Abstract
The eukaryotic endomembrane system (ES) is served by hundreds of dedicated proteins. Experimental characterization of the ES-associated molecular machinery in several model eukaryotes complemented by a recent progress in phylogenomics and comparative genomics have revealed a conserved complex core of the machinery that appears to have been established before the last eukaryotic common ancestor (LECA). At the same time, modern eukaryotes exhibit a huge variation in the ES resulting from a multitude of evolutionary processes operating along the ever-branching paths from the LECA to its descendants. The most important source of evolutionary novelty in the ES functioning has undoubtedly been gene duplication followed by divergence of the gene copies, responsible not only for the pre-LECA establishment of many multi-paralog families of proteins in the very core of the ES-associated machinery, but also for post-LECA lineage-specific elaborations via family expansions and the origin of novel components. Extreme sequence divergence has obscured actual homologous relationships between potentially many components of the machinery, even between orthologous proteins, as illustrated by the yeast Vps51 subunit of the vesicle tethering complex GARP hypothesized here to be a highly modified ortholog of a conserved eukaryotic family typified by the zebrafish Fat-free (Ffr) protein. A dynamic evolution of many ES-associated proteins, especially those centred around RAB and ARF GTPases, seems to take place at the level of their domain architectures. Finally, reductive evolution and recurrent gene loss are emerging as pervasive factors shaping the ES in all phylogenetic lineages.
Introduction
Although some prokaryotic cells are known to house relatively complex arrangements of internal membranes with more eukaryote-like features that we would acknowledge a few years ago (Fuerst Citation2005, Lonhienne et al. Citation2010), none can perhaps compete in complexity with the elaborated endomembrane system (ES) of eukaryotes. Microscopical, biochemical and genetic investigations by several generations of biologists have yielded an integrated view of the general organization and functioning of the eukaryotic ES. Embodied in innumerable textbook schemes, this view shows the ES as a series of intracellular compartments delimited by a unit membrane and interconnected by traffic of membranous vesicles budding off and fusing with the compartments and the plasma membrane (for general reviews on the molecular aspects of membrane trafficking see Bonifacino and Glick Citation2004, Cai et al. Citation2007, Pfeffer Citation2007, Stenmark Citation2009, and the other reviews in this volume). Importantly, the compartments can often be homologized across the whole span of the eukaryotic diversity, indicating that they differentiated early in eukaryotic evolution and have since maintained their identity over myriads of cell generations (Cavalier-Smith Citation2004). Compartments generally considered to be a part of the canonical Bauplan of the eukaryotic cell include the endoplasmic reticulum (ER) comprising the nuclear envelope (NE) as its special domain, the Golgi complex typically organized as a stack of flat cisternae (the dictyosome), the trans-Golgi network (TGN), the early (sorting) endosomes, the late endosomes (also called the multivesicular bodies), and the lysosomes/vacuoles. The reach of the ES may, however, be substantially broader. First, after decades of discussions it now seems established that peroxisomes (and their variants) are both evolutionarily and physically connected with the ES, specifically with the ER (Tabak et al. Citation2008, Gabaldón Citation2010). Second, it has been recently unveiled that a specialized and perhaps ancient branch of the secretory pathway is used for bringing proteins and membranes to the cilium (flagellum) (Baldari and Rosenbaum Citation2010, Rohatgi and Snell Citation2010), possibly with the help of the newly described vesicle coat BBSome (Jin et al. Citation2010). Third, compartments of the ES usually neglected by generalized schemes can occur quite widely in eukaryotes, such as acidocalcisomes (Moreno and Docampo Citation2009, Docampo et al. Citation2010) or autophagosomes (Hughes and Rusten Citation2007, Kiel Citation2010). Fourth, there seems to be continuity between the outer membrane of mitochondria and the ES, at least during the biogenesis of the autophagosome (Hailey et al. Citation2010). Likewise, plastids are also actually linked with the ES, be they primary plastids, which seems to receive some of their proteins via an incompletely understood pathway traversing the Golgi (Nanjo et al. Citation2006, Kitajima et al. Citation2009), or secondary plastids, which in some algal taxa (e.g., euglenophytes and dinoflagellates) or in apicomplexans (such as the malaria parasite Plasmodium falciparum) occupy an independent compartment communicating with the rest of the ES (Nassoury et al. Citation2003, Sláviková et al. Citation2005, Tonkin et al. Citation2008). Furthermore, different eukaryotic lineages depart to various extents from the generalized textbook version by possessing novel specialized compartments, such as diverse types of extrusomes (Rosati and Modeo Citation2003), cortical alveoli (Gould et al. Citation2008), contractile vacuoles (Allen and Naitoh Citation2002), or the cell plate in dividing plant cells (Jürgens Citation2005).
The attention paid to the eukaryotic ES has predominantly concerned its functional aspects, especially in a handful of ‘model organisms’ like the yeast Saccharomyces cerevisiae, some metazoans, or the plant Arabidopsis thaliana. However, advances in reconstructing the eukaryotic phylogeny and a rapidly growing list of sequenced genomes of phylogenetically diverse eukaryotes have recently fuelled interest in the diversity of the ES and its evolutionary sources. Studying these questions is not only intellectually appealing per se, but provides a valuable context and inspiration of functional research, too. Indeed, transferring the knowledge gained from yeast or metazoan cells onto the ES of other eukaryotic lineages, typically via bioinformatic identification of homologs of established metazoan or yeast components of the ES-associated molecular machinery and their subsequent experimental characterization guided by prior expectations about their properties and function, has proven extremely productive and led to an amazing improvement of our understanding of the ES functioning at the molecular level in groups such as plants (Bassham et al. Citation2008), trypanosomatids (Engstler et al. Citation2007), or ciliates (Plattner Citation2010). This is per se a good indication that the ES and the associated molecular machinery are widely conserved over the eukaryotic kingdom. However, taking evolution into consideration sheds light on inherent limits that are associated with using metazoan or yeast cells as general eukaryotic models. For example, acknowledging the relatively close phylogenetic position of Metazoa and Fungi (both belonging to the same eukaryotic branch, the Opisthokonta; Adl et al. Citation2005) should teach us that features of the ES (or any other cellular system) ‘conserved from yeast to man’ may simply be opisthokont-specific evolutionary innovations irrelevant to non-opisthokont taxa. Indeed, through the operation of a multitude of evolutionary processes each phylogenetic lineage of eukaryotes is expected to bear its own idiosyncratic features of the ES, not always accessible via a comparative approach.
There have recently been a number of reviews and theoretical discussions published dealing with various evolutionary aspects of the ES (Dacks and Field Citation2007, Field et al. Citation2007, Gurkan et al. Citation2007, Hughes and Rusten Citation2007, Jékely Citation2007, Mironov et al. Citation2007, Dacks et al. Citation2009, Field and Dacks Citation2009, Brighouse et al. Citation2010, Cavalier-Smith Citation2010a). The purpose of the present paper is to provide an updated perspective on ES evolution, with special attention to characteristic evolutionary patterns and processes discussed in the context of recent advances in phylogenomics and comparative genomics. The review starts with a summary of current ideas about the phylogenetic tree of eukaryotes and the position of its root, only to use it as a framework for drawing a picture of the ES of current eukaryotes as an evolutionary derivative of a complex ES of the last eukaryotic common ancestor (LECA) through the action of innovative and reductive evolutionary processes. The rest of the review is devoted to discussing aspects of these evolutionary processes shaping the eukaryotic ES at its molecular level, with particular emphasis on the membrane-trafficking apparatus. Discussion on the origins of the eukaryotic ES, certainly a crucial question of the ES evolution, had to be omitted for the sake of focus (see Jékely Citation2007, Cavalier-Smith Citation2010a, for recent treatments of this subject).
The phylogeny of eukaryotes: A framework for understanding evolution of the ES
Key to a full understanding of evolution of the ES, like of any other aspect of the eukaryotic cell, is a robust phylogenetic scheme bringing an order into the vast diversity of eukaryotes. A thorough review of eukaryotic diversity from the perspective of cell organization (ultrastructural identity) revealed about 70 distinct groups that could not be arranged in any higher-order taxa (Patterson Citation1999), indicating limited utility of morphological characters for inferring the global eukaryotic phylogeny. The recent progress in reconstructing the eukaryotic phylogenetic tree has thus relied almost exclusively on molecular phylogenetics, especially on phylogenomics boosted by important developments in the theory of phylogenetic inference and a rapid growth of genome-level sequence data available for diverse eukaryotes thanks to genome sequencing projects or EST surveys (Delsuc et al. Citation2005). Given the limited space available for this review, readers are encouraged to consult any of the excellent recent papers to learn details of this subject (see Simpson and Roger Citation2004, Keeling et al. Citation2005, Parfrey et al. Citation2006, Brinkmann and Philippe Citation2007, Burki et al. Citation2007, Dacks et al. Citation2008a, Hampl et al. Citation2009, Parfrey et al. Citation2010). A current estimate of the global eukaryotic phylogeny is shown at and a detailed hierarchical taxonomy of eukaryotes (especially protists) largely reflecting the known phylogeny can be found in a work of Adl and colleagues (Citation2005). In the current scheme, a series of some 15 or so clearly monophyletic lineages consistently supported by multi-gene or even single-gene phylogenetic analyses can be recognized in eukaryotes, most of them dominated by diverse and often poorly studied protists.
Figure 1. A current estimate of the eukaryotic phylogenetic tree. The scheme is a consensus of recent phylogenetic and phylogenomic analyses [Burki et al. Citation2007, Citation2008, Citation2009, Hampl et al. Citation2009, Baurain et al. Citation2010, Glücksman et al. Citation2010, Parfrey et al. Citation2010, Yabuki et al. Citation2010]; relationships with inconsistent support are indicated by dashed branches. The limited molecular data available for Collodictyonida, Ancyromonadida, and Micronuclearia podoventralis attest to their independence on other lineages, but do not allow for their more precise placement in the tree. Following a recent proposal [Cavalier-Smith Citation2010b], the name ‘Chromista’ is here applied in a broader sense as compared to the original meaning of the name; the newly defined Chromista comprise ‘chromalveolates’ expanded by inclusion of Rhizaria and some minor previously unplaced lineages (centroheliozoans and telonemids). Three alternative positions of the root of the tree discussed in the text are marked by wedges. Note that the root R1 as indicated in the tree only approximately corresponds to the ‘unikont-bikont’ rooting previously advocated by Stechmann and Cavalier-Smith (Citation2003) and Richards and Cavalier-Smith (Citation2005), since the latter assumed Apusomonadida to be ‘bikonts’, while more recent evidence and the present scheme shows these two groups branching off on the ‘unikont’ side. Two alternative scenarios on the evolutionary path of the GTPase RAB24 implied by the alternative roots R2 and R3 are shown by green and red arrows, respectively (a scenario for the R1 root is omitted for clarity). Note that the R2 scenario assumes the presence of RAB24 at the root (i.e., already in the LECA) and secondary loss in the Discoba lineage (among other lineages), whereas the R3 scenario implies post-LECA emergence of RAB4 and its primary absence in the Discoba.
![Figure 1. A current estimate of the eukaryotic phylogenetic tree. The scheme is a consensus of recent phylogenetic and phylogenomic analyses [Burki et al. Citation2007, Citation2008, Citation2009, Hampl et al. Citation2009, Baurain et al. Citation2010, Glücksman et al. Citation2010, Parfrey et al. Citation2010, Yabuki et al. Citation2010]; relationships with inconsistent support are indicated by dashed branches. The limited molecular data available for Collodictyonida, Ancyromonadida, and Micronuclearia podoventralis attest to their independence on other lineages, but do not allow for their more precise placement in the tree. Following a recent proposal [Cavalier-Smith Citation2010b], the name ‘Chromista’ is here applied in a broader sense as compared to the original meaning of the name; the newly defined Chromista comprise ‘chromalveolates’ expanded by inclusion of Rhizaria and some minor previously unplaced lineages (centroheliozoans and telonemids). Three alternative positions of the root of the tree discussed in the text are marked by wedges. Note that the root R1 as indicated in the tree only approximately corresponds to the ‘unikont-bikont’ rooting previously advocated by Stechmann and Cavalier-Smith (Citation2003) and Richards and Cavalier-Smith (Citation2005), since the latter assumed Apusomonadida to be ‘bikonts’, while more recent evidence and the present scheme shows these two groups branching off on the ‘unikont’ side. Two alternative scenarios on the evolutionary path of the GTPase RAB24 implied by the alternative roots R2 and R3 are shown by green and red arrows, respectively (a scenario for the R1 root is omitted for clarity). Note that the R2 scenario assumes the presence of RAB24 at the root (i.e., already in the LECA) and secondary loss in the Discoba lineage (among other lineages), whereas the R3 scenario implies post-LECA emergence of RAB4 and its primary absence in the Discoba.](/cms/asset/1cf485f6-9152-4e10-8980-4dbf501df9a8/imbc_a_521201_f0001_b.jpg)
Some specific higher-order groupings (‘supergroups’) of these major lineages have been proposed (), but they remain contentious at best. Green algae and plants (Chloroplastida), red algae (Rhodophyta), and glaucophytes sharing a prominent cellular feature – a primary plastid – are hypothesized to form a monophyletic group called the Archaeplastida (or Plantae) (Adl et al. Citation2005), but such a group, if recovered at all, generally lacks significant statistical support, even in multi-gene phylogenomic analyses (Burki et al. Citation2007, Citation2008, Yoon et al. Citation2008, Hampl et al. Citation2009, Nozaki et al. Citation2009). Three protist lineages, Metamonada (anaerobic flagellates such as diplomonads and parabasalids), Discoba (Euglenozoa, Heterolobosea, and Jakobida), and malawimonads may form a common supergroup (Excavata) with a synapomorphic feeding ventral groove and associated characteristic arrangement of the microtubular cytoskeleton (actually retained only by some members of Metamonada and Discoba; Simpson Citation2003). However, the monophyly of excavates as a whole has not been convincingly demonstrated even in the most recent phylogenomic analysis concentrated specifically on this issue (Hampl et al. Citation2009). A bulk of protist species may belong to a huge hypothetical assemblage called Chromalveolata or Chromista (Adl et al. Citation2005, Cavalier-Smith Citation2010b), which is hypothesized to stem from one or potentially two consecutive endosymbiotic fusions of a host eukaryotic cell with a red and potentially also a green alga (Elias and Archibald Citation2009a, Moustafa et al. Citation2009, Cavalier-Smith Citation2010b). This idea has aroused a lot of controversy and evidence from phylogenomics and comparative genomics, speaking for or against it, has been levelled by different camps (Keeling Citation2008, Burki et al. Citation2009, Bodył et al. Citation2009, Baurain et al. Citation2010). Regardless the uncertainties in the actual history of endosymbioses in chromist (chromalveolate) taxa and the monophyly of the whole group, solid evidence exists for a common descent of three major subgroups – stramenopiles (= heterokonts), alveolates, and Rhizaria (the latter previously considered as an independent ‘supergroup’), constituting the so-called SAR clade (Burki et al. Citation2007), recently dubbed the ‘Harosa’ (Cavalier-Smith Citation2010b). The remaining chromists (haptophytes, cryptomonads, and a few additional minor protist groups) may belong to another clade called the Hacrobia (Burki et al. Citation2009, Okamoto et al. Citation2009), although it receives less consistent support in phylogenomic analyses that the SAR clade (Burki et al. Citation2009, Parfrey et al. Citation2010, Baurain et al. Citation2010).
From the point of interpreting evolution of the ES, the uncertainties about monophyly of some hypothetical ‘supergroups’ may be less critical than an unresolved position of the root of the eukaryotic tree (). Knowing the root position is a prerequisite for distinguishing ancestral and derived character states in extant eukaryotes, and hence for ordering in time the evolutionary events that affected different cellular systems in individual eukaryotic lineages since the last eukaryotic common ancestor (LECA). An apparently attractive idea of the root lying in a presumably basal and primitively amitochondrial eukaryotic kingdom ‘Archezoa’ (Cavalier-Smith Citation1993, Sogin et al. Citation1996) has been dismissed due to the discovery remnant or highly modified mitochondria in all ‘archezoan’ lineages (Hampl and Simpson Citation2008, Hjort et al. Citation2010) and attributing the basal position of these lineages in some phylogenies to artefacts of phylogenetic inference (Philippe Citation2000, Brinkmann et al. Citation2005). A hypothesis that has also been widely adopted (again perhaps too uncritically) by the biological community is that placing the root between two hypothetical principal eukaryotic groups, the unikonts and the bikonts (Stechmann and Cavalier-Smith Citation2003, Richards and Cavalier-Smith Citation2005). I refrain from repeating the arguments behind this hypothesis and only state that the evidence for the monophyly of both the unikonts and the bikonts has dissolved with new results from phylogenetics and comparative genomics, as excellently discussed elsewhere (Roger and Simpson Citation2009).
Given this situation, Cavalier-Smith has very recently offered a new idea arguing that the root lies between Euglenozoa (euglenoids, kinetoplastids, and two less well known lineages of marine flagellates) and remaining eukaryotes (Cavalier-Smith Citation2010b). This hypothesis is based on interpreting the phylogenetic distribution of some molecular characters, but since it is in conflict with some other evidence (e.g., a synapomorphic insertion in a ribosomal protein shared by Euglenozoa and other Discoba; Rodríguez-Ezpeleta et al. Citation2007), and because the characters of Euglenozoa placing them aside other eukaryotes may well be secondary modifications, I am personally rather sceptical as to the possibility of this hypothesis surviving further scrutiny. Rogozin et al. (Citation2009) recently took a different and a more systematic approach to locate the eukaryotic root by analysing a large collection of insertions/deletions (indels) and rare substitutions in alignments of proteins sequences conserved across several distantly related eukaryotic species. The analysis suffers from absence or poor representation of some critical lineages in the dataset analyzed, but the results are interesting and promising for the future. Specifically, the authors concluded that the root probably lies between Plantae (= Archaeplastida) and the remaining eukaryotes, although the taxon sampling employed actually could not have excluded the root from within Archaeplastida.
The ES of extant eukaryotes derives from a surprisingly complex ES of the LECA
The uncertainties about the actual relationships among major eukaryotic lineages and about the position of the root notwithstanding, the recent achievements of eukaryotic phylogenetics allows for far more accurate interpretations of evolutionary trends relating to various cellular systems, including the ES, than it was possible a decade ago. One crucial insight is that it is misleading to distinguish ‘lower’ and ‘higher’ extant eukaryotes, as there are probably no living eukaryotes that would primitively lack major characteristic features of the eukaryotic cell such as the mitochondrion, the Golgi apparatus, or the peroxisome (Dacks et al. Citation2008a). We should rather view each eukaryotic species as a mixture of primitive (i.e., ancestral) and derived characters, with the latter including results of ‘innovative’ as well as reductive evolution. Indeed, the lineages previously considered primitive, e.g., the ‘archezoans’ Mircosporidia or diplomonads, seem to actually belong among the most advanced eukaryotes, in terms of the number of evolutionary events that have occurred in these lineages since the LECA. A high number of evolutionary changes have impacted also the ES of these two groups, including the loss of the peroxisome (Gabaldón Citation2010) and an extreme modification of their Golgi (Beznoussenko et al. Citation2007, Abodeely et al. Citation2009, Mowbrey and Dacks Citation2009, Stefanic et al. Citation2009).
The second and related implication of the current view of the eukaryotic phylogeny is a strikingly complex nature of the LECA, which was very likely a fully-fledged eukaryote with a characteristically organized genome, cytoskeleton and the ES, with a mitochondrion and the cilium, and with a modern-like complexity of the associated molecular machinery and regulatory circuits (Fritz-Laylin et al. Citation2010, Koonin Citation2010a, Citation2010b, Cavalier-Smith Citation2010a). The notion of a genetically complex LECA comes from comparative genomics revealing that there are thousands of orthologous genes with phylogenetically wide, though sometimes punctuated, distribution indicating an ancient origin of these genes in early eukaryotes. Considering specifically LECA's ES, it seems to have resembled the ES of modern eukaryotes in embracing the complete array of the ‘standard’ compartments and transport pathways underpinned by a molecular machinery including several vesicle coat complexes and the related nuclear pore complex (NPC), small GTPases of the RAB and ARF/SAR1 families (master regulators of vesicle budding and fusion) and their specific regulators (GEFs and GAPs), several different tethering complexes (devices involved in tethering vesicles to the membrane of their target compartments), SNAREs (a family of membrane proteins directly responsible for membrane fusion), SM family proteins (regulators of SNARE function), ESCRT I to ESCRT III complexes (involved in sorting of proteins destined for degradation and in formation of the MVB), and numerous other components (Dacks and Field Citation2004, Citation2007, Kloepper et al. Citation2007, Koumandou et al. Citation2007, Field and Dacks Citation2009, DeGrasse et al. Citation2009, Brighouse et al. Citation2010). It is now also clear that the LECA very likely exhibited a Golgi apparatus in the typical form of stacked cisternae, which has been retained in the vast majority of extant eukaryotes but secondarily modified in at least eight independent lineages exemplified by the yeast S. cerevisiae or the whole group of Heterolobosea (Mowbrey and Dacks Citation2009).
Naturally, assuming different position of the root of the eukaryotic tree leads to different versions of the LECA inferred, especially concerning the set of its genes and proteins. For example, the RAB24 paralog of the RAB family (probably involved in the membrane traffic accompanying autophagy; Munafó and Colombo Citation2002) appears to exhibit a scattered distribution across eukaryotic phyla (; M. Elias, J. B. Dacks, M. C. Field, unpublished work). Such a distribution at any rate implies a number of independent losses, but whether RAB24 was present already in LECA (and its absence from modern organisms is always due to loss) or whether it was invented in a post-LECA ancestor of only a subgroup of extant eukaryotes (meaning that some eukaryotes lack RAB24 primarily) depends on the actual position of the root. Thus, the ‘unikont-bikont’ or the ‘Archaeplastida-first’ root hypotheses are consistent with the former possibility, whereas the ‘Euglenozoa-first’ rooting leads to the inference of a post-LECA origin of RAB24 and its primary absence in the whole (paraphyletic) Discoba group (). However, one should be very careful with claims about absence of a particular gene in any eukaryotic lineage, since these may need revision with a single new genome sequenced. Consider, for instance, the case of the SNARE of the Npsn type, which had seemed to be missing from opisthokonts despite many opisthokont genome sequences available, only to be eventually found in chytrid fungi (Kienle et al. Citation2009). Indeed, as will be argued below, gene loss has been one of the major processes of post-LECA evolution. Hence, two avenues of future research must be followed to enable reconstruction of the LECA and its ES precise in detail: (1) Clarifying the position of the root of the eukaryotic tree (which may prove to be a real challenge), and (2) establishing patterns of presence/absence of individual genes in extant eukaryotes based on a much denser sampling of genome sequences. Nevertheless, my personal bet is that the current view of the LECA as a complex creature with modern-like gene repertoire will withstand the test of time.
The emerging complexity of the LECA and its ES does not diminish the magnitude of evolutionary changes that have affected the ES in each eukaryotic lineage. While we are aware of the ES diversity resulting from all this intricate history, we are currently hardly able to establish precisely the order of individual evolutionary steps and map them onto the eukaryotic phylogeny. Putting aside inherent limitations of any historical analysis, this is largely because of our poor knowledge or even complete ignorance about molecular details of the ES in most species. For whole major lineages, including Rhizaria, Preaxostyla, cryptomonads, glaucophytes, jakobids, or ancyromonads, there are even no reference genome sequences currently available. In contrast, the major eukaryotic group Opisthokonta, constituted by multicellular animals (Metazoa), fungi (including yeasts), and their protist relatives (e.g., choanoflagellates), contains most species serving as models for studies on molecular processes underlying the function of the ES (above all, S. cerevisiae, mammalian cells, Drosophila melanogaster, Caenorhabditis elegans). Not surprisingly, it has also been showered by attention paid to it by genome sequencers. Opisthokonts thus offer a plenty of examples of ES-associated novelties, sometimes with a well-understood relation to organismal phylogeny (). These examples comprise a range of categories, including:
Figure 2. Examples of evolutionary events affecting the ES-associated machinery in the eukaryotic ‘supergroup’ Opisthokonta. The scheme shows a non-exhaustive selection of events with a well-understood relation to the organismal phylogeny. The events are mapped on a schematic opisthokont phylogeny according to the following published surveys: SNARE proteins (Vam7, Npsn, Sec22, Syx6, Syx7, SN47, and Gs15) – Kloepper et al. (Citation2008), Kienle et al. (Citation2009), Sec2 – Elias (Citation2008), BBSome – Hodges et al. (Citation2010), GGA, Epsin, Eps15, caveolins, stonins – Field et al. (Citation2007), ESCRT 0 – Leung et al. (Citation2008), RAB7 – Mackiewicz and Wyroba (2007). The change affecting Vps51 is reported below in this review, the remaining events in the fungal branch are shown according to analyses reported in Ma et al. (Citation2009). The fate of RAB40 and AP-4 in metazoans is based on my own BLASTP searches against the non-redundant protein database at NCBI.
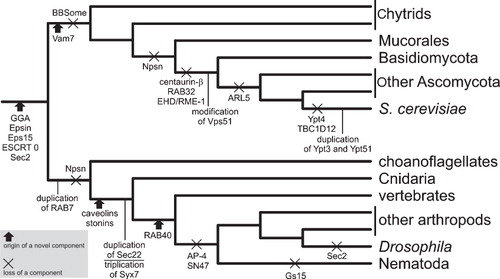
(1) The emergence of new paralogs by gene duplication, such as the RAB9 GTPase (participating in transport from late endosomes to the TGN) that evolved in the Holozoa clade (Metazoa and their closest protist relatives) from a duplicated RAB7 gene (Mackiewicz and Wyroba Citation2009); this is probably the most prevalent type of evolutionary novelties associated with the ES.
(2) The origin of proteins with novel domain architectures, exemplified by the vacuole-associated Vam7 SNARE protein, which arose in the stem lineage of fungi as a new paralog modified by adding an N-terminal phospholipid-binding PX domain and deleting the C-terminal trans-membrane region (Kienle et al. Citation2009).
(3) The origin of ‘novel’ proteins, actually representing proteins that have diverged substantially from their relatives up to the point where recognition of homologs may be difficult or impossible; good examples are metazoa-specific proteins of the stonin family of sorting adaptors functioning in endocytosis, which still bear signs of their probable origin from the ancient and ubiquitous μ subunit of the AP-2 adaptin complex (Field et al. Citation2007, Maritzen et al. Citation2010), or the caveolae-associated caveolin family, for which homologs outside Metazoa could not be found (Field et al. Citation2007; see also the Caveolin Pfam entry, http://pfam.sanger.ac.uk/family/PF01146].
(4) Simplification of the ES due to loss of its components, e.g., of centaurin-beta (an ArfGAP protein) in the lineage leading to dikaryan fungi (Ma et al. Citation2009). Other examples of evolutionary changes in the ES-associated machinery having happened in the course of the opisthokont phylogeny are shown in .
Interestingly, a growing list of ES-associated features appear to be novel for opisthokonts as a whole, that is shared by at least some metazoans and fungi (the two principal opisthokont lineages) but missing outside opisthokonts. This category includes the GGA family (monomeric clathrin adaptors that mediate the sorting of mannose-6-phosphate receptors between the trans-Golgi network and endosomes) or Epsin and Eps15 (adaptor proteins important in clathrin-mediated endocytosis) (Field et al. Citation2007), the heterodimeric complex ESCRT 0 initiating the assembly of the whole ESCRT machinery at the nascent MVB by recognising ubiquitylated cargo (Leung et al. Citation2008), or the family of Sec2-related guanine exchange factors (GEFs), acting together with certain RAB GTPases to regulate exocytosis (Elias Citation2008). These observations, although needing to be tested with a far better sampling of non-opisthokonts genomes to reduce the possibility of pre-opisthokont origin of the proteins listed above, suggest that a wealth of novel features of the ES might have evolved in stem opisthokonts (). This would be a significant conclusion, since nothing is know that would define the opisthokont ES at the level of morphology or overall organization, indicating that there might be a lot of ‘hidden’ evolutionary novelty associated with the emergence of other major eukaryotic groups.
Hence, the analysis of opisthokont ES evolution provides important lessons about general evolutionary processes moulding the ES in eukaryotes. Indeed, operation of both main processes, i.e. inventing of novel components of the ES-associated proteome and its reduction, has been demonstrated in other eukaryotic lineages, too. The rest of this review offers rumination on different facets of these processes using selected illustrative examples (mainly related to the membrane-trafficking machinery).
Gene duplication as the major source of ES elaboration
Ever since the seminal book by Susumu Ohno on the role of gene duplication in evolution (Ohno Citation1970), this process is seen as one of the most important sources of evolutionary novelty (Koonin Citation2009). This is especially the case when one of the paralogs resulting from a gene duplication event gains a new function, which can be accompanied by dramatic divergence of its sequence up to an extent leading to the emergence of a ‘new gene’. Extensive gene duplications marked the origin of eukaryotes (Makarova et al. Citation2005, Koonin Citation2010a, Zhou et al. Citation2010) and footprints of this duplication burst are readily apparent on the ES-associated protein machinery as well (Dacks et al. Citation2009, Field and Dacks Citation2009). It is impossible to enumerate here all building blocks of the ES-associated machinery exhibiting ancient paralogous relationships, but beside those commonly mentioned in this context, such as RAB and ARF/SAR1 families, SNAREs, SM proteins, some ESCRT III subunits, or several families of NPC, COP I, COP II and AP-clathrin components (Dacks et al. Citation2009, Field and Dacks Citation2009), the following can also be noted: (1) The Bet3 and the sedlin family, each comprising several subunits of the vesicle tethering complex TRAPP (Sacher et al. Citation2008); (2) the REP/GDI family comprising two ancient paralogs critical for a proper RAB function (see above; Hála et al. Citation2005); or (3) the two ancient paralogous ArfGEFs, BIG/SEC7 and GBF/GEA (Cox et al. Citation2004). Other families of paralogous proteins or proteins sharing paralogous domains, such as the Pra1/Yip3 family of the putative RAB-specific GDI displacement factors or ArfGAP and RabGAP (TBC) domain proteins can each also have many members in individual species in diverse eukaryotic lineages (Bernards Citation2003, Vernoud et al. Citation2003, Alvim Kamei et al. Citation2008, Field and O'Reilly Citation2008), but it has yet to be investigated whether this diversity at least partly tracks back to multiple paralogs differentiated already in the LECA or whether it stems solely from post-LECA lineage-specific duplications. Finally, weak sequence similarity was detected among subunits of the tethering complexes COG, GARP, Exocyst, and Dsl1 (Whyte and Munro Citation2002, Koumandou et al. Citation2007) and interpreted as either evidence for common descent (Whyte and Munro Citation2002, Pei et al. Citation2009) or convergent evolution (Koumandou et al. Citation2007). Recent structural studies on several COG, Exocyst and Dsl1 subunits (Croteau et al. Citation2009, Richardson et al. Citation2009, Tripathi et al. Citation2009) provide strong evidence for true evolutionary ties between at least some subunits (or their parts) of these three complexes (no subunit of the GARP complex has been characterized yet).
As crucial as gene duplication was in laying the foundations of the archetypal eukaryotic ES, it has been just as important and pervasive in post-LECA lineage-specific ES elaboration (Dacks and Field Citation2007, Dacks et al. Citation2008b, Citation2009), meaning that any attempt of an exhaustive review of all the duplication events is doomed to fail. The sheer volume of duplications affecting the ES-associated protein machinery has also serious implications towards reconstructing the actual history of the duplications, especially those accompanied by extensive sequence divergence. For instance, we succeeded in showing the existence of a novel RAB paralog (Rab1A) shared by the three groups of the SAR (Harosa) clade of chromists, but this was only thanks to very careful phylogenetic analyses designed in a way mitigating adverse effects of divergent sequences (Elias et al. Citation2009). Even the care taken to the analyses, however, did not help decide whether a certain divergent cryptomonad RAB protein is, or is not, an ortholog of Rab1A in the SAR clade. So it is presently unclear whether Rab1A is a synapomorphy of the SAR clade or a more ancient invention predating the divergence of the SAR clade and cryptomonads. Such lack of resolution is probably rather a rule than an exception in analyses of complex gene families like RABs or SNAREs, and novel methodological approaches combined with a much better genome sampling might be needed for overcoming these limitations.
Looking more closely on the distribution of duplication rates across the different components of the membrane-trafficking machinery, we see an obvious pattern with some categories systematically more prone to duplicate than others. This pattern actually has quasi-fractal properties: It is true that the RAB GTPase family exhibits far greater propensity toward duplication than the ARF GTPase family, but the same distinction applies on different lineages within the RAB family. To document this, I present a phylogenetic tree of a set of RAB sequences from three ancient subfamilies, RAB11, RAB18, and RAB23, each stemming from a separate paralog differentiated before the LECA (). It is evident that the RAB23 paralog, which seems to have a role in trafficking to the cilium (Wang et al. Citation2006, Boehlke et al. Citation2010), does not form post-LECA in-paralogs. The RAB18 subfamily, with (at least in mammalian cells) a role in the ER-Golgi trafficking (Dejgaard et al. Citation2008) and lipid droplets functioning (Martin et al. Citation2005), remains in a single gene in most species, with the exception of the green alga Chlamydomonas reinhardtii and the moss Physcomitrella patens, which each acquired separately multiple RAB18 in-paralogs. Finally, the RAB11 subfamily, which is probably generally implicated in the post-Golgi trafficking (Chow et al. Citation2008), has experienced multiple independent lineage-specific duplications resulting in multiple in-paralogs in most species (some of them quite divergent, as witnessed by their long branches in the tree at ). The same trend, i.e., the lack of duplications of the RAB23 paralog, duplications of the RAB18 paralog restricted to only some lineages, and widespread duplications of the RAB11 paralog, holds even if a wider sample of taxa is analyzed (M. Elias, J. B. Dacks, M. C. Field, unpublished work).
Figure 3. Ancestral RAB paralogs exhibit different propensity to generating lineage-specific in-paralogs. The tree was inferred using the maximum likelihood method (WAG + Γ + I substitution model) as implemented in RAxML-HPC (7.2.6) run at the CIPRES portal (http://www.phylo.org/portal2/). Bootstrap values (based on 100 replicates) only higher than 50 are shown. Note the lack of significant bootstrap support for the RAB11 clade, probably due to the inclusion of several rather divergent sequences (exhibiting long branches in the tree). Sequences selected for the analysis come from nine phylogenetically diverse eukaryotes (Homo sapiens, the chytrid fungus Spizellomyces punctatus, the amoebozoan Dictyostelium discoideum, the heterolobosean Naegleria gruberi, the metamonad Trichomonas vaginalis, the moss Physcomitrella patens, the green alga Chlamydomonas reinhardtii, the oomycete Phytophthora sojae, and the haptophyte Emiliania huxleyi) and represent members (in-paralogs) of three particular ancestral paralogous RAB groups (RAB11, RAB18, RAB23; the assignment of the sequences is based on large-scale phylogenetic analyses of the RAB family, M. Elias, J. B. Dacks, M. C. Field, unpublished work). Sequences from the same species are highlighted in the same colour. The sequence identifiers provided refer to the NCBI protein database (http://www.ncbi.nlm.nih.gov/), except Phytophthora and Spizellomyces, where the sequences come from the JGI (http://genome.jgi-psf.org/Physo1_1/Physo1_1.home.html) and Broad Institute (http://www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html) databases, respectively.
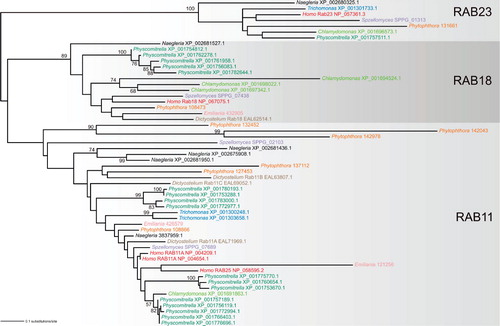
Despite the pervasiveness of such a highly uneven rate of duplications across genes, we are rather ignorant about its causes. One factor that might generally affect the propensity towards duplication is whether the protein is or is not a part of a stable heteromeric complex with a defined stoichiometry of its subunits. The gene dosage balance hypothesis predicts that genes coding for subunits of such complexes are less likely to duplicate, since duplication of one of them causes imbalance among the component with possible deleterious effects on fitness (Papp et al. Citation2003, Liang and Fernandez Citation2008). Predictions of this hypothesis seem to fit quite well the actual situation of the membrane-trafficking machinery, since genome surveys indeed indicate infrequent duplication of genes coding for subunits of complexes such as the various multisubunit tethering factors (Koumandou et al. Citation2007) or ESCRT complexes (Leung et al. Citation2008), at least relatively to the extent of duplication within categories of proteins engaged in only transient complexes (such as RABs or SNAREs). However, exceptions do occur, including an unprecedented case of an extreme lineage-specific duplication burst of a component of the membrane-trafficking machinery documented from plants. The Exo70 subunit of the exocytosis-specific tethering factor Exocyst is generally encoded by a single gene throughout eukaryotes (Koumandou et al. Citation2007), but several rounds of duplications of the ancestral gene encoding the Exo70 subunit occurred in the land plant lineage, resulting in 23 Exo70 paralogs in Arabidopsis and up to 41 in rice (Synek et al. Citation2006, Chong et al. Citation2010). Evolutionary forces driving this duplication and its functional consequences remain a mystery pending functional characterization of the proteins encoded by the expanded plant EXO70 gene family.
The pattern of expansion of the membrane-trafficking machinery is punctuated also along the axis of the organismal phylogeny, with different lineages exhibiting different duplication activity in different times of their history. Let us take just a few examples. Phylogenetic analyses of the SNARE superfamily revealed its significant expansion in metazoans and land plants, suggesting potential connection to multicellularity (Sanderfoot Citation2007, Kloepper et al. Citation2008). However, multicellular fungi do well with a relatively simple complement of SNARE proteins conserved throughout the whole fungal kingdom (Kienle et al. Citation2009). A similar pattern seems to be followed also by the RAB family, being highly expanded in metazoans and land plants but not in multicellular fungi or the stramenopile seaweed Ectocarpus siliculosus (Pereira-Leal and Seabra Citation2001, Pereira-Leal Citation2008, Cock et al. Citation2010, M. Elias, J. B. Dacks, M. C. Field, unpublished work). Multicellular fungi or Ectocarpus are arguably simpler organisms than metazoans or land plants with multiple tissue types, but these observations still suggest that multicellular body organization per se is not necessarily dependent on an exceptionally expanded membrane-trafficking machinery (see also Dacks and Field Citation2007). Indeed, the complexity of this machinery in some unicellular eukaryotes can exceed that of multicellular groups, consider, for instance, the parasitic parabasalid Trichomonas vaginalis with its expanded set of proteins of the membrane-trafficking machinery including the largest complement of RAB GTPases (some 300) ever reported for a single organism (Carlton et al. Citation2007). The amoebozoan genera Entamoeba and Dictyostelium are remarkable for each possessing an array of RAB genes that is not only highly expanded, but also evolutionarily dynamic, as shown by genomic comparisons of different species within the genera (E. histolytica and E. invades, D. discoideum and D. purpureum) revealing an accumulation of many species-specific paralogs, perhaps mainly due to RAB gene duplications postdating the evolutionary divergence of the species (Nakada-Tsukui et al. Citation2010, Sucgang et al. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum, under revision). Given the morphological uniformity of both genera, such differences are unexpected and raise a question on what their evolutionary causes (or functional implications) might be.
Sequence divergence: The limits of the BLAST algorithm and the case of obscure orthologs of the yeast GARP complex subunit Vps51
It is trivial to say that the basic sources of evolutionary change are mutations of DNA sequences resulting in divergence of corresponding protein sequences. Theoretical modelling and empirical research on real protein sequence data over the past few years have contributed greatly to understanding the general principles governing protein sequence evolution (Goldstein Citation2008, Zeldovich and Shakhnovich Citation2008, Lobkovsky et al. Citation2010, Povolotskaya and Kondrashov Citation2010, Sleator Citation2010). Sequence evolution of proteins of the ES-associated machinery probably does not obey any special rules. One important point is that divergence with functional consequences probably more often accompanies evolution of duplicated genes (discussed in the previous section) than of orthologs in different species. Indeed, much of the sequence variation between orthologous proteins probably results from neutral evolution and is functionally silent (Dolinski and Botstein Citation2007, Koonin Citation2009), as evidenced by countless instances of successful complementation of a disrupted gene in one organism by an orthologous gene from even a distantly related organism. Thus, a mutation in a yeast gene coding for a RAB-specific GDP dissociation inhibitor (GDI) that is essential for recycling RAB proteins between ES compartments can be complemented by a GDI ortholog from Arabidopsis (Zárský et al. Citation1997), despite the only 52% amino acid identity of the yeast and Arabidopsis GDI and despite the fact that the most recent ancestor shared by the yeast and Arabidopsis may well have been the LECA itself (see above). On the other hand, a mutation in a yeast gene encoding a GDI paralog called REP (RAB escort protein required for geranylgeranylation of RAB GTPases) cannot be complemented by its Arabidopsis ortholog (Hála et al. Citation2005). However, this is not due simply to a lower similarity between the yeast and Arabidopsis REPs (26%), but because of a unique plant-specific substitution of a single amino acid residue probably abrogating interaction of the Arabidopsis REP protein with the yeast geranylgeranyl transferase. Changing this position in the Arabidopsis REP back to the ancestral state makes the Arabidopsis REP capable of complementing the function of the mutated yeast REP (Hála et al. Citation2005).
Instead of delving further into sequence divergence between orthologs in functional terms, I would like to contemplate its implications towards the central exercise of any evolutionary analysis, that is, identification of sequence homology. Extensive sequence divergence can pose serious challenges to this task. BLAST searches, as the most widely employed method for identification of homologous genes and proteins, become ineffective beyond some degree of sequence divergence between homologs. This may lead to erroneous claims about the ‘absence’ of true homologs in particular species. Fortunately, more sensitive methods have been developed enabling recognition at the sequence level of less obvious cases of homology, including PSI-BLAST (position-specific iterated BLAST; Altschul et al. Citation1997), comparing sequences with sequence profile HMMs (hidden Markov models; Eddy Citation1998) or HMM-HMM comparisons (Söding et al. Citation2005) (not to mention comparing protein tertiary structures as an obvious pinnacle of remote homology-detection methods). The power of these methods, still perhaps too much neglected for answering evolutionary questions related to the ES, can be demonstrated, e.g., by the detection of previously unrealized homology between the Munc13/Unc13 family of proteins important for exocytic neurotransmitter release and some subunits of the vesicle tethering complexes GARP, Exocyst, COG, and Dsl1, revealing their common evolutionary origin (Pei et al. Citation2009).
Let me document the usefulness of ‘post-BLAST’ approaches on my own example of the Vps51 subunit of the GARP protein complex, which works as a vesicle tethering factor at the late Golgi/TGN (Conibear and Stevens Citation2000, Oka and Krieger Citation2005). The complex was originally described in the yeast S. cerevisiae, where it consists of four subunits (Vps51, Vps52, Vps53, and Vps54). While orthologs of three subunits have been easily identified in diverse eukaryotic lineages (Koumandou et al. Citation2007), and at least in mammals shown to form a complex functionally equivalent to the yeast GARP complex (Liewen et al. Citation2005, Pérez-Victoria et al. Citation2008, Pérez-Victoria and Bonifacino Citation2009), no Vps51p has been reported outside yeasts [note that the human Vps51 ortholog claimed to exist by Koumandou et al. (Citation2007) has eventually turned out to be a false BLAST hit; J. B. Dacks, personal communication]. Indeed, when a standard BLASTP search is conducted against the non-redundant protein database at NCBI (National Center for Biotechnology Information; http://blast.ncbi.nlm.nih.gov/) with the S. cerevisiae Vps51p sequence as a query (using the default setting of search parameters), only hits representing protein sequences from some other yeasts (Saccharomycetales) receive E-values low enough (<0.01) to be considered as probable Vps51p homologs. However, the yeast Vps51p has been proposed to contain a region homologous to some other subunits (Cog1 and Exo84) of the related vesicle tethering complexes COG and the Exocyst (see below), and this region is annotated as the Vps51 domain (PF08700) in the Pfam database (http://pfam.sanger.ac.uk/family/PF08700; Finn et al. Citation2010). Using the S. cerevisiae Vps51p as a query in a PSI-BLAST search against the NCBI protein database (with the default PSI-BLAST threshold of 0.005) yields with the second iteration additional hits with the E-value < 0.005, including proteins from other yeast species and some non-yeast fungi. With the third iteration a large number of additional hits from diverse eukaryotes is recovered with E-values < 0.005. Closer inspection of these hits (by searching against the Pfam database) reveals that the region identified by the PSI-BLAST search as homologous to the Vps51p query corresponds to the Vps51 domain (). Importantly, neither of these proteins appears to be an ortholog of Cog1 or Exo84 and the only characterized entry among them is a protein from the zebrafish Danio rerio (NP_001036200.1) encoded by the locus fat-free (ffr).
Figure 4. A multiple alignment of the Vps51 domain sequences of putative Vps51 orthologs. The Vps51 domain of representative sequences identified by PSI-BLAST and BLASTP searches with the yeast Vps51p (NP_012945) and the zebrafish Ffr (NP_001036200), respectively, as a query (see text for details), were aligned using the hmmalign program of the HMMER 2.3.2 package and the Pfam Vps51 (PF08700) profile alignment as a template. Sequence IDs correspond to the NCBI protein database, with the exception of the sequences from Batrachochytrium, Mucor, and Emiliania taken from the respective databases at JGI (http://genome.jgi-psf.org/Batde5/Batde5.home.html, http://genome.jgi-psf.org/Mucci2/Mucci2.home.html, http://genome.jgi-psf.org/Emihu1/Emihu1.home.html).
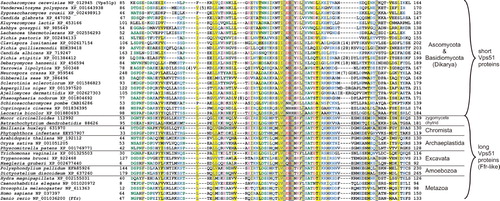
Experimental characterization of the zebrafish Ffr protein revealed that it is localized to the perinuclear region and TGN and is implicated in vesicle trafficking and protein sorting at the Golgi (Ho et al. Citation2006). The authors noted similarity of the Ffr protein to the COG complex subunit Cog8, which in fact contains a domain (Dor1) classified by Pfam in the same clan (CL0295) as the Vps51 domain. My BLASTP analyses show that the Ffr protein belongs to a family of orthologs readily identifiable in a wide array of eukaryotes, including chytrid and zygomycete fungi, and is characterized by a Vps51 domain close to the N-terminus followed by much longer region (600–700 residues) with good conservation across the whole breadth of eukaryotic diversity ( and Figure S1 in Supplementary data, available online). This Ffr-like family actually comprises the hits identified with the third PSI-BLAST iteration with the S. cerevisiae Vps51p as a query. The zebrafish Ffr also gives significant BLASTP matches (E-value of ≥1e-15) to proteins from ascomycete (except Saccharomycetales) and basidiomycete fungi (collectively called Dikarya), which are also among these identified by PSI-BLAST with the yeast Vps51p, but in this case the region of homology is restricted only to the Vps51 domain, since these fungal proteins (and also Vps51p homologs in Saccharomycetales) have only a very short (0–150 residues) and poorly conserved region C-terminal to the Vps51 domain. The only candidate Ffr homolog suggested by BLASTP in Saccharomycetales (E-value of 0.002) is a Yarrowia lipolytica protein also found in the PSI-BLAST search with Vps51p. When the regions of the Ffr protein downstream to the Vps51 domain is used as a query in PSI-BLAST searches, potential homology to the Exocyst subunit Sec5 and the COG complex subunit Cog8 is detected. In light of the functional characterization of the zebrafish Ffr and the results of sequence analyses described above, I suggest that the Ffr protein and its homologs in other eukaryotes are Vps51p orthologs most probably serving as subunits of the GARP complex in these species. Next, it seems that the Vps51 protein underwent a drastic modification in the lineage leading to Dikarya due to loss of the region C-terminal to the Vps51 domain. Third, even the sequence of the Vps51 domain itself has diverged in the Saccharomycetales lineage to an extent making recognition of the actual homology impossible with BLASTP. Ffr-like proteins outside dikaryan fungi thus represent the ‘prototypical’ eukaryotic form of the Vps51 subunit, more similar to the ancestral form of the protein.
Evolution of the ES at the layer of protein domain architecture
Proteins do not evolve only by accumulation of local mutations in corresponding genes, but also by shuffling larger portions of DNA sequences, typically coding for structurally and functionally independent protein domains (Basu et al. Citation2008, Moore et al. Citation2008, Chothia and Gough Citation2009). Proteins associated with the ES are no exception, although a systematic analysis of domain evolution in this protein cohort is wanting. One point that is quite clear is a very different propensity towards experimenting with the domain architecture exhibited by different protein classes. Indeed, some ES-associated proteins are build from multiple modules (domains) that combined before LECA and have remained in the same arrangement up to our time, for instance some components of coat complexes and the NPC represented by a conserved combination of an N-terminal β-propeller domain with a C-terminal α-solenoid domain (Devos et al. Citation2004, Field and Dacks Citation2009).
Even such ancient and seemingly rigid mergers can however occasionally show readiness to evolutionary change. SNARE proteins from the longin group have a conserved architecture with the longin domain (actually occurring in many other ES-associated proteins; Kinch and Grishin Citation2006) N-terminally fused to the SNARE core region (Rossi et al. Citation2004). Three longin paralogs (Ykt6, VAMP7 and Sec22) inherited from the LECA has retained this original domain architecture, but a new paralog (‘phytolongins’) emerged within the VAMP7 paralog in the land plant lineage by extreme divergence of the SNARE region or its replacement by another domain (; Vedovato et al. Citation2009). Two fused domains – an ATPase related to the dynamin GTPase superfamily and the EH domain – define the EHD/RME-1 family of proteins with an important role in the endocytic transport (Grant and Caplan Citation2008). Whereas metazoan EHD proteins exhibit the ATPase-EH domain order, a reverse domain order (EH-ATPase) was found in flowering plants (Bar et al. Citation2008). Extending the survey (M. Elias, unpublished work) reveals that the ATPase-EH type is widespread across diverse eukaryotic lineages, whereas the EH-ATPase type is exclusive for plants and green algae (Chloroplastida; ). The most likely evolutionary scenario is that that the ATPase-EH type is ancestral and the EH-ATPase type arose by a rearrangement of the domains in the stem lineage of Chloroplastida (this would then be one of the first known ES-associated features synapomorphic for the whole Chloroplastida group).
Figure 5. Domain architecture of the membrane-trafficking machinery. (A) Origin of the land plant-specific phytolongin proteins from a SNARE protein of the VAMP7 family via replacement or extreme sequence divergence of the SNARE region (TMD – trans-membrane domain). Adopted from Vedovato et al. (Citation2009). (B) Two different domain architectures of the EHD/RME-1 protein family. (C) Sequence features and domain organization of ‘standard’ RAB proteins and a unique RAB from the cryptomonad Guillardia theta. Note the two geranylgeranyl moieties attached to two cysteine residues occurring (in various arrangements) near the C-terminus of the standard RABs. The Guillardia RAB lacks these C-terminal cysteine residues and instead has an N-terminal extension with a pair of MSP domains. (D) The diversity of taxon-specific domain architectures of proteins serving as regulators of RAB and ARF GTPases. The cartoon shows examples of proteins with ArfGAP, SEC7 (= ArfGEF), TBC (= RabGAP), or VPS9 (= GEF for RAB5-related RABs) domains combined with any of an array of ‘promiscuous’ domains reoccurring in many other proteins. The domain architecture of the proteins shown was determined using SMART (http://smart.embl-heidelberg.de/), Pfam (http://pfam.sanger.ac.uk/search), and CDD (http://www.ncbi.nlm.nih.gov/cdd) search tools. Sequence IDs correspond to the NCBI protein database, with the exception of the protein from Emiliania huxleyi taken from the Emiliania JGI genome database (http://genome.jgi-psf.org/Emihu1/Emihu1.home.html).
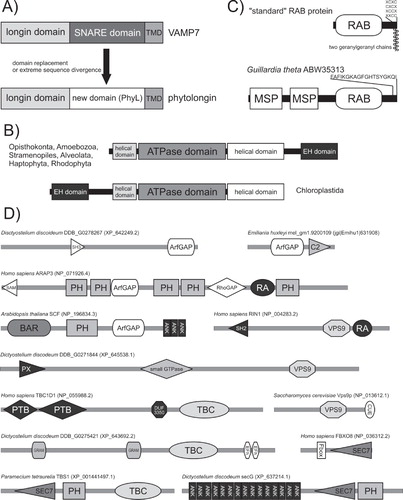
A frequent result of evolutionary domain shuffling is accretion of accessory domains, here exemplified with two cases from the RAB family. RAB proteins typically represent just a GTPase domain with an unstructured hypervariable C-terminal tail modified by one or two geranylgeranyl residues. However, occasionally this core has been decorated by extra domains, as in an unusual RAB of the cryptomonad Guillardia theta that seems to have orthologs in other chromists but differs from them by the absence of a C-terminal geranylgeranylation motif and the presence of a unique N-terminal projection including two MSP (major sperm protein) domains (; Elias et al. Citation2009). The MSP domain is known as a protein-protein interaction device (Tarr and Scott Citation2005). It is, therefore, conceivable that the pair of the MSP domains mediates an expected membrane association of the G. theta RAB via binding to a membrane-anchored protein(s), opportunistically providing an alternative solution for RAB membrane attachment compensating the missing C-terminal geranylgeranylation. Another notable case is the human RAB45 and related RABs so far found only in Metazoa (Shintani et al. Citation2007; M. Elias, unpublished work). Here the RAB domain represents just a C-terminal end of a larger protein including also one or two EF-hand domains near the N-terminus and a coiled-coil region in the middle. Given its distribution, this RAB subfamily very likely represents a metazoan-specific innovation that came into being through two consecutive processes – duplication and divergence of a RAB gene (the RAB part of the proteins seems to be a novel Metazoa-specific paralog) and fusion with a DNA segment encoding the extra N-terminal regions (whose ultimate origin remains obscure).
The RAB45 subfamily is also interesting in that it directly combines a module from the conserved core of the membrane-trafficking machinery (the RAB domain) with a module (EF-hand) from an extensive category of promiscuous domains re-occurring in diverse functional contexts as devices sensing inputs from cellular regulatory circuits and signalling pathways (Basu et al. Citation2008, Citation2009). Although no comprehensive analyses have yet been published, preliminary surveys (M. Elias, unpublished work) suggest that, in the context of the membrane-trafficking machinery, such domains most often occur in combinations with domains regulating the function of RAB and ARF GTPases, i.e., GEFs (GDP/GTP exchange factors such as the VPS9 and SEC7 domain) and GAPs (GTPase-activating proteins, i.e., TBC and ArfGAP domains). Moreover, it seems that evolution of these proteins may be very dynamic with a plethora of lineage-specific domain architectures. For instance, beside the universal and highly conserved ArfGEFs of the BIG/SEC7 and GBF/GEA subfamilies, other types of ArfGEF proteins combining the SEC7 domain with various other domains (PH, F-box, Ankyrin repeat) were found restricted to particular eukaryotic lineages (Cox et al. Citation2004; M. Elias, unpublished work). An ArfGEF subfamily (TBS) so far specific for ciliates is interesting in bringing together the SEC7 domain and a TBC (RabGAP) domain, physically demonstrating a cross-talk between ARF- and RAB-mediated regulation of membrane trafficking (Mouratou et al. Citation2005). shows a number of other proteins with the GTPase-regulating domains combined with other domains in an arrangement potentially representing evolutionary innovations of particular eukaryotic groups (pinpointing the origin of these architectures will need a careful analysis of a comprehensive set of eukaryotic genomes).
The omnipresent reductive evolution
Scattered phylogenetic distribution of homologous genes may be indicative of horizontal gene transfer (HGT), but such a pattern may be also generated by gene loss (Elias and Archibald Citation2009b), an important but perhaps underestimated evolutionary process. Reductive evolution is often being associated with parasitic organisms, but parasites can actually display hypertrophy, rather than reduction, of their cellular systems (see above the case of T. vaginalis and E. histolytica). On the other hand, there are a growing number of examples demonstrating the pervasiveness of gene loss in non-parasitic organisms. Loss of ancestral features of the eukaryotic ES has accompanied, for example, evolution of the fungal kingdom, particularly the yeast lineage including S. cerevisiae (); components of the ES secondarily missing in the budding yeast due to losses at different points of the fungal evolutionary history include, for example, the adaptin complex AP4, the BBSome coat complex, several paralogs of the RAB family, at least one ArfGEF and ArfGAP, several RabGAP proteins, the Golgi-associated ARL5 GTPase, the EHD/RME-1 family, or subunits of the BLOC-1, BLOC-2 and BLOC3 complexes implicated in biogenesis of lysosomes-related organelles (Boehm and Bonifacino Citation2001, Pereira-Leal Citation2008, Ma et al. Citation2009, Cheli and Dell'Angelica Citation2010, Hodges et al. Citation2010). Such reductions have also occurred throughout Metazoa, as exemplified, for example, by the absence of the AP4 adaptin complex and the endocytic protein Eps15 in C. elegans and D. melanogaster (Boehm and Bonifacino Citation2001, Field et al. Citation2007), by numerous losses of different SNARE proteins in many metazoan lineages (Kloepper et al. Citation2008), or by the loss of the Sec2 RabGEF from dipteran insects (Elias Citation2008). Interestingly, there is a paucity of reported ES-affecting secondary losses specific for the human lineage, suggesting that the human ES may be a quite ‘complete’ derivate of the archetypal eukaryotic ES. However, our own unpublished analyses revealed at least one ancestral RAB paralog secondarily missing from the human genome (M. Elias, J. B. Dacks, M. C. Field, unpublished work) and future investigations will undoubtedly uncover additional reductive events in the human lineage.
A notable aspect of reductive evolution is that the same event can happen recurrently, that is independently in different lineages, as exemplified by multiple independent losses of phagocytosis (Cavalier-Smith Citation2002). This pattern seems to hold true also for many individual proteins or protein complexes. For example, of the four paralogous adaptin complexes, AP1 is conserved in all eukaryotes investigated, while AP2, AP3, and AP4 complexes have been lost each independently in several eukaryotic lineages (Field et al. Citation2007, Dacks et al. Citation2008b); multiple independent losses of AP3 are evident even within the single group of Apicomplexa (Nevin and Dacks Citation2009). Recurrent loss was also noted for a component of the machinery mediating macroautophagy (Rigden et al. Citation2009), and it would be possible to cite many other examples (see also above the case of RAB24), but I add just one more. Atlastins form a group of GTPases of the dynamin superfamily recently shown to be involved in generating tubular ER network and homotypic fusion of ER membranes in metazoan cells (Hu et al. Citation2009, Orso et al. Citation2009, Park and Blackstone Citation2010). Atlastins were claimed to be restricted to metazoans while having functional orthologs represented in all other eukaryotes by a related yet apparently distinct group of dynamin-related GTPases of the Sey1/RHD3 family (Hu et al. Citation2009). Although no explicit evolutionary scenario was raised by the authors, it would be tempting to speculate that atlastins evolved through a radical sequence modifications in the metazoan stem lineage from the broadly occurring and hence probably ancestral Sey1/RHD3 family. However, this idea is very likely incorrect, since our recent investigation revealed that atlastin orthologs exist in at least one non-metazoan lineage – the distantly related stramenopiles, with at least one stramenopile (the brown alga E. siliculosus) possessing both an atlastin and a Sey1/RHD3 gene (Cock et al. Citation2010). Provided that atlastins were not exchanged between the metazoan and stramenopile lineages via HGT, it is likely that atlastins and Sey1/RHD3 are paralogs (rather than ‘functional orthologs’) that probably separated early in the evolutionary history of eukaryotes (potentially already in the LECA) and have been later selectively retained or lost by most species with the exceptions like Ectocarpus.
In contrast to the proteins experiencing recurrent losses through the eukaryotic phylogeny, there are categories of protein more recalcitrant to loss, probably because they are more tightly interwoven within the cellular fabric. In the very core of the ES-associated machinery there are proteins that even have probably never been lost, since such an event would be fatal. For example, the GTPase Sar1 regulating the assembly of the COP II coat complex in the course of vesicle budding at the ER can be found encoded by every single draft or complete eukaryotic genome sequence available (M. Elias, unpublished work). Based on published surveys of eukaryotic genomes (Dacks and Field Citation2004, Field et al. Citation2007, Kloepper et al. 207, Koumandou et al. Citation2007, Dacks et al. Citation2008b), other candidates for components of the essential core of the ES-associated machinery include, for example, some of the coat complexes (COP I, COP II, AP1/clathrin), some RABs and the ARF GTPase, the tethering complex TRAPP I, the four paralogs of the SM (Sec1-related) family and multiple SNARE proteins. However, the diversity of life exceeds our imagination and one can never be certain that an organism lacking either of these components is found one day. As a good example, take the GTPase SRβ, a eukaryote-specific membrane-anchored subunit of the signal recognition particle (SRP) receptor that arose early in eukaryotic evolution and recruits the soluble SRα subunit to the ER (Schwartz Citation2007). SRβ is readily identified in genomes of all sequenced eukaryotes, with the striking exception of Microsporidia (M. Elias, unpublished work), an extremely divergent and reduced parasitic group of the former ‘Archezoa’ actually representing highly derived fungi (Corradi and Keeling Citation2009). No microsporidian SRβ orthologs are discernible even with the use of sensitive PSI-BLAST searches, indicating that the absence may be genuine and raising the question as to how microsporidia have managed to modify their apparatus for protein import into the ER keeping it functional without the SRβ subunit.
Further perspectives
The evolutionary history of the endomembrane system is far richer than I could have expounded in this essay, having been limited by the lack of space, expertise, or sufficient information available. For example, a growing body of evidence points to the importance of HGT and endosymbiotic gene transfer (EGT) in eukaryotic evolution (Timmis et al. Citation2004, Lane and Archibald Citation2008, Andersson Citation2009, Keeling Citation2009), but apart a few anecdotal and inconclusive cases (e.g., a hypothetical HGT or EGT event to explain the presence of the plant-type vacuolar sorting receptor in stramenopiles and alveolates; Becker and Hoef-Emden Citation2009), the real impact of these processes on the ES is essentially unknown. Another neglected aspect of evolution of the ES is the role of convergence, a pervasive pattern of evolution in general (Conway Morris Citation2003). A form of evolutionary convergence is the recurrent loss of some of the ES-associated components discussed above. However, ‘positive’ convergence seems to affect the ES as well, as argued for the multiple independent origins of dense core granules (specialized secretory vesicles) in diverse eukaryotic lineages (Elde et al. Citation2007). Another example might be potentially independent recruitment of dynamin-superfamily proteins to clathrin-mediated endocytosis, originally thought to be unique for metazoans, but then found in ciliates (Elde et al. Citation2005) and plants (Fujimoto et al. Citation2010). Finally, the characteristic behaviour of the nuclear envelope during mitosis, i.e., its breakdown by vesiculation at prophase and reassembly at telophase by vesicle fusion (open mitosis), may not be the prototypical property of the eukaryotic cell, but a feature convergently evolved in several independent lineages (metazoans or streptophyte plants, among others) from the presumably ancestral state represented by closed mitosis (Cavalier-Smith Citation2010a). It would be extremely interesting to compare the molecular determinants of open mitosis in different lineages to see whether the morphological convergence transpires down to the molecular level.
Evolution is not only about mutations in DNA or changing allele frequencies within a population (Jablonka and Lamb Citation2005). This statement is especially pertinent to the evolution of the ES (Cavalier-Smith Citation2004), since the ES may have something like a DNA-independent heredity, i.e., irreducible information built-in to the spatial organization of membranes and proteins and maintained through a self-sustaining network of interactions. If this view is correct, it is then a question how much of the evolutionary change of the ES is triggered by physical reconfigurations of this network rather than by the ‘standard’ evolutionary mode of mutation in a gene encoding an ES-associated protein. This brings us to a more general question of evolutionary patterns and processes at the systems-biology level. Indeed, proteins function embedded in a network of interactions with other proteins, so evolution of any cellular system cannot be understood fully without touching questions such as: What are the evolutionary dynamics and phylogenetic pattern of physical interactions between components of the ES-associated machinery? How the whole system reacts (in evolutionary terms) on the appearance of a new component? What is the extent of correlation in loss of interacting components of the same complexes or modules? We essentially do not know yet, but the progress in genome sequencing and proteomics, captured by the emerging discipline of evolutionary systems biology (Koonin and Wolf Citation2006), brings hopes for answers coming soon.
Note
The homology of the Ffr-related proteins to the yeast Vps51p subunit of the GARP complex, suggested in this paper on the basis of bioinformatic analyses, has been very recently demonstrated experimentally, see Pérez-Victoria et al. Ang2/Fat-free is a Conserved Subunit of the Golgi-associated Retrograde Protein (GARP) Complex. Mol Biol Cell, 21:3386–3395. In addition, the very recently solved structure of the C-terminal fragment of the Vps53 subunit of the GARP complex (Vasan et al. Structure of a C-terminal fragment of its Vps53 subunit suggests similarity of Golgi-associated retrograde protein [GARP] complex to a family of tethering complexes. Proc Natl Acad Sci USA 107, 14176–14181) brings further support for the idea of a common evolutionary origin of the GARP, COG, Dsl1 and Exocyst complexes.
Acknowledgements
I would like to thank J. Malcolm East for inviting me to write this piece. I am indebted to Joel B. Dacks for recommending my participation in the thematic issue. J. B. Dacks, Fatima Cvrčková and two anonymous reviewers provided highly valuable comments on the manuscript. I apologise to all colleagues whose work on the evolution of the endomembrane system could not be cited due to space limitations. My work is supported by the P305/10/0205 grant from the Czech Science Fundation and by research project 21620828 of Czech Ministry of Education.
Declaration of interest : The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Abodeely M, DuBois KN, Hehl A, Stefanic S, Sajid M, DeSouza W, Attias M, Engel JC, Hsieh I, Fetter RD, McKerrow JH. 2009. A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryot Cell 8:1665–1676.
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52:399–451.
- Allen RD, Naitoh Y. 2002. Osmoregulation and contractile vacuoles of protozoa. Int Rev Cytol 215:351–394.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25:3389–402.
- Alvim Kamei CL, Boruc J, Vandepoele K, Van den Daele H, Maes S, Russinova E, Inzé D, De Veylder L. 2008. The PRA1 gene family in Arabidopsis. Plant Physiol 147:1735–1749.
- Andersson JO. 2009. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol 63:177–193.
- Baldari CT, Rosenbaum J. 2010. Intraflagellar transport: It's not just for cilia anymore. Curr Opin Cell Biol 22:75–80.
- Bar M, Aharon M, Benjamin S, Rotblat B, Horowitz M, Avni A. 2008. AtEHDs, novel Arabidopsis EH-domain-containing proteins involved in endocytosis. Plant J 55:1025–1038.
- Bassham DC, Brandizzi F, Otegui MS, Sanderfoot, AA. 2008. The secretory system of Arabidopsis. In: The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists. Available from the website: http://www.aspb.org/publications/arabidopsis/.
- Basu MK, Carmel L, Rogozin IB, Koonin EV. 2008. Evolution of protein domain promiscuity in eukaryotes. Genome Res 18:449–461.
- Basu MK, Poliakov E, Rogozin IB. 2009. Domain mobility in proteins: Functional and evolutionary implications. Brief Bioinform 10:205–216.
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. 2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes and stramenopiles. Mol Biol Evol 27:1698–1709.
- Becker B, Hoef-Emden K. 2009. Evolution of vacuolar targeting in algae. Bot Mar 52:117–128.
- Bernards A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta 1603:47–82.
- Beznoussenko GV, Dolgikh VV, Seliverstova EV, Semenov PB, Tokarev YS, Trucco A, Micaroni M, Di Giandomenico D, Auinger P, Senderskiy IV, Skarlato SO, Snigirevskaya ES, Komissarchik YY, Pavelka M, De Matteis MA, Luini A, Sokolova YY, Mironov AA. 2007. Analogs of the Golgi complex in microsporidia: Structure and avesicular mechanisms of function. J Cell Sci 120:1288–1298.
- Bodył A, Stiller JW, Mackiewicz P. 2009. Chromalveolate plastids: Direct descent or multiple endosymbioses? Trends Ecol Evol 24:119–121.
- Boehlke C, Bashkurov M, Buescher A, Krick T, John AK, Nitschke R, Walz G, Kuehn EW. 2010. Differential role of Rab proteins in ciliary trafficking: Rab23 regulates smoothened levels. J Cell Sci 123:1460–1467.
- Boehm M, Bonifacino JS. 2001. Adaptins: The final recount. Mol Biol Cell 12:2907–2920.
- Bonifacino JS, Glick BS. 2004. The mechanisms of vesicle budding and fusion. Cell 116:153–166.
- Brighouse A, Dacks JB, Field MC. 2010. Rab protein evolution and the history of the eukaryotic endomembrane system. Cell Mol Life Sci 67:3449–3465.
- Brinkmann H, Philippe H. 2007. The diversity of eukaryotes and the root of the eukaryotic tree. Adv Exp Med Biol 607:20–37.
- Brinkmann H, van der Giezen M, Zhou Y, De Raucourt GP, Philippe H. 2005. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol 54:743–757.
- Burki F, Inagaki Y, Bråte J, Archibald JM, Keeling PJ, Cavalier-Smith T, Sakaguchi M, Hashimoto T, Horak A, Kumar S, Klaveness D, Jakobsen KS, Pawlowski J, Shalchian-Tabrizi K. 2009. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol 2009:231–238.
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One 2:e790.
- Burki F, Shalchian-Tabrizi K, Pawlowski J. 2008. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett 4:366–369.
- Cai H, Reinisch K, Ferro-Novick S. 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12:671–682.
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Müller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M, Okumura CY, Schneider R, Smith AJ, Vanacova S, Villalvazo M, Haas BJ, Pertea M, Feldblyum TV, Utterback TR, Shu CL, Osoegawa K, de Jong PJ, Hrdy I, Horvathova L, Zubacova Z, Dolezal P, Malik SB, Logsdon JM Jr, Henze K, Gupta A, Wang CC, Dunne RL, Upcroft JA, Upcroft P, White O, Salzberg SL, Tang P, Chiu CH, Lee YS, Embley TM, Coombs GH, Mottram JC, Tachezy J, Fraser-Liggett CM, Johnson PJ. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212.
- Cavalier-Smith T. 1993. Kingdom protozoa and its 18 phyla. Microbiol Rev 57:953–994.
- Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52:297–354.
- Cavalier-Smith T. 2004. Membranome and membrane heredity in development and evolution. In: Hirt RP, Horner DS, editors. Organelles, genomes and eukaryote phylogeny. An evolutionary synthesis in the age of genomics. Boca Raton, FL: The Systematic Association, CRC Press. pp 335–351.
- Cavalier-Smith T. 2010a. Origin of the cell nucleus, mitosis and sex: Roles of intracellular coevolution. Biol Direct 5:7.
- Cavalier-Smith T. 2010b. Kingdoms protozoa and chromista and the eozoan root of the eukaryotic tree. Biol Lett 6:342–345.
- Cheli VT, Dell'Angelica EC. 2010. Early origin of genes encoding subunits of biogenesis of lysosome-related organelles complex-1, -2 and -3. Traffic 11:579–586.
- Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, Goring DR. 2010. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol 185:401–419.
- Chothia C, Gough J. 2009. Genomic and structural aspects of protein evolution. Biochem J 419:15–28.
- Chow CM, Neto H, Foucart C, Moore I. 2008. Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20:101–123.
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GJ, Coelho SM, Collén J, Corre E, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CMM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Bail AL, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collén P, Peters AF, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder D, Ségurens B, Strittmatter M, Tonon T, Tregear J, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y and Wincker P. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465:617–621.
- Conibear E, Stevens TH. 2000. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell 11:305–323.
- Conway Morris S. 2003. Life's solution. Inevitable humans in a lonely universe. New York: Cambridge University Press.
- Corradi N, Keeling PJ. 2009. Microsporidia: A journey through radical taxonomical revisions. Fung Biol Rev 23:1–8.
- Cox R, Mason-Gamer RJ, Jackson CL, Segev N. 2004. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell 15:1487–1505.
- Croteau NJ, Furgason ML, Devos D, Munson M. 2009. Conservation of helical bundle structure between the exocyst subunits. PLoS One 4:e4443.
- Dacks JB, Field MC. 2004. Eukaryotic cell evolution from a comparative genomic perspective: The endomembrane system. In: Hirt RP, Horner DS, editors. Organelles, genomes and eukaryote phylogeny. An evolutionary synthesis in the age of genomics. Boca Raton, FL: The Systematic Association, CRC Press. pp 309–334.
- Dacks JB, Field MC. 2007. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J Cell Sci 120:2977–2985.
- Dacks JB, Peden AA, Field MC. 2009. Evolution of specificity in the eukaryotic endomembrane system. Int J Biochem Cell Biol 41:330–340.
- Dacks JB, Walker G, Field MC. 2008a. Implications of the new eukaryotic systematics for parasitologists. Parasitol Int 57:97–104.
- Dacks JB, Poon PP, Field MC. 2008b. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci USA 105:588–593.
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8:2119–2130.
- Dejgaard SY, Murshid A, Erman A, Kizilay O, Verbich D, Lodge R, Dejgaard K, Ly-Hartig TB, Pepperkok R, Simpson JC, Presley JF. 2008. Rab18 and Rab43 have key roles in ER-Golgi trafficking. J Cell Sci 121:2768–2781.
- Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6:361–375.
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2:e380.
- Docampo R, Ulrich P, Moreno SN. 2010. Evolution of acidocalcisomes and their role in polyphosphate storage and osmoregulation in eukaryotic microbes. Philos Trans R Soc Lond B Biol Sci 365:775–784.
- Dolinski K, Botstein D. 2007. Orthology and functional conservation in eukaryotes. Annu Rev Genet 41:465–507.
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763.
- Elde NC, Long M, Turkewitz AP. 2007. A role for convergent evolution in the secretory life of cells. Trends Cell Biol 17:157–164.
- Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. 2005. Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet 1:e52.
- Elias M, Archibald JM. 2009a. Sizing up the genomic footprint of endosymbiosis. Bioessays 31:1273–1279.
- Elias M, Archibald JM. 2009b. The RJL family of small GTPases is an ancient eukaryotic invention probably functionally associated with the flagellar apparatus. Gene 442:63–72.
- Elias M, Patron NJ, Keeling PJ. 2009. The RAB family GTPase Rab1A from Plasmodium falciparum defines a unique paralog shared by chromalveolates and rhizaria. J Eukaryot Microbiol 56:348–356.
- Elias M. 2008. The guanine nucleotide exchange factors Sec2 and PRONE: Candidate synapomorphies for the Opisthokonta and the Archaeplastida. Mol Biol Evol 25:1526–1529.
- Engstler M, Bangs JD, Field MC. 2007. Intracellular transport systems in trypanosomes: Function, evolution, and virulence. In: Barry JD, Mottram J, McCulloch R, Acosta-Serrano A, editors. Trypanosomes – after the genome. Norfolk: Horizon Bioscience. pp 281–318.
- Field MC, Dacks JB. 2009. First and last ancestors: Reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol 21:4–13.
- Field MC, Gabernet-Castello C, Dacks JB. 2007. Reconstructing the evolution of the endocytic system: Insights from genomics and molecular cell biology. Adv Exp Med Biol 607:84–96.
- Field MC, O'Reilly AJ. 2008. How complex is GTPase signaling in trypanosomes? Trends Parasitol 24:253–257.
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res 38:D211–222.
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642.
- Fuerst JA. 2005. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol 59:299–328.
- Fujimoto M, Arimura S, Ueda T, Takanashi H, Hayashi Y, Nakano A, Tsutsumi N. 2010. Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc Natl Acad Sci USA 107:6094–6099.
- Gabaldón T. 2010. Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci 365:765–773.
- Glücksman E, Snell EA, Berney C, Bass D, Cavalier-Smith T. 2010. The novel marine gliding zooflagellate genus Mantamonas (Mantamonadida ord.n.:Apusozoa). Available from the website: http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B7GX3-514G4J6-1&_user=1490772&_coverDate=09%2F29%2F2010&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000053052&_version=1&_urlVersion=0&_userid=1490772&md5=8e77ac1a369a23ae7f2a40698b1caeae&searchtype=a. doi:10.1016/j.protis.2010.06.004.
- Goldstein RA. 2008. The structure of protein evolution and the evolution of protein structure. Curr Opin Struct Biol 18:170–177.
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. 2008. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol 25:1219–1230.
- Grant BD, Caplan S. 2008. Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic 9:2043–2052.
- Gurkan C, Koulov AV, Balch WE. 2007. An evolutionary perspective on eukaryotic membrane trafficking. Adv Exp Med Biol 607:73–83.
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141:656–667.
- Hála M, Eliás M, Zárský V. 2005. A specific feature of the angiosperm Rab escort protein (REP) and evolution of the REP/GDI superfamily. J Mol Biol 348:1299–1313.
- Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc Natl Acad Sci USA 106:3859–3864.
- Hampl V, Simpson AGB. 2008. Possible mitochondria-related organelles in poorly-studied ‘amitochondriate’ eukaryotes. In: Tachezy J, editor. Hydrogenosomes and mitosomes: Mitochondria of anaerobic eukaryotes. Microbiology Monographs 9. Berlin, Heidelberg: Springer Verlag. pp 265–282.
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. 2010. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci 365:713–727.
- Ho SY, Lorent K, Pack M, Farber SA. 2006. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab 3:289–300.
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. 2010. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123:1407–1413.
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138:549–561.
- Hughes T, Rusten TE. 2007. Origin and evolution of self-consumption: Autophagy. Adv Exp Med Biol 607:111–118.
- Jablonka E, Lamb MJ. 2005. Evolution in four dimensions: Genetic, epigenetic, behavioral, and symbolic variation in the history of life. Cambridge: The MIT Press.
- Jékely G. 2007. Origin of eukaryotic endomembranes: A critical evaluation of different model scenarios. Adv Exp Med Biol 607:38–51.
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141:1208–1219.
- Jürgens G. 2005. Plant cytokinesis: Fission by fusion. Trends Cell Biol 15:277–283.
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. 2005. The tree of eukaryotes. Trends Ecol Evol 20:670–676.
- Keeling PJ. 2008. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol 56:1–8.
- Keeling PJ. 2009. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev 19:613–619.
- Kiel JA. 2010. Autophagy in unicellular eukaryotes. Philos Trans R Soc Lond B Biol Sci 365:819–830.
- Kienle N, Kloepper TH, Fasshauer D. 2009. Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol 9:19.
- Kinch LN, Grishin NV. 2006. Longin-like folds identified in CHiPS and DUF254 proteins: Vesicle trafficking complexes conserved in eukaryotic evolution. Protein Sci 15:2669–2674.
- Kitajima A, Asatsuma S, Okada H, Hamada Y, Kaneko K, Nanjo Y, Kawagoe Y, Toyooka K, Matsuoka K, Takeuchi M, Nakano A, Mitsui T. 2009. The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21:2844–2858.
- Kloepper TH, Kienle CN, Fasshauer D. 2007. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell 18:3463–3471.
- Kloepper TH, Kienle CN, Fasshauer D. 2008. SNAREing the basis of multicellularity: Consequences of protein family expansion during evolution. Mol Biol Evol 25:2055–2068.
- Koonin EV, Wolf YI. 2006. Evolutionary systems biology: Links between gene evolution and function. Curr Opin Biotechnol 17:481–487.
- Koonin EV. 2009. Darwinian evolution in the light of genomics. Nucleic Acids Res 37:1011–1034.
- Koonin EV. 2010a. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11:209.
- Koonin EV. 2010b. Preview. The incredible expanding ancestor of eukaryotes. Cell 140:606–608.
- Koumandou VL, Dacks JB, Coulson RM, Field MC. 2007. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 7:29.
- Lane CE, Archibald JM. 2008. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol 23:268–275.
- Leung KF, Dacks JB, Field MC. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716.
- Liang H, Fernandez A. 2008. Evolutionary constraints imposed by gene dosage balance. Front Biosci 13:4373–4378.
- Liewen H, Meinhold-Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, Jung M, Pfreundschuh M, Stenner-Liewen F. 2005. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res 306:24–34.
- Lobkovsky AE, Wolf YI, Koonin EV. 2010. Universal distribution of protein evolution rates as a consequence of protein folding physics. Proc Natl Acad Sci USA 107:2983–2988.
- Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA 107:12883–12888.
- Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, Elias M, Idnurm A, Lang BF, Sone T, Abe A, Calvo SE, Corrochano LM, Engels R, Fu J, Hansberg W, Kim JM, Kodira CD, Koehrsen MJ, Liu B, Miranda-Saavedra D, O'Leary S, Ortiz-Castellanos L, Poulter R, Rodriguez-Romero J, Ruiz-Herrera J, Shen YQ, Zeng Q, Galagan J, Birren BW, Cuomo CA, Wickes BL. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet 5:e1000549.
- Mackiewicz P, Wyroba E. 2009. Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol Biol 9:101.
- Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. 2005. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res 33:4626–4638.
- Maritzen T, Podufall J, Haucke V. 2010. Stonins – specialized adaptors for synaptic vesicle recycling and beyond? Traffic 11:8–15.
- Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: Effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem 280:42325–42335.
- Mironov AA, Banin VV, Sesorova IS, Dolgikh VV, Luini A, Beznoussenko GV. 2007. Evolution of the endoplasmic reticulum and the Golgi complex. Adv Exp Med Biol 607:61–72.
- Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. 2008. Arrangements in the modular evolution of proteins. Trends Biochem Sci 33:444–451.
- Moreno SN, Docampo R. 2009. The role of acidocalcisomes in parasitic protists. J Eukaryot Microbiol 56:208–213.
- Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jürgens G, Melançon P, Cherfils J. 2005. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics 6:20.
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. 2009. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726.
- Mowbrey K, Dacks JB. 2009. Evolution and diversity of the Golgi body. FEBS Lett 583:3738–3745.
- Munafó DB, Colombo MI. 2002. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 3:472–482.
- Nakada-Tsukui K, Saito-Nakano Y, Husain A, Nozaki T. 2010. Conservation and function of Rab small GTPases in Entamoeba: Annotation of E. invadens Rab and its use for the understanding of Entamoeba biology. Exp Parasitol 126:337–347.
- Nanjo Y, Oka H, Ikarashi N, Kaneko K, Kitajima A, Mitsui T, Muñoz FJ, Rodríguez-López M, Baroja-Fernández E, Pozueta-Romero J. 2006. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell 18:2582–2592.
- Nassoury N, Cappadocia M, Morse D. 2003. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci 116:2867–2874.
- Nevin WD, Dacks JB. 2009. Repeated secondary loss of adaptin complex genes in the Apicomplexa. Parasitol Int 58:86–94.
- Nozaki H, Maruyama S, Matsuzaki M, Nakada T, Kato S, Misawa K. 2009. Phylogenetic positions of Glaucophyta, green plants (Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol 53:872–880.
- Ohno S. 1970. Evolution by gene duplication. New York: Springer-Verlag.
- Oka T, Krieger M. 2005. Multi-component protein complexes and Golgi membrane trafficking. J Biochem 137:109–114.
- Okamoto N, Chantangsi C, Horák A, Leander BS, Keeling PJ. 2009. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS One 4:e7080.
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, Daga A. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460:978–983.
- Papp B, Pál C, Hurst LD. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194–197.
- Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. 2006. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet 2:e220.
- Parfrey LW, Grant JR, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 59:518–533.
- Park SH, Blackstone C. 2010. Further assembly required: Construction and dynamics of the endoplasmic reticulum network. EMBO Rep 11:515–521.
- Patterson DJ. 1999. The diversity of eukaryotes. Am Nat 154:S96–124.
- Pei J, Ma C, Rizo J, Grishin NV. 2009. Remote homology between Munc13 MUN domain and vesicle tethering complexes. J Mol Biol 391:509–517.
- Pereira-Leal JB, Seabra MC. 2001. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313:889–901.
- Pereira-Leal JB. 2008. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 9:27–38.
- Pérez-Victoria FJ, Bonifacino JS. 2009. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol 29:5251–5263.
- Pérez-Victoria FJ, Mardones GA, Bonifacino JS. 2008. Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell 19:2350–2362.
- Pfeffer SR. 2007. Unsolved mysteries in membrane traffic. Annu Rev Biochem 76:629–645.
- Philippe H. 2000. Opinion: Long branch attraction and protist phylogeny. Protist 151:307–316.
- Plattner H. 2010. Membrane trafficking in protozoa: SNARE proteins, H+-ATPase, actin, and other key players in ciliates. Int Rev Cell Mol Biol 280:79–184.
- Povolotskaya IS, Kondrashov FA. 2010. Sequence space and the ongoing expansion of the protein universe. Nature 465:922–926.
- Richards TA, Cavalier-Smith T. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–1118.
- Richardson BC, Smith RD, Ungar D, Nakamura A, Jeffrey PD, Lupashin VV, Hughson FM. 2009. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA 106:13329–13334.
- Rigden DJ, Michels PA, Ginger ML. 2009. Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 5:784–794.
- Rodríguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. 2007. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Curr Biol 17:1420–1425.
- Roger AJ, Simpson AG. 2009. Evolution: Revisiting the root of the eukaryote tree. Curr Biol 19:R165–167.
- Rogozin IB, Basu MK, Csürös M, Koonin EV. 2009. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol 2009:99–113.
- Rohatgi R, Snell WJ. 2010. The ciliary membrane. Curr Opin Cell Biol 22:541–546.
- Rosati G, Modeo L. 2003. Extrusomes in ciliates: Diversification, distribution, and phylogenetic implications. J Eukaryot Microbiol 50:383–402.
- Rossi V, Banfield DK, Vacca M, Dietrich LE, Ungermann C, D'Esposito M, Galli T, Filippini F. 2004. Longins and their longin domains: Regulated SNAREs and multifunctional SNARE regulators. Trends Biochem Sci 29:682–688.
- Sacher M, Kim YG, Lavie A, Oh BH, Segev N. 2008. The TRAPP complex: Insights into its architecture and function. Traffic 9:2032–2042.
- Sanderfoot A. 2007. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol 144:6–17.
- Schwartz TU. 2007. Origins and evolution of cotranslational transport to the ER. Adv Exp Med Biol 607:52–60.
- Shintani M, Tada M, Kobayashi T, Kajiho H, Kontani K, Katada T. 2007. Characterization of Rab45/RASEF containing EF-hand domain and a coiled-coil motif as a self-associating GTPase. Biochem Biophys Res Commun 357:661–667.
- Simpson AG, Roger AJ. 2004. The real ‘kingdoms’ of eukaryotes. Curr Biol 14:R693–696.
- Simpson AG. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int J Syst Evol Microbiol 53:1759–1777.
- Sláviková S, Vacula R, Fang Z, Ehara T, Osafune T, Schwartzbach SD. 2005. Homologous and heterologous reconstitution of Golgi to chloroplast transport and protein import into the complex chloroplasts of Euglena. J Cell Sci 118:1651–1661.
- Sleator RD. 2010. An overview of the processes shaping protein evolution. Sci Prog 93:1–6.
- Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–248.
- Sogin ML, Morrison HG, Hinkle G, Silberman JD. 1996. Ancestral relationships of the major eukaryotic lineages. Microbiologia 12:17–28.
- Stechmann A, Cavalier-Smith T. 2003. The root of the eukaryote tree pinpointed. Curr Biol 13:R665–666.
- Stefanic S, Morf L, Kulangara C, Regös A, Sonda S, Schraner E, Spycher C, Wild P, Hehl AB. 2009. Neogenesis and maturation of transient Golgi-like cisternae in a simple eukaryote. J Cell Sci 122:2846–2856.
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10:513–525.
- Synek L, Schlager N, Eliás M, Quentin M, Hauser MT, Zárský V. 2006. AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J 48:54–72.
- Tabak HF, van der Zand A, Braakman I. 2008. Peroxisomes: Minted by the ER. Curr Opin Cell Biol 20:393–400.
- Tarr DE, Scott AL. 2005. MSP domain proteins. Trends Parasitol 21:224–231.
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5:123–135.
- Tonkin CJ, Kalanon M, McFadden GI. 2008. Protein targeting to the malaria parasite plastid. Traffic 9:166–175.
- Tripathi A, Ren Y, Jeffrey PD, Hughson FM. 2009. Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat Struct Mol Biol 16:114–123.
- Vedovato M, Rossi V, Dacks JB, Filippini F. 2009. Comparative analysis of plant genomes allows the definition of the ‘Phytolongins’: A novel non-SNARE longin domain protein family. BMC Genomics 10:510.
- Vernoud V, Horton AC, Yang Z, Nielsen E. 2003. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131:1191–1208.
- Wang Y, Ng EL, Tang BL. 2006. Rab23: What exactly does it traffic? Traffic 7:746–750.
- Whyte JR, Munro S. 2002. Vesicle tethering complexes in membrane traffic. J Cell Sci 115:2627–2637.
- Yabuki A, Inagaki Y, Ishida KI. 2010. Palpitomonas bilix gen. et sp. nov.: A novel deep-branching heterotroph possibly related to Archaeplastida or Hacrobia. Protist 161:523–538.
- Yoon HS, Grant J, Tekle YI, Wu M, Chaon BC, Cole JC, Logsdon JM Jr, Patterson DJ, Bhattacharya D, Katz LA. 2008. Broadly sampled multigene trees of eukaryotes. BMC Evol Biol 8:14.
- Zárský V, Cvrcková F, Bischoff F, Palme K. 1997. At-GDI1 from Arabidopsis thaliana encodes a rab-specific GDP dissociation inhibitor that complements the sec19 mutation of Saccharomyces cerevisiae. FEBS Lett 403:303–308.
- Zeldovich KB, Shakhnovich EI. 2008. Understanding protein evolution: From protein physics to Darwinian selection. Annu Rev Phys Chem 59:105–127.
- Zhou X, Lin Z, Ma H. 2010. Phylogenetic detection of numerous gene duplications shared by animals, fungi and plants. Genome Biol 11:R38.