Abstract
Aim: The relationship between the methylenetetrahydrofolate reductase (MTHFR) A1298C polymorphism and the susceptibility of diabetes remains inclusive or controversial. For better understanding of the influence of MTHFR A1298C polymorphism on diabetes risk, we performed this meta-analysis. Methods: All related articles were identified through a search of PubMed, Embase, Chinese Biomedical Literature Database (CBM, Chinese), China National Knowledge Infrastructure (CNKI), and Wangfang Database (Chinese). The relationship between the MTHFR A1298C polymorphism and diabetes susceptibility was conducted by odds ratios (ORs) and 95% confidence intervals. Results: Total of six studies with 897 cases and 852 controls were included in our meta-analysis. Overall, the significance associated was found between MTHFR A1298C polymorphism and the susceptibility of diabetes under recessive model (CC vs. AC/AA: OR = 1.70, 95% CI = 1.18–2.45, p = 0.004). On the subgroup analysis according to ethnicity, the results indicated that MTHFR A1298C polymorphism has a significant association with diabetes in Asian population under dominant model (CC/AC vs. AA: OR = 1.31, 95% CI = 1.003–1.72, p = 0.047). However, there was no association found between MTHFR A1298C polymorphism and diabetes susceptibility in Caucasians. Conclusions: The results indicated that the MTHFR A1298C polymorphism is a dangerous factor for diabetes, especially for Asians.
Introduction
The prevalence of diabetes increased rapidly all over the world in recent years. It was estimated by the world health organization that the global number of diabetes mellitus was 173 million in 2002 and this figure is likely to be double by 2030.Citation1,Citation2 Diabetes is a group of metabolic disease, which is characterized as high blood sugar; it is caused by the defects in insulin secretion and (or) insulin disorder. Two factors can usually caused diabetes, one is a lack of insulin that lead to type 1 diabetes (T1D), another one is blockage in the insulin signaling pathway that causes type 2 diabetes (T2D).Citation3 T1D and T2D are two major types of diabetes. Microbial exposures and sex hormones together with lifestyle factors have been shown to play an important role in the development of this complex disease.Citation4,Citation5 Besides environmental factors, many genetic factors also contribute to its development and progression, such as PPAR,Citation6 IL-2,Citation7 IL-10,Citation8 AdiponectinCitation9, and so on.
Methylenetetrahydrofolate reductase (MTHFR) is a key enzyme for intracellular folate homeostasis and metabolism. It catalyses the irreversible transformation of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is the primary circulating form of folate and provides methyl groups for the methylation of homocysteine to methionine.Citation10 A1298C (rs1801131) is one of the most popular research polymorphism of MTHFR, a replace of A to C changing the encoded amino acid from Glu to Ala, this polymorphism has been considered to affect the enzyme activity of MTHFR.Citation11,Citation12 Several original studies have reported the role of MTHFR A1298C polymorphism in diabetes susceptibility,Citation13–18 but the results are contradictory.
In order to get more accurate results, we performed a meta-analysis. In this study, we intend to explore the possible relationship between the MTHFR A1298C polymorphism and the susceptibility of diabetes. To our knowledge, this is the most comprehensive meta-analysis conducted to date with regard to the relationship between the MTHFR A1298C polymorphism with the susceptibility of diabetes.
Materials and methods
Search strategy
We conducted a comprehensive search strategy towards the electronic databases, such as PubMed, Embase, Chinese Biomedical Literature Database (CBM, Chinese), China National Knowledge Infrastructure (CNKI, Chinese), and Wangfang Database (Chinese) with search terms including ‘‘diabetes’’ together with ‘‘MTHFR’’ or “methylenetetrahydrofolate reductase “or ‘‘polymorphism’’ or ‘‘variants’’ or “mutation’’. There were no limitations to the language of publications.
Inclusion and exclusion criteria
The selection of articles in this meta-analysis was abided by the criteria as follows: (a) case-control studies; (b) the publications were referring to the relationship between MTHFR A1298C polymorphism and diabetes susceptibility; (c) selected studies have sufficient data to calculate ORs with the corresponding 95% CIs; (d) the studies were published. The following exclusion criteria were used for excluding studies: (a) reviews or case reports; (b) studies contained duplicate data; (c) no usable data reported; (d) the distribution of genotypes among controls are not in the Hardy–Weinberg equilibrium.
Data extraction
Data were carefully extracted by two authors, independently from each study, based on the inclusion criteria mentioned above. If encountered the conflicting evaluations, an agreement was reached following a discussion; if could not reached agreement, then a third author was consulted to resolve the debate. The following information was extracted: name of first author, year of publication, country of origin, ethnicity, genotyping method, source of controls, and sample size. Ethnicity was categorized as “Caucasian”, “Asian”, and “Mix”. We also evaluated whether the genotype distributions were in the Hardy–Weinberg equilibrium.
Statistical analysis
Crude odds ratios (ORs) together with their corresponding 95% CIs were used to assess the strength of association between the MTHFR A1298C polymorphism and diabetes risk. The pooled ORs were performed for allele contrast (C vs. A), homozygous (CC vs. AA), heterozygous (CA vs. AA), dominant model (CC/AC vs. AA), and recessive model (CC vs. AC/AA). The heterogeneity was assessed by a Chi-square based Q-statistic test. The influence of heterogeneity was quantified by using I2 value as well as p-value.Citation19 If I2 value > 50% or p < 0.10, that suggest obvious heterogeneity was existed, ORs were pooled by a random-effects model (Der Simonian and Laird method).Citation20 Otherwise, the fixed-effects model (the Mantel–Haenszel method) was used.Citation21 We also performed subgroup analysis by ethnicity (Caucasian, Asian, and Mix).
A professional web-based program was used to assess the Hardy–Weinberg equilibriumCitation22 of controls (http://ihg2.helmholtz-muenchen.de/cgibin/hw/hwa1.pl), if the p > 0.05 suggests the controls was followed the HWE balance. The sensitivity analysis was performed to assess the stability of the results. It was performed by excluding a single study each time.Citation23 When the Hardy–Weinberg equilibrium disequilibrium existed (p < 0.05 was considered statistically significant), the sensitivity analysis was also conducted. Publication bias was estimated by Egger's test (p < 0.05 was considered representative of statistically significant publication bias)Citation24 and visual observation of the funnel plot.Citation25 All statistical tests were performed with STATA Software (version 9.2, Stata Corp., College Station, TX).
Results
Search results and study characteristics
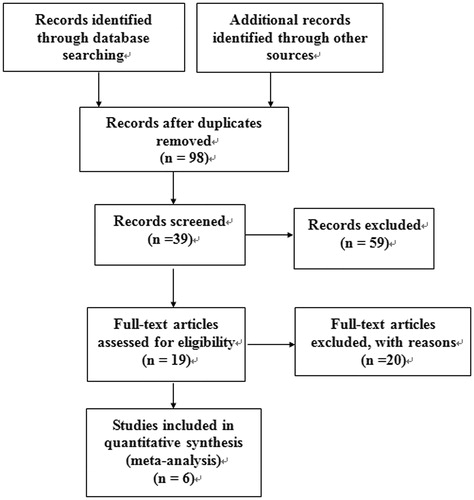
After a careful examination based on the inclusion criteria listed above, totally six studiesCitation13–18 with 897 cases and 852 controls were included in our meta-analysis (flowchart shown in ). Among them, three studies were for Caucasian, two studies were for Asian and one was for Mix. The genotypes distribution in the controls group of all the studies included in this meta-analysis were consistent with HWE (all p > 0.05). The detailed information is shown in .
Table 1. General characteristics of studies included in the meta-analysis.
Quantitative synthesis of data
The main results of the meta-analysis are listed in . We found that the MTHFR A1298C polymorphism was significantly associated with the diabetes susceptibility in overall population only under the recessive model (CC vs. AC/AA: OR = 1.70, 95% CI = 1.18–2.45, p = 0.004, ), while there was no association found between MTHFR A1298C polymorphism and diabetes risk in the others models (C vs. A: OR = 0.90, 95% CI = 0.56–1.43, p = 0.649; CC vs. AA: OR = 1.45, 95% CI = 0.81–2.59, p = 0.208; AC vs. AA: OR = 1.03, 95% CI = 0.81–1.31, p = 0.838; CC/AC vs. AA: OR = 1.05, 95% CI = 0.83–1.32, p = 0.688. Figure not shown).
Figure 2. (A) The forest plot describing the meta-analysis under recessive model for the association between MTHFR A1298C polymorphism and the risk of diabetes (CC vs. CA + AA). (B) The Begg-funnel plot for publication bias test for the association between MTHFR A1298C polymorphism and the risk of diabetes under recessive model (CC vs. CA + AA). Each point represents a separate study for the indicated association. log [OR], natural logarithm of OR. Horizontal line means effect size.
![Figure 2. (A) The forest plot describing the meta-analysis under recessive model for the association between MTHFR A1298C polymorphism and the risk of diabetes (CC vs. CA + AA). (B) The Begg-funnel plot for publication bias test for the association between MTHFR A1298C polymorphism and the risk of diabetes under recessive model (CC vs. CA + AA). Each point represents a separate study for the indicated association. log [OR], natural logarithm of OR. Horizontal line means effect size.](/cms/asset/8b8af01b-d61b-4a81-9eae-0aef7eba17f0/irnf_a_917429_f0002_b.jpg)
Table 2. Results of meta-analysis for MTHFR A1298C polymorphism and diabetes risk.
In the sub-group analysis according to ethnicity, the results suggested that MTHFR A1298C polymorphism was not associated with diabetes risk in Caucasians population (). However, the significant association was found in Asian population under the dominant model (CC/AC vs. AA: OR = 1.31, 95% CI = 1.003–1.72, p = 0.047, ).
Heterogeneity analysis and sensitive analysis
There was a significant heterogeneity for all genetic models in the overall population. To examine the source of heterogeneity, we assessed the dominant model (CC/AC vs. AA) by ethnicity (Caucasian, Asian, or Mix) and sample size (≤200 subjects or >200 subjects). As a result, ethnicity (p = 0.006) but not sample size (p > 0.05) was found to contribute to substantial heterogeneity. Sensitivity analysis was used to reflect the influence of the individual data set to the pooled ORs by sequential omission of individual studies. In this meta-analysis, results of the sensitive analysis show that there is no other study which could influence the overall results, indicating the results of this meta-analysis are robust and reliable.
Publication bias
The funnel plot and Egger's test were used to test the publication bias. The funnel plot is relatively straightforward to observe whether the publication bias is present, and Egger's test was performed to provide statistical evidence of symmetries of the plots. As showed in , the shape of the funnel plot did not show obvious asymmetry. Similarly, the results of Egger's test shows no publication bias (all p > 0.05, data not shown).
Discussion
It is well recognized that there is a variation in individual susceptibility to the diabetes even with the same environmental exposure. Host factors, including polymorphisms of genes involved in this formation of diabetes might have been responsible for this difference. Therefore, genetic susceptibility to diabetes has been a research focus in scientific community. Recently, genetic polymorphisms of the MTHFR A1298C gene associated with diabetes have drawn increasing attention. Previous results of the research on the relationship between MTHFR A1298C polymorphism and diabetes risk were contradictory. These inconsistent results are possibly because of a small effect of the polymorphism on diabetes risk or the relatively small sample size of the published studies. A meta-analysis is a powerful strategy because it potentially investigates a large number of individuals and can estimate the effect of a genetic factor on the risk of the disease.Citation26–28
The present meta-analysis, including six studies with 897 cases and 852 controls, explored the association between the MTHFR A1298C polymorphism and diabetes risk. To our knowledge, this is the first meta-analysis of prospective cohort studies on MTHFR A1298C polymorphism and the risk of diabetes. Results of the present analysis suggested that the CC gene in MTHFR A1298C is a risk factor for diabetes (CC vs. AC/AA: OR = 1.70, 95% CI = 1.18–2.45, p = 0.004). Subgroup analysis based on ethnicity indicated that MTHFR A1298C polymorphism was a risk factor for diabetes in Asian population (CC/AC vs. AA: OR = 1.31, 95% CI = 1.003–1.72, p = 0.047).
The heterogeneity plays an important role when performing meta-analysis and finding the source of heterogeneity is very important for the final result of meta-analysis. In the current study, obvious heterogeneity between study was found in the overall population. The heterogeneity cannot be explained by several possible source of heterogeneity, such as ethnicity and sample size (≤200 subjects or >200 subjects). By conducting the meta-regression, we found the ethnicity was the major source of the high heterogeneity in our meta-analysis, which could be explained by the race-specific effect of MTHFR A1298C polymorphism on the susceptibility to diabetes, because different countries may have different genetic backgrounds and life styles. However, the ethnicity did not explain all heterogeneity in this meta-analysis and other sources need further investigating. It is possible that other limitations of recruited studies may partially contribute to the observed heterogeneity. Publication bias is another important aspect which may have a negative effect on a meta-analysis. In our meta-analysis, the funnel plot and Egger's test were used to test the publication bias of the included studies. Both the shape of the funnel plot and statistical results show no obvious publication bias, this suggests that the results of our meta-analysis are relatively stable.
Although comprehensive analysis was conducted to show the association between MTHFR A1298C polymorphism and risk of diabetes, there are still some limitations that need to be pointed out. Firstly, the number of studies and the number of samples included in the meta-analysis were relatively small. Secondly, publishing bias might exist in this meta-analysis since only publications in English and Chinese were included in the current meta-analysis. Thirdly, the effects of genetic factors on diabetes risk were confounded by life style and others genes. This means that there will not be a single gene or single environmental factor that has larger effects on diabetes susceptibility. So, studies with more gene–gene and gene–environment interactions should be performed.
In conclusion, the results suggest that CC mutant gene of the MTHFR A1298C polymorphism was significantly related to diabetes risk. In the sub-group analysis based on ethnicity, we found significant association was existed between MTHFR A1298C polymorphism and the susceptibility of diabetes in Asian population. Considering the limited studies and sample size included in the meta-analysis, further large scaled and well-designed studies are needed to confirm our results.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Acknowledgements
We thank our teachers and classmates, who have provided their assistance and support during this study.
References
- Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:4–14
- Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053
- Tang L, Wang L, Liao Q, et al. Genetic associations with diabetes: meta-analyses of 10 candidate polymorphisms. PLoS One. 2013;8:e70301
- Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088
- Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell. 2012;148:1160–1171
- Wang L, Teng Z, Cai S, Wang D, Zhao X, Yu K. The association between the ppargamma2 pro12ala polymorphism and nephropathy susceptibility in type 2 diabetes: a meta-analysis based on 9,176 subjects. Diagn Pathol. 2013;8:118–124
- Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–366
- Hua Y, Shen J, Song Y, Xing Y, Ye X. Interleukin-10 -592c/a, -819c/t and -1082a/g polymorphisms with risk of type 2 diabetes mellitus: a huge review and meta-analysis. PLoS One. 2013;8:e66568
- Li YY, Yang ZJ, Zhou CW, et al. Adiponectin-11377cg gene polymorphism and type 2 diabetes mellitus in the chinese population: a meta-analysis of 6425 subjects. PLoS One. 2013;8:e61153
- Rosenblatt DS. Methylenetetrahydrofolate reductase. Clin Invest Med. 2001;24:56–59
- van der Put NM, Gabreels F, Stevens EM, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051
- Weisberg IS, Jacques PF, Selhub J, et al. The 1298a–>c polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis. 2001;156:409–415
- Mello AL, Cunha SF, Foss-Freitas MC, Vannucchi H. Evaluation of plasma homocysteine level according to the c677t and a1298c polymorphism of the enzyme MTHRF in type 2 diabetic adults. Arq Bras Endocrinol Metabol. 2012;56:429–434
- Benrahma H, Abidi O, Melouk L, et al. Association of the c677t polymorphism in the human methylenetetrahydrofolate reductase (MTHFR) gene with the genetic predisposition for type 2 diabetes mellitus in a Moroccan population. Genet Test Mol Biomarkers. 2012;16:383–387
- Chang YH, Fu WM, Wu YH, Yeh CJ, Huang CN, Shiau MY. Prevalence of methylenetetrahydrofolate reductase c677t and a1298c polymorphisms in Taiwanese patients with type 2 diabetic mellitus. Clin Biochem. 2011;44:1370–1374
- Mtiraoui N, Ezzidi I, Chaieb M, et al. MTHFR c677t and a1298c gene polymorphisms and hyperhomocysteinemia as risk factors of diabetic nephropathy in type 2 diabetes patients. Diabetes Res Clin Pract. 2007;75:99–106
- Dinleyici EC, Kirel B, Alatas O, Muslumanoglu H, Kilic Z, Dogruel N. Plasma total homocysteine levels in children with type 1 diabetes: relationship with vitamin status, methylene tetrahydrofolate reductase genotype, disease parameters and coronary risk factors. J Trop Pediatr. 2006;52:260–266
- Liu D, Fan X, Yu D. Methylenetetrahydrofolate reductase 677c → t and 1298a → c polymorphism and cardiovascular disease associated with diabetes. Chin J Diabetes. 2005;13:219–222
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748
- Zamora-Ros R, Rothwell JA, Scalbert A, et al. Dietary intakes and food sources of phenolic acids in the European prospective investigation into cancer and nutrition (epic) study. Br J Nutr. 2013;110:1500–1511
- Tobias A, Campbell MJ. Modelling influenza epidemics in the relation between black smoke and total mortality. A sensitivity analysis. J Epidemiol Community Health. 1999;53:583–584
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101
- Fujisawa T, Ikegami H, Kawaguchi Y, Ogihara T. Meta-analysis of the association of trp64arg polymorphism of beta 3-adrenergic receptor gene with body mass index. J Clin Endocrinol Metab. 1998;83:2441–2444
- Liwei L, Chunyu L, Ruifa H. Association between manganese superoxide dismutase gene polymorphism and risk of prostate cancer: a meta-analysis. Urology. 2009;74:884–888
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108