Folate treatment and unbalanced methylation and changes of allelic expression induced by hyperhomocysteinemia in patients with uremia. Lancet. 2003;361:1693–1699.
- Ingrosso D, Perna AF. Epigenetics in hyperhomocysteinemic states. A special focus on uremia. Biochim Biophys Acta. 2009;1790:892–899.
- Castro R, Rivera I, Struys EA, . Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296.
- Nanayakkara PW, Kiefte-de Jong JC, Stehouwer CD, . Association between global leukocyte DNA methylation, renal function, carotid intima-media thickness and plasma homocysteine in patients with stage 2–4 chronic kidney disease. Nephrol Dial Transplant. 2008;23:2586–2592.
- Stenvinkel P, Karimi M, Johansson S, . Impact of inflammation on epigenetic DNA methylation—a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499.
- Wu IW, Hsu KH, Lee CC, p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant. 2011;26:938–947.
- Balada E, Ordi-Ros J, Serrano-Acedo S, Martinez-Lostao L, Rosa-Leyva M, Vilardell-Tarrés M. Transcript levels of DNA methyltransferases DNMT1, DNMT3A and DNMT3B in CD4+ T cells from patients with systemic lupus erythematosus. Immunology. 2008;124:339–347.
- Zhao M, Gao F, Wu X, Tang J, Lu Q. Abnormal DNA methylation in peripheral blood mononuclear cells from patients with vitiligo. Br J Dermatol. 2010;163:736–742.
- Hodge DR, Xiao W, Clausen PA, . Interleukin-6 regulation of the human DNA metyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001;276: 39508–39511.
- Montes de Oca A, Madueño JA, Martinez-Moreno JM, . High-phosphate-induced calcification is related to SM22α promoter methylation in vascular smooth muscle cells. J Bone Miner Res. 2010;25:1996–2005.
- Kaye M. The anemia associated with renal disease. J Lab Clin Med. 1958;52:83–100.
- Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66:729–735.