Abstract
CitS of Klebsiella pneumoniae and GltS of Escherichia coli are Na+-dependent secondary transporters from different families that are believed to share the same fold and quaternary structure. A 10 kDa protein tag (Biotin Acceptor Domain [BAD]) was fused to the N-terminus of both proteins (CitS-BAD1 and GltS-BAD1, respectively) and inserted in the central cytoplasmic loop that connects the two halves of the proteins (CitS-BAD260 and GltS-BAD206). Both CitS constructs and GltS-BAD206 were produced and shown to be active transporters, but GltS-BAD1 could not be detected in the membrane. Distance relationships in the complexes were studied by cross-linking studies. Both CitS constructs were shown to be in the dimeric state after purification in detergent by cross-linking with glutaraldehyde. The concentration of glutaraldehyde resulting in 50% cross-linking was significantly higher for CitS-BAD1 than for CitS and CitS-BAD260. Remarkably, GltS and GltS-BAD260 were not cross-linked by glutaraldehyde because of the lack of productive reactive sites. Cross-linking of GltS was observed when the N-terminal 46 residues of CitS with or without BAD at the N-terminus were added to the N-terminus of GltS. The stretch of 46 residues contains the first transmembrane segment of CitS that is missing in the GltS structure. The data support an orientation of the monomers in the dimer with the N-termini close to the dimer interface and the central cytoplasmic loops far away at the ends of the long axis of the dimer structure in a view perpendicular to the membrane.
| Abbreviations: | ||
| TMS, | = | transmembrane segment |
| BAD, | = | biotin acceptor domain |
| NEM, | = | N-ethylmaleimide |
| DDM, | = | β-dodecyl maltoside |
| RSO, | = | right-side-out |
| PMS, | = | phenazine methosulfate |
Introduction
CitS of Klebsiella pneumoniae and GltS of Escherichia coli are secondary transporters catalyzing Na+ symport that belong to different families. The Na+-citrate transporter CitS belongs to the 2-hydroxycarboxylate transporter (2HCT; TC 2.A.24 [Saier Citation2000]) family (Sobczak and Lolkema Citation2005) while the Na+-glutamate transporter GltS belongs to the Glutamate Sodium Symporter (ESS; TC 2.A.27) family. Members of the 2HCT and ESS families are found exclusively in bacteria. No sequence homology can be detected between members from the two families, but both families are found in the same structural class ST[3] of the MemGen classification (Lolkema and Slotboom Citation1998, Citation2003, Citation2008) implying that the members are structurally homologous, i.e., they share the same core structure. MemGen class ST[3] contains 32 families of secondary transporters and includes the Ion Transporter (IT) superfamily (Prakash et al. Citation2003). A high resolution 3D structure is not available for any member of class ST[3].
Insight into the structure and function of class ST[3] proteins comes in addition to sequence analysis mainly from studies of CitS and GltS. The structural model of the transporters () shows a core of two homologous domains consisting of five transmembrane segments (TMS) each that are connected by a large cytoplasmic loop region (Van Geest and Lolkema Citation1999, Citation2000, Lolkema et al. Citation2005, Dobrowolski et al. Citation2007). The CitS protein and all members of the 2HCT family have an additional TMS at the N-terminal end of the core structure, placing the N-terminus in the cytoplasm. Members of the ESS family including GltS do not have this additional segment and their structure corresponds to the core structure that has the N-terminus in the periplasm. Other families in class ST[3] are characterized by additional TMSs at the C-terminal end. Because of the odd number of helices in the two domains that form the core structure, they have opposite orientations in the membrane (‘inverted topology’), a structural motif that is frequently observed in membrane proteins. In between the 4th and 5th TMS in each domain, the connecting loop folds back in between the TMSs to form a so-called ‘pore loop’ or ‘reentrant loop’ (Sobczak and Lolkema Citation2004, Dobrowolski and Lolkema Citation2009). The reentrant loop in the N-terminal domain enters the membrane from the periplasmic side, the one in the C-terminal domain from the cytoplasm. Sequence motifs GGXG present in the transporters of both the 2HCT and ESS families are at the vertex of the reentrant loops and were demonstrated to be crucial for the activity of the CitS and GltS proteins (Dobrowolski and Lolkema Citation2009). Recently, evidence was presented suggesting that in the 3D structure, the pore-loops of the two domains are in close vicinity and overlapping at the interface of the two domains (Dobrowolski and Lolkema 2010). The two domains would fold upon each other as the two halves of a clam shell with the central cytoplasmic loop forming the hinge at the closed side and the N- and C-termini at the open side. It is believed that the two pore loops form the translocation pore and that translocation proceeds through an alternate access mechanism (Sobczak and Lolkema Citation2003, Citation2004), involving movement of the two domains relative to one another (Dobrowolski and Lolkema 2010a).
Figure 1. Structural model of class ST[3] transporters. Cylinders represent transmembrane segments. The homologous N and C domains were indicated in dashed boxes. VB and XA represent reentrant loops. Arrows point at the insertion/fusion sites of the BAD domain in the indicated hybrid proteins. Shown is the model for CitS of K. pneumoniae. GltS of E. coli lacks the first transmembrane segment. The N terminus of CitS is in the cytoplasm.
![Figure 1. Structural model of class ST[3] transporters. Cylinders represent transmembrane segments. The homologous N and C domains were indicated in dashed boxes. VB and XA represent reentrant loops. Arrows point at the insertion/fusion sites of the BAD domain in the indicated hybrid proteins. Shown is the model for CitS of K. pneumoniae. GltS of E. coli lacks the first transmembrane segment. The N terminus of CitS is in the cytoplasm.](/cms/asset/f745a21b-530a-4df5-86a6-19c108805d08/imbc_a_581252_f0001_b.jpg)
In addition to a similar 3D structure, the CitS and GltS proteins also share the same quaternary structure. Several techniques were used to demonstrate that the proteins are dimeric in the detergent solubilized state (Pos et al. Citation1994, Veenhoff et al. Citation2001, Kästner et al. Citation2003, Mościcka et al. Citation2009). Single-particle electron microscopy yielded projection maps of the dimers showing in top view biscuit-shaped particles of dimension 160 × 84 Å and 145 × 84 Å for CitS and GltS, respectively. A side view projection of CitS revealed asymmetry in the shape of the protein relative to the plane of the membrane (Mościcka et al. Citation2009). In this study, proximity relations of the two monomers in the dimeric structure was studied by site-specific tagging of the monomers with the 10 kDa Biotin Acceptor Domain (BAD) of the oxaloacetate decarboxylase of Klebsiella pneumoniae (Bott Citation1997) followed by cross-linking with glutaraldehyde. The approach takes advantage of the observation that the GltS monomers could not be cross-linked with glutaraldehyde in the dimeric complex. The results place the cytoplasmic loop that connects the two domains of the monomers at the end of the long axis of the dimer structure, while the N-terminus is in the neighborhood of the dimer interface.
Methods
Materials
Ampicilin was obtained from Roche Diagnostic GmbH, Mannheim, Germany, arabinose from Sigma-Aldrich GmbH, Steinheim, Germany, and [1,5-14C]-citrate and L-[14C]-glutamate from Amersham Pharmacia, Roosendaal, The Netherlands.
Growth conditions and membrane preparation
CitS and GltS transporters were expressed in Escherichia coli strain DH5α under control of the arabinose promoter using pBAD24 (Invitrogen) derived plasmids. Freshly transformed bacteria were used to inoculate an overnight preculture. Twenty-five ml of preculture was added to 1 l of Lysogeny broth (LB) medium containing 50 μg/ml ampicilin in a 5-l flask at 37°C that was under continuous shaking at 200 rpm. At an OD660 of 0.6, 0.1% arabinose (Sigma-Aldrich GmbH, Steinheim, Germany) was added to induce expression of the transporters after which the culture was allowed to grow for an additional hour. Cells were harvested by spinning at 8000 rpm at 4°C for 10 min, washed with 50 ml 50 mM KPi, pH 7 at 4°C and resuspended in the same buffer. Cells from a 1-l culture resuspended in 5 ml of buffer containing 1 μg/ml DNAse were passed three times through a pressure-cell-disrupter at 13.5 MPa at 4°C. Following a low spin at 8,000 rpm for 10 min at 4°C to remove debris and unbroken cells, the supernatant was centrifuged at 80,000 rpm for 25 min at 4°C (high spin). The membrane fraction was resuspended and washed once with 50 mM KPi pH 7 buffer containing 1 M NaCl. The membranes were resuspended in 1 ml of 50 mM KPi pH 7 buffer and stored in liquid N2 until use.
Construction of plasmids
All genetic manipulations were done in E. coli DH5α using standard techniques. Plasmids pBADCitS and pSB260 were described before (Van Geest and Lolkema Citation1996, Sobczak and Lolkema Citation2003). pBADCitS encodes the CitS protein extended with a N-terminal His6-tag and an enterokinase cleavage site. pSB260 is a pBlueScript II (Stratagene) derivative containing the citS gene with the gene encoding BAD inserted in a NcoI restriction site at a position corresponding to amino acid residue 260 in the CitS sequence. The BAD encoding sequence was excised from pSB260 and ligated into the unique NcoI site of pBADCitS situated in between the enterokinase site and CitS encoding sequences yielding plasmid pCitS-BAD1. Plasmid pCitS-BAD260 was obtained by replacing a Bpu1102I-XbaI fragment of pBADCitS with the corresponding fragment including the BAD encoding sequence of pSB260.
Plasmids pBADGlts and pN356C encoding the GltS protein and GltS N356C mutant, respectively, both extended with a N-terminal His6-tag, were described before (Dobrowolski et al. Citation2007, Dobrowolski and Lolkema Citation2009). Plasmid pGltS-BAD1 was obtained by ligation of the BAD encoding fragment with NcoI overhangs into the NcoI site of pN356C situated in between the His-tag and GltS encoding sequences. Plasmid pGltS-BAD206 was obtained in a similar way after introduction of a unique NcoI cleavage site at the position of the 206th amino acid in GltS. Plasmid pTMSI-GltS was obtain by introducing an additional NcoI site at the position of the 46th amino acid in the citS gene, cutting the NcoI fragment from the plasmid and ligating the fragment into the NcoI site of pN356C. The second NcoI site was mutated to a NdeI site to introduce a TEV cleavage site in the encoded protein by ligating two hybridized complementary synthetic primers encoding the TEV site and with NdeI overhangs. Plasmid pBAD-TMSI-GltS was obtained by insertion of the BAD encoding sequence into the remaining NcoI site of pTMSI-GltS. The GltS derivatives containing the TEV protease sites could not be cleaved with the TEV enzyme. This feature was not further used in this study.
Plasmid pBAD was produced by introduction of a NcoI site just in front of the stop codon in pGltS-BAD1 followed by restriction with NcoI and relegation. pBAD encodes the BAD protein extended with a His-tag at the N-terminus.
Purification of CitS and GltS derivatives
CitS and GltS derivatives were purified to homogeneity as described before (Mościcka et al. Citation2009). Briefly, transporter proteins were solubilized from the membranes by partial extraction. Membranes from a 1-l culture incubated for 1 h at 4°C and under continuous shaking in 50 mM KPi pH 7 buffer containing 400 mM NaCl, 10% glycerol and 0.5% β-dodecyl maltoside (DDM). Undissolved membrane material which contained the transporter proteins was recovered by spinning at 80,000 rpm for 25 min at 4°C. The extraction procedure was repeated with the same buffer containing 1% Triton X-100 instead of DDM. The transporter proteins were recovered from the supernatant after spinning. The supernatant was passed through a 0.2 μm filter and applied to a 1 ml bed volume Ni-NTA column (HisTrap HP) in an ÄKTA FPLC system. Before, the column was conditioned for 5 min at 1 ml/min flow rate of the carrier buffer (50 mM KPi pH 8, 600 mM NaCl, 10% glycerol, 0.03% DDM, 20 mM imidazole). The loaded column was washed with 10 ml of 6% of the elution buffer (50 mM KPi pH 7, 600 mM NaCl, 10% glycerol, 0.03% DDM, 500 mM imidazole). The proteins were eluted in a linear gradient of 6–80% of the elution buffer. The OD280 of the eluate was measured continuously, after which the eluate was collected in fractions of 0.5 ml.
Expression levels of CitS and GltS derivatives in right-side-out (RSO) membrane vesicles used in the transport assays were measured by a batch wise partial purification procedure (Dobrowolski and Lolkema Citation2009). RSO membranes were solubilized in 1 ml of 50 mM KPi pH 7 buffer containing 400 mM NaCl, 10% glycerol and 1% Triton X-100 for 1 h at 4°C while shaking. The solubilized material was recovered by spinning the samples in an ultracentrifuge at 80000 rpm for 25 min at 4°C. The supernatant fractions were mixed with Ni-NTA beads (50 μl bed volume) preconditioned in 50 mM KPi pH 8, 600 mM NaCl, 10% glycerol, 0.1% Triton X-100 20 mM imidazole and left shaking at 4°C for 12 h. The Ni-NTA resin was recovered by brief spinning at 4,000 rpm at room temperature using a table top centrifuge and washed three times with the equilibration buffer and once with the same buffer containing 40 mM imidazole. Proteins were eluted with 30 μl of elution buffer (50 mM KPi pH 7, 600 mM NaCl, 10% glycerol, 0.03% DDM, 250 mM imidazole) and analyzed by SDS-PAGE.
Transport studies
Right-side-out (RSO) membranes were prepared from a 1-l culture by the osmotic shock procedure as described (Kaback Citation1974). RSO membranes were resuspended in 50 mM KPi pH 7.0 and aliquoted in 150 μl portions that were stored at −80°C. Membrane protein concentration in the samples was determined by the DC protein assay kit (BioRad Laboratories, Hercules, CA, USA).
RSO membrane vesicles were diluted to 1 mg/ml in 50 mM KPi pH 6 containing 100 mM NaCl. When indicated RSO membranes containing the N356C mutant of GltS were treated with 1 mM N-ethylmaleimide (NEM) for 10 min. The treatment was stopped by adding an equimolar concentration of dithiothreitol. Aliquots of 100 μl were incubated for 2 min at 30°C with 10 mM K-ascorbate and 100 μM phenazine methosulfate (PMS) under a flow of water-saturated air and continuous stirring with a magnetic bar. At t = 0, [1,5-14C]-citrate or L-[14C]glutamate was added to final concentrations of 4.4 and 2.9 μM, respectively. Uptake was stopped at 5, 10, 20, 40, 60 sec by the addition of 2 ml ice-cold 0.1 M LiCl followed by immediate filtering over cellulose nitrate filter (0.45 μm pore size) and washing of the filter with an additional 2 ml of the LiCl solution. Filters were collected and their radioactivity was measured in a liquid scintillation counter.
Cross-linking studies
For each experiment, a 50 mM glutaraldehyde solution in water was prepared freshly from a stock solution (Grade I, 25% aqueous glutaraldehyde solution, Sigma-Aldrich GmbH, Steinheim, Germany) stored at −20°C. Unless otherwise indicated, protein samples were treated at 2 mM glutaraldehyde concentration for 10 min at room temperature. The reaction was stopped by making the solution 100 mM in Tris-HCl pH 7 and incubating the sample for another 10 min at room temperature after which the samples were analyzed by SDS-PAGE (Laemmli Citation1970) using a 12% polyacrylamide gel and staining with Coomassie Brilliant Blue. The intensity of the protein bands was measured using a Fuji Imager LAS-4000.
Results
Biotin Acceptor Domain (BAD) tagged CitS
The Biotin Acceptor Domain (BAD) domain of the oxaloacetate decarboxylase of Klebsiella pneumoniae is a ∼10 kDa protein moiety. The gene coding for BAD was fused at the 5′ end and inserted into the middle of the Na+-citrate transporter gene citS yielding plasmids pCitS-BAD1 and pCitS-BAD260, respectively. The plasmids code for hybrid proteins consisting of the CitS protein with BAD fused at the N-terminus and with BAD inserted in the central cytoplasmic loop at position 260 in between the two putative domains of CitS, respectively. All constructs contain a His6-tag at the N-terminus ().
Figure 2. BAD-tagged CitS transporters. (A) Schematic representation of CitS-BAD hybrids. h, His6-tag; CitSN, N terminal domain of CitS; CitSC, C terminal domain of CitS. (B) Uptake of 14C-citrate by RSO membranes containing CitS (•), CitS-BAD1 (▴) and CitS-BAD260 (▪). Membrane protein concentrations were 1 mg/ml. Citrate uptake was expressed in nmol per mg of membrane protein in the sample. (C) SDS-PAGE of partially purified CitS moieties from the RSO membrane vesicles used in the uptake experiments shown in panel B. Lane 1, CitS, lane 2, CitS-BAD1, lane 3, CitS-BAD260, left lane, marker proteins. Molecular masses were indicated in kDa. Arrows at the right (from top to bottom) point at the BAD-tagged CitS moieties and CitS, respectively. (D) SDS-PAGE analysis after cross-linking of CitS moieties with glutaraldehyde. Samples of purified CitS, CitS-BAD1 and CitS-BAD260 were treated with 2 mM glutaraldehyde (GA) for 20 min in the presence and absence of 1% SDS as indicated at the bottom. Left lane, marker proteins with the molecular masses indicated in kDa. Arrows at the right (from top to bottom) points at dimeric CitS-BAD hybrids, dimeric CitS, monomeric CitS-BAD hybrids, and monmeric CitS.
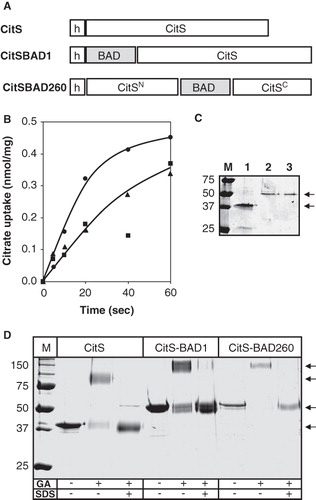
Membrane vesicles with a right-side-out (RSO) orientation were prepared from E. coli DH5α cells expressing wild-type CitS and the two hybrid proteins and were assayed for citrate uptake activity. Membrane vesicles prepared from the host strain are completely devoid of citrate uptake activity because of the lack of expression of citrate transporters during aerobic growth of E. coli (not shown; Lolkema et al. Citation1994). The initial rate of uptake of citrate driven by a proton motive force imposed by the artificial electron donor system K-ascorbate/PMS of the RSO membranes containing the two hybrid proteins was roughly half of the rate observed for those containing wild-type CitS (). The level of expression was determined by purifying the proteins from the membranes by Ni-NTA affinity chromatography using a batch-wise protocol. SDS-PAGE analysis of the partially purified proteins revealed the two hybrid proteins with apparent molecular masses of 50 kDa (, lanes 2 and 3) which is about 10 kDa more than the untagged CitS protein as expected (lane 1). The intensity of the bands of the two hybrid proteins was similar, but both were clearly less intense than observed for wild-type CitS. The lower expression level of the two hybrid proteins largely explains the difference in citrate uptake activity of the different RSO membrane preparations in . Introduction of BAD at the N-terminus and at position 260 of the CitS protein does not seem to significantly affect the transport activity of the CitS moieties. The result is in line with the positive phenotype of E. coli cells expressing the hybrid proteins on Simmons agar indicator plates (Van Geest and Lolkema Citation1996, Citation2000).
Cross-linking of CitS and tagged CitS moieties
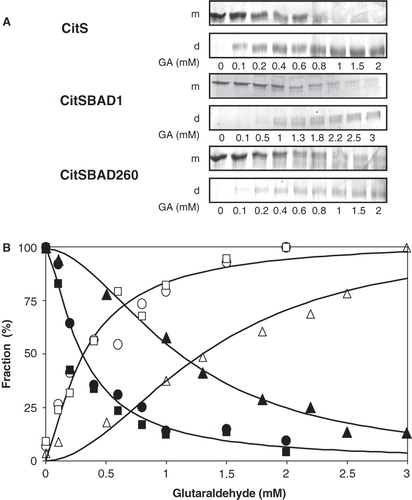
The CitS protein was shown before to be a dimer in the detergent solubilized state by cross-linking with glutaraldehyde (Dobrowolski and Lolkema 2010a). In agreement, treatment of purified CitS with 2 mM glutaraldehyde for 10 min shifted the protein band on SDS-PAGE up to a protein with an apparent molecular mass of 100 kDa (). Pretreatment of the sample with SDS prevented the up shift demonstrating that cross-linking in the native state is not the result of collisions between monomeric particles. The two hybrid proteins were purified to homogeneity. Similarly, as observed for the CitS protein, treatment of the hybrid proteins with glutaraldehyde resulted in an upshift of the proteins on SDS-PAGE that was not observed with the unfolded proteins (). For all cases, treatment with glutaraldehyde resulted in more diffuse protein bands, most likely due to the heterogeneity caused by random labeling of the protein and/or intramolecular cross-linking. It follows that the introduction of BAD at the N-terminus and at position 260 of the CitS protein does not interfere with the dimeric state of the CitS moieties.
Using the same conditions, cross-linking of CitS-BAD1 appeared to be less efficient than observed for CitS and CitS-BAD260. While the monomeric protein band had essentially disappeared in the case of the latter two, a significant band was still visible in case of the former (see ). The observation was further substantiated by titration of the cross-linking reaction with increasing concentrations of glutaraldehyde. Densitometric analysis of the monomeric and dimeric protein bands () showed that within the accuracy of the experiment, the behavior was similar for CitS and CitS-BAD260 with a concentration of 0.29 ± 0.05 mM glutaraldehyde giving 50% cross-linking. In marked contrast, the concentration of glutaraldehyde required for 50% cross-linking of CitS-BAD1 was significantly higher, 1.2 ± 0.1 mM (). It follows that the presence of BAD at the N-terminus of CitS negatively affects the cross-linking efficiency while BAD inserted in the central loop has no significant effect on cross-linking.
Figure 3. Titration of cross-linking of CitS and BAD-tagged CitS with glutaraldehyde. (A) SDS-PAGE analysis of purified CitS, CitS-BAD1 and CitS-BAD260 treated with the indicated concentration of glutaraldehyde (GA) for 10 min. m, monomeric protein bands; d, dimeric protein bands. (B) Densitometric analysis of the protein bands corresponding to monomeric and dimeric CitS (•,○), CitS-BAD1 (▴,Δ), CitS-BAD260 (▪,□). The data points were fitted to a sigmoidal curve after which the ‘begin’ and ‘end’ points of the curve were set to 100%.
Biotin Acceptor Domain (BAD) tagged GltS
A similar set of hybrid proteins as described above for the Na+-citrate transporter CitS was constructed with the Na+-glutamate transporter GltS (). The hybrids were constructed using the N356C mutant of GltS which was shown to be equally active in accumulating glutamate in RSO membranes but completely inactive after treatment with NEM (Dobrowolski and Lolkema Citation2009). The N356C mutant allows for the estimation of the background activity by endogenous glutamate transporters encoded on the chromosome of the host cells. GltS-BAD1 has BAD fused to the N-terminus and GltS-BAD206 has BAD inserted in the cytoplasmic loop of GltS at position 206 which corresponds to position 260 in CitS. In addition, a control plasmid was constructed coding for the BAD domain with an N-terminal His6-tag.
Figure 4. BAD-tagged GltS transporters. (A) Schematic representation of GltS-BAD hybrids. h, His6-tag. GltSN, N terminal domain of GltS. GltSC, C terminal domain of GltS. The star indicates the N356C mutation in GltS. (B) Uptake of 14C-glutamate by RSO membranes containing GltS (•), GltS-BAD206 (▴) and GltS-BAD1 (▪,□) before (▪) and after (□) treatment of the membranes with 1 mM NEM for 10 min. Membrane protein concentrations were 1 mg/ml. Glutamate uptake was expressed in nmol per mg of membrane protein in the sample. (C) SDS-PAGE of partial purified GltS moieties from the RSO membrane vesicles used in the uptake experiment shown in panel B. Lane 1, GltS, lane 2, GltS-BAD1, lane 3, GltS-BAD206, left lane, marker proteins. Molecular masses were indicated in kDa. Arrows at the right (from top to bottom) point at the BAD-tagged GltS moieties and GltS, respectively. (D) SDS-PAGE analysis after treatment of purified GltS (left), GltS-BAD206 (middle) and the BAD protein (right) with 2 mM glutaraldehyde (GA) for 20 min in the presence and absence of 1% SDS as indicated at the bottom. Left lanes, marker proteins with the molecular masses indicated in kDa. Arrows point at the monomeric protein bands.
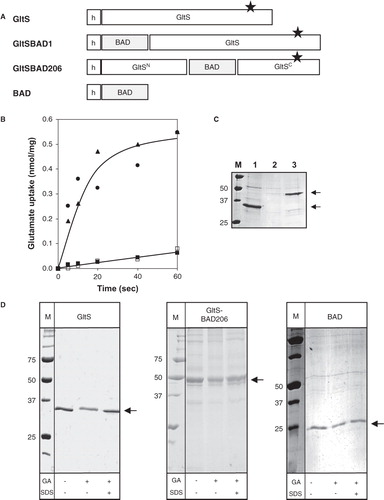
Membrane vesicles with a right-side-out (RSO) orientation were prepared from E. coli DH5α cells expressing the GltS mutant N356C and the hybrid proteins and were assayed for glutamate uptake activity. Mutant N356C and GltS-BAD206 showed similar uptake activities (). Pretreatment of the membranes with NEM reduced the uptake to the activity of endogenous glutamate transporters (Dobrowolski and Lolkema Citation2009). In contrast, membranes containing GltS-BAD1 revealed no significant activity above the level that was observed after treatment of the same membranes with NEM. SDS-PAGE analysis of the partially purified proteins revealed that the lack of activity of GltS-BAD1 was due to an expression/insertion problem. While the N356C mutant and GltS-BAD206 showed up as clear protein bands with apparent molecular masses of 35 and 45 kDa, respectively, as expected (, lanes 1 and 3) (Mościcka et al. Citation2009), no band could be observed at the expected size for GltS-BAD1 (lane 2). It follows that, similarly as observed for CitS, insertion of BAD in the central cytoplasmic loop of the GltS protein did not significantly affect the transport activity of the protein, but, in contrast to CitS, BAD fused at the N-terminus of GltS in GltS-BAD1 decreased the expression level of the hybrid protein. It should be noted that the N-termini of GltS and CitS are located at opposite sites of the membrane, in the periplasm and cytoplasm, respectively, and, consequently, the BAD moiety in GltS-BAD1 has to be translocated across the membrane during biogenesis.
Cross-linking of GltS and tagged GltS moieties
The GltS protein was shown to be dimeric by binding affinity to Ni-NTA, BN-PAGE, and EM analysis (Mościcka et al. Citation2009) but not by cross-linking with glutaraldehyde. Contrary to expectations, treatment of purified GltS protein with 2 mM of glutaraldehyde for 10 min did not result in the appearance of dimeric protein on SDS-PAGE (, left panel). Some reduction in the intensity of the monomeric protein band was observed which was due to a small fraction of aggregated protein that following treatment with the cross-linker did not enter the gel. Pretreatment of the sample with 1% of SDS before cross-linking resolved the aggregates. The cross-linking reaction was repeated with glutaraldehyde concentrations in the range of 0.1–3 mM, but under none of these conditions cross-linking was observed. The lack of cross-linking of GltS was considered to be due to the lack of suitable cross-linking sites on the protein which might be provided by the BAD moiety added to GltS in the hybrid GltS-BAD206 protein. To exclude the possibility that BAD would drive dimerization of the hybrid, the domain was purified and treated with glutaraldehyde. Under the conditions of the experiment, BAD was a monomeric protein (, right panel). Treatment of purified GltS-BAD206 with glutaraldehyde under the same conditions did not show any cross-linking to the dimer. Like for untagged GltS, a slight reduction of the intensity of the monomer was observed and aggregated material appeared (, middle panel). In all cases, the aggregates resolved in the presence of 0.1% SDS. It follows that insertion of the 10 kDa BAD domain in the central loop connecting the N and C domains of GltS does not promote cross-linking of the GltS complex.
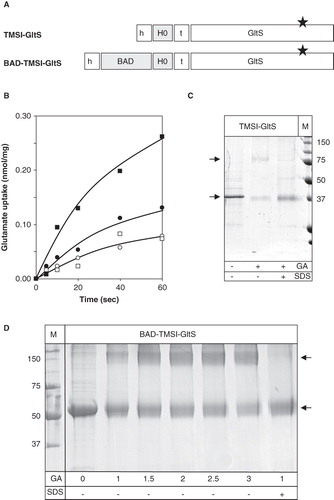
The citrate transporter CitS carries an extra TMS at the N-terminus which is not present in the glutamate transporter GltS. In construct TMSI-GltS, the first TMS of CitS including the adjacent loop regions were fused to the N-terminus of the N356C mutant of GltS (). Construct BAD-TMSI-GltS has, in addition, the BAD domain fused to the N-terminus and corresponds to the GltS version of the CitS-BAD1 construct. Transport assays in RSO membranes containing the TMSI-GltS construct showed a significantly higher uptake activity than after treatment of the same membranes with NEM (). The activity was ∼20% of membranes containing wild-type GltS which was mainly due to a lower level of expression (not shown). In contrast to wild-type GltS, a significant fraction of the protein could be cross-linked to the dimeric form by treatment with glutaraldehyde (). The BAD-TMSI-GltS construct revealed ∼50% of the activity of wild type GltS in RSO membranes () with a comparable level of expression (not shown). Treatment of the hybrid protein with glutaraldehyde resulted in the appearance of a cross-linked product (). Titration of the reaction with increasing concentration of glutaraldehyde indicated that the concentration required to give 50% cross-linking was between 1 and 1.5 mM which is in the same range as observed for cross-linking of CitS-BAD1 (). The cross-linked product has an apparent molecular mass that is more than twice the apparent mass of the monomeric species. However, since in the titration no intermediate bands were observed it is concluded that the cross-linked product is dimeric. The slow mobility must be a consequence of aberrant behavior of the cross-linked dimer on SDS-PAGE. It follows that the addition of the first TMS of CitS with or without a fused BAD moiety at the N-terminus of GltS allows for the cross-linking of the GltS monomers in the dimeric complex.
Figure 5. Fusions of TMSI of CitS to the GltS transporter. (A) Schematic representation of the TMSI-GltS and BAD-TMSI-GltS hybrids. h, His6-tag; BAD, BAD domain; H0, N-terminal plus TMSI of CitS; t, TEV protease cleavage site. The star indicates the N356C mutation in GltS. (B) Uptake of 14C-glutamate by RSO membranes containing TMSI-GltS (•,○) and BAD-TMSI-GltS (▪,□) before (•,▪;) and after (○,□) treatment of the membranes with 1 mM NEM for 10 min. Membrane protein concentrations were 1 mg/ml. Glutamate uptake was expressed in nmol per mg of membrane protein in the sample. (C) SDS-PAGE analysis after treatment of partially purified TMSI-GltS with 2 mM glutaraldehyde (GA) for 20 min in the presence and absence of 1% SDS as indicated at the bottom. Right lane, marker proteins with the molecular masses indicated in kDa. (D) SDS-PAGE analysis of partially purified BAD-TMSI-GltS treated with the indicated concentrations of glutaraldehyde (GA) for 10 min in the presence and absence of 1% SDS as indicated at the bottom. Left lane, marker proteins. Arrows from top to bottom point at the dimeric and monomeric protein bands, respectively.
Discussion
The projection structure of the secondary transporters CitS and GltS obtained by electron microscopy and single particle averaging reported before gave several new insights into the architecture of these relatively small proteins (Mościcka et al. Citation2009). EM analysis of purified CitS of K. pneumoniae revealed two dominant classes of 2D projection maps which were interpreted as representing top and side views of the protein. The top view showed a biscuit-shaped particle with a long axis of about twice the length of the short axis (, top). In side view, CitS was kidney-shaped. The shapes and sizes were consistent with a homodimeric particle with an indentation at one side of the membrane at the interface of the subunits that would be formed by the short axis. The EM images of the GltS protein of E. coli revealed the same overall shape as observed for CitS but no clear discrimination between top and side views was obtained. The aim of the present study was to determine the relative orientation of the monomers in the dimeric structure by determining the position of the N-termini and the central loops that connect the two putative domains in the monomers. Distance relationships were inferred by tagging the N-terminus and central loop with the biotin acceptor domain (BAD) of the oxaloacetate decarboxylase of K. pneumoniae and determining their effect on cross-linking of the monomers in the dimers by glutaraldehyde. Hybrids CitS-BAD1 (N-terminus), CitS-BAD260 (central loop) and GltS-BAD206 (central loop) were active in Na+-dependent transport ( and ) indicating that the transporter parts of the proteins had retained their native structure. The data support a relative orientation of the monomers with the N-termini close to the dimer interface and the central loops distant at the ends of the long axis (, bottom).
Figure 6. Relative orientation of the subunits in dimeric CitS and GltS. Schematic top view structures of dimeric wild type CitS (top), CitS-BAD206 (bottom left) and CitS-BAD1 (bottom right). Top view was defined as perpendicular to the membrane. The ‘closed’ and ‘open’ sides of the clam shell model of one subunit were indicated. The connection at the ‘closed’ side is formed by the central cytoplasmic loop. The N- and C-termini are positioned at the ‘open’ side. Dimensions are based upon EM projection structures. Vertical and horizontal dashed lines indicate subunit and domain interfaces, respectively. Circles in grey represent the BAD domains.
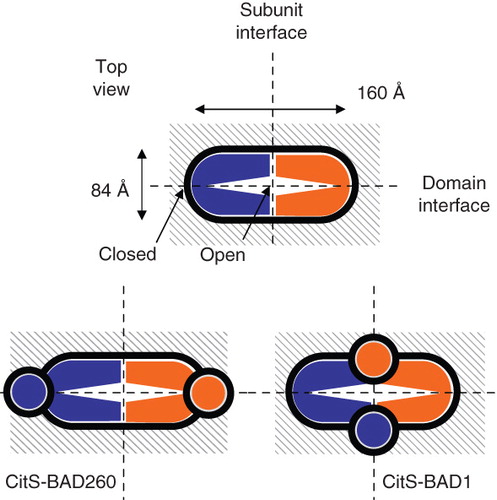
Support for the relative positions of the central loop and N-terminus in the dimeric complex was obtained from the cross-linking studies of CitS and GltS in several ways. Monomeric glutaraldehyde is a bivalent unspecific cross-linker that in proteins preferentially reacts with the ϵ-NH2 group of lysine residues, but, for instance, also with the N atom of peptide bonds (Wine et al. Citation2007). The CitS dimer solubilized in detergent solution was readily cross-linked by glutaraldehyde (Dobrowolski and Lolkema 2010a), but, surprisingly, GltS was not while its dimeric state was demonstrated using other techniques (Mościcka et al. Citation2009); this study). The different cross-linking behavior of GltS and CitS may reflect the different amino acid sequences of the two proteins, but also be related to the extra TMS and adjacent cytoplasmic N-terminus present at the N-terminal side of the core structure of CitS that is absent in GltS (see ). Titration of the cross-linking of CitS by increasing concentrations of glutaraldehyde suggested the latter view. The concentration of glutaraldehyde needed to obtain 50% cross-linking was raised by a factor of four when BAD was fused to the N-terminus of CitS (). Clearly, the presence of the BAD domain hinders the cross-linking, most likely by covering up the sites in CitS that result in cross-linking in the absence of BAD. The cross-linking sites of the untagged CitS would have to be close to BAD, i.e., close to the N-terminus. The cross-linking of GltS when BAD plus the first TMS of CitS was fused at its N-terminus, a chimera that was active in glutamate transport, is in agreement with this view (). The similar concentration of glutaraldehyde required to obtain 50% cross-linking suggests a similar relative position of the two BAD moieties as in the CitS-BAD1 dimer (). Further evidence for close proximity of the N-termini in the dimer was obtained by the cross-linking of the GltS dimer when TMS1 of CitS was fused at the N-terminus which places the N-terminus of the construct in the cytoplasm (). The latter construct expressed poorly but significant activity of the TMSI-GltS hybrid could be demonstrated. The complementary experiment in which TMS1 plus the N-terminal residues of CitS were deleted failed because no expression of the deletion mutant was obtained (not shown). Nevertheless, together these experiments suggest that the N-termini of CitS and TMSI-GltS are close to the dimeric interface allowing cross-linking of the monomers by glutaraldehyde. Equally important was the lack of effect on cross-linking when BAD was inserted in the central cytoplasmic loop. No significant difference was observed between the concentrations of glutaraldehyde to obtain 50% cross-linking for CitS and CitS-BAD260, suggesting no contribution of BAD to the cross-linking (). Also no cross-linking of GltS was observed when BAD was inserted in the central loop yielding GltS-BAD206. Apparently, the BAD moieties in the central loop are to far away to result in cross-linking.
The structural model for the monomeric subunits of CitS and GltS consists of two homologous domains with the substrate and co-ion translocation sites at the interface of the two domains (Sobczak and Lolkema Citation2005). Strong support for the model was recently obtained by demonstrating cross-linking between the pore-loops situated between the 4th and 5th TMS in each domain (Dobrowolski and Lolkema 2010a). In a simple three-dimensional model, the two domains would fold upon each other like a clam shell with the central loop that connects the two domains forming the hinge at the ‘closed’ side of the monomer and the N- and C-terminal ends at the opposite ‘open’ side. In the dimer structure, the monomers could be oriented ‘closed-to-closed’ or ‘open’ to ‘open’. The present results favor the latter orientation with the ‘closed’ sides of the monomers in the EM projection structure positioned at the end of the long axis to far away to contribute to cross-linking and the ‘open’ sides in close vicinity at the dimer interface (see the model in , bottom).
Recently, we constructed an artificial GltS transporter (GltSswap) in which the two domains in the monomer were swapped by disconnecting the central loop and reconnecting the domains by a linker between TMS1 and TMS10 (Dobrowolski and Lolkema 2010b). The new ‘central loop’ (i.e., the linker) resides in the periplasm rather than the cytoplasm. GltSswap with a 19 residue long linker was equally active as wild type GltS, but, in the context of the present study, even more surprisingly, the protein was readily cross-linked by glutaraldehyde (Dobrowolski and Lolkema 2010) while wild type GltS is not. This strongly suggests that cross-linking proceeds via the linker sequence in agreement with the above conclusion that the N- and C-termini would be close to the dimer interface. Most likely, peptide bond N atoms provided the sites for cross-linking since the linker did not contain lysine residues. In the structural model, the periplasmic linker of GltSswap and the endogenous central loop of wild type GltS would be at the same relative positions in a top view projection structure but at different sides of the membrane. The marked difference in contribution to cross-linking may reflect: (i) The structural asymmetry in the dimer relative to the plane of the membrane (side view; Mościcka et al. Citation2009), or (ii) a defined structure of the wild type central loop keeping the loop in a fixed position and hiding potential reactive sites.
Conclusion
The two domains of monomeric CitS and GltS fold upon each other like a clam shell with the connecting cytoplasmic loop forming the hinge region at the ‘closed’ side. The CitS monomers in the dimer were readily cross-linked by glutaraldehyde, in contrast to GltS that was not cross-linked at all. Tagging the cytoplasmic loop with BAD had no effect on cross-linking efficiency, suggesting that the cytoplasmic loops (the ‘hinges’) are located at opposite sides of the dimer. Tagging the N-terminus of CitS with BAD or GltS with TMS1 of CitS (with or without BAD) affected cross-linking of both transporters; CitS by covering up suitable sites, thus decreasing cross-linking efficiency and GltS by supplying suitable sites, thus making cross-linking possible. This suggests that the N-termini are located in close vicinity and that they are at the ‘open’ sides of the monomers that form the dimer interface. Together, the distances of the two cytoplasmic loops and the two N-termini indicate that the relative orientation of the monomers in the dimer is ‘open-to-open’.
Declaration of interest: This work was supported by grants from the Dutch Organization for Scientific Research (NWO-ALW). (Grant number: 814.02.008). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bott M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol 167:78–88.
- Dobrowolski AJ, Lolkema JS. 2009. Functional importance of GGXG sequence motifs in putative reentrant loops of 2HCT and ESS transport proteins. Biochemistry 48:7448–7456.
- Dobrowolski AJ, Lolkema JS. 2010a. Cross-linking of trans re-entrant loops in the Na+-citrate transporter CitS of Klebsiella pneumoniae. Biochemistry 49:4509–4515.
- Dobrowolski AJ, Lolkema JS. 2010b. Evolution of antiparallel two-domain membrane proteins. Swapping domains in the glutamate transporter GltS. Biochemistry 49:5972–5974.
- Dobrowolski AJ, Sobczak-Elbourne I, Lolkema JS. 2007. Membrane topology prediction by hydropathy profile alignment: Membrane topology of the Na+-glutamate transporter GltS. Biochemistry 46:2326–2332.
- Kaback HR. 1974. Transport in isolated bacterial membrane vesicles. Methods Enzymol 31:698–709.
- Kästner CN, Prummer M, Sick B, Renn A, Wild UP, Dimroth P. 2003. The citrate carrier CitS probed by single-molecule fluorescence spectroscopy. Biophys J 84:1651–1659.
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685.
- Lolkema JS, Slotboom DJ. 1998. Estimation of structural similarity of membrane proteins by hydropathy profile alignment. Mol Membr Biol 15:33–42.
- Lolkema JS, Slotboom DJ. 2003. Classification of 29 families of secondary transport proteins into a single structural class using hydropathy profile analysis. J Mol Biol 327:901–909.
- Lolkema JS, Slotboom DJ. 2008. The major amino acid transporter superfamily has a similar core structure as Na+-galactose and Na+-leucine transporters. Mol Membr Biol 25:567–570.
- Lolkema JS, Enequist H, van der Rest ME. 1994. Transport of citrate catalyzed by the sodium-dependent citrate carrier of Klebsiella pneumoniae is obligatorily coupled to the transport of two sodium ions. Eur J Biochem 220:469–475.
- Lolkema JS, Sobczak I, Slotboom DJ. 2005. Secondary transporters of the 2HCT family contain two homologous domains with inverted membrane topology and trans re-entrant loops. FEBS J 272:2334–2344.
- Mościcka KB, Krupnik T, Boekema EJ, Lolkema JS. 2009. Projection structure by single-particle electron microscopy of secondary transport proteins GltT, CitS, and GltS. Biochemistry 48:6618–6623.
- Pos KM, Bott M, Dimroth P. 1994. Purification of two active fusion proteins of the Na+-dependent citrate carrier of Klebsiella pneumoniae. FEBS Lett 347:37–41.
- Prakash S, Cooper G, Singhi S, Saier MH. 2003. The ion transporter superfamily. Biochim Biophys Acta 1618:79–92.
- Saier MH Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 64:354–411.
- Sobczak I, Lolkema JS. 2003. Accessibility of cysteine residues in a cytoplasmic loop of CitS of Klebsiella pneumoniae is controlled by the catalytic state of the transporter. Biochemistry 42:9789–9796.
- Sobczak I, Lolkema JS. 2004. Alternating access and a pore-loop structure in the Na+-citrate transporter CitS of Klebsiella pneumoniae. J Biol Chem 279:31113–31120.
- Sobczak I, Lolkema JS. 2005. The 2-hydroxycarboxylate transporter family: physiology, structure and mechanism. Microbiol Mol Biol Rev 69:665–695.
- Van Geest M, Lolkema JS. 1996. Membrane topology of the Na+ ion dependent citrate carrier of Klebsiella pneumoniae. J Biol Chem 271:25582–25589.
- Van Geest M, Lolkema JS. 1999. Transmembrane segment (TMS) VIII of the Na+/Citrate transporter CitS requires downstream TMS IX for insertion in the Escherichia coli membrane. J Biol Chem 274:29705–29711.
- Van Geest M, Lolkema JS. 2000. Membrane topology of the Na+/citrate transporter CitS of Klebsiella pneumoniae by insertion mutagenesis. Biochim Biophys Acta 1466:328–338.
- Veenhoff LM, Heuberger EHML, Duurkens HH, Poolman B. 2001. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J Mol Biol 317:591–600.
- Wine Y, Cohen-Hadar N, Freeman A, Frolow F. 2007. Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis. Biotechnol Bioeng 98:711–718.