Primary congenital glaucoma (PCG) is a major cause of childhood blindness if untreated, and is the second most common cause of blindness worldwide.Citation1 PCG can be treated successfully with surgery or medication if it is detected early, resulting in a good prognosis in 80–90% of patients.Citation2 PCG occurs as a result of the maldevelopment of the trabecular meshwork and the anterior angle of the eye, resulting in an improper development of the drainage angle of the anterior chamber.Citation3 Mutations in the cytochrome P450 1B1 (CYP1B1) gene have been reported in numerous individuals with PCG. There are over 100 CYP1B1 mutations reported in the Human Gene Mutation Database (HGMD professional 2011.1). Mutations have been identified throughout the gene, including missense mutations and mutations resulting in a truncated CYP1B1 product which have been shown to result in PCG.
We present an 11-month-old male Caucasian patient clinically diagnosed with PCG. The patient had congenital bilateral glaucoma, elevated intraocular pressure, and enlargement of the globe, corneal edema/clouding, and photophobia. The patient has a sister with a diagnosis of congenital glaucoma. The family history was otherwise non-contributory, and the patient’s parents denied consanguinity. Written informed consent was obtained from the patients’ parents. This study was approved by the University of Utah Institutional Review Board (IRB#35637).
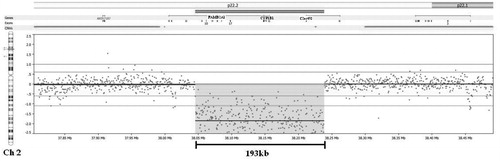
The patient’s sample had been submitted to ARUP Laboratories for CYP1B1 sequencing for molecular diagnosis. Genomic DNA was extracted via automated Magna Pure (Roche Diagnostics, Indianapolis, IN) from whole blood. The CYP1B1 gene consists of three exons, with a 5.2 kb transcript, with the first exon being non-coding. CYP1B1 specific primers were used to amplify the three exons of the CYP1B1 gene, and the resulting products, if obtained, were run on a 2% agarose gel. After several PCR failures from the patient’s DNA with different primer sets and a successful PCR result with a different DNA sample, we investigated large gene deletions on this patient. Multiplex ligation-dependent probe amplification (MLPA) (MRC-Holland, kit P128-B1, Amsterdam, Netherlands) was used to identify the suspected gene deletion, with MLPA probes located in exons 1 and 3. Since MLPA result showed the homozygous deletion, we used Chromosome 2 Comparative Genomic Hybridization (CGH) array (Nimblegen, Madison, WI) to confirm the homozygous deletion and to define the breakpoints. We have identified the homozygous deletion of the entire CYP1B1, as well as portions of neighboring genes. Exons 1–2 of the C2orf58 (chromosome 2 open reading frame 58, MGC34824), and exons 3–11 of the FAM82A1 (Family with sequence similarity 82, member A1) were also deleted (). This 193kb deletion encompasses the chromosomal region 38,045,327--38,238,757. Assembly: March 2006 build (NCB136/hg18). We identified the same homozygous deletion in the patient’s affected sister.
Figure 1. Homozygous deletion and breakpoints in chromosome 2, encompassing the entire CYP1B1 gene, including exons 1–2 of the C2orf58 (chromosome 2 open reading frame 58, MGC34824) gene, and exons 3–11 of the FAM82A1 (Family with sequence similarity 82, member A1). CNV regions are indicated by the pink bars. The double red bars indicate the area of the 193 kb deletion. Exons of the genes involved are indicated by marks below each gene name. Probes are indicated by dots. The median probe spacing for this array is 575 bp. The area of the deletion is indicated by the pink highlighted area encompassing the probes which occur on the scale below the normal variation, indicative of a homozygous deletion.
Only one gross gene deletion has been previously reported in the CYP1B1gene in association with PCG, encompassing 146 kb.Citation4 In addition, a 140 kb contiguous gene deletion including CYP1B1 was reported in a patient, his mother, and grandmother as part of a study interrogating candidate genes associated with cleft lip and/or palate.Citation5 No association was made with primary congenital glaucoma in reference to this family. The breakpoints of the homozygous deletion in our siblings are different from those reported by Shi et al.Citation5 and Kelberman et al.Citation4 Parental DNA samples were not available to determine their carrier status. In order to look for an indirect evidence of consanguinity of the parents, we ran short tandem repeat (STR) markers on the siblings (AmpFLSTR Identifier® PCR Amplification Kit, Applied Biosystems, Foster City, CA), which excluded the possibility of consanguinity (data not shown). This case highlights the importance for deletion analysis of CYP1B1 in clinical testing to aid in a molecular diagnosis when an early diagnosis is crucial for patient treatment and care.
ACKNOWLEDGeMENTS
We gratefully thank the family for their participation in this study. We would also like to thank the University of Iowa Hospitals and Clinics.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Thylefors B, Negrel A. The global impact of glaucoma. Bull WHO 1994;72:323–326
- Demetrios Vavvas, Cynthia Grosskreutz, Louis Pasquale. Congenital glaucoma (childhood). Digital J Ophthalmol 2012. www.djo.harvard.edu/site.php?url=/patients/pi/416
- Traboilsi EI. Genetic diseases of the eye. New York: Oxford University Press; 1998
- Kelberman D, Islam, L, Jacques, TJ, et al CYP1B1-related anterior segment developmental anomalies. Ophthalmology 2011;118:1865–1873
- Shi M, Mostowska A, Jugessur A, et al Identification of microdeletions in candidate genes for cleft lip and/or palate. Birth Defects Res Part A: Clin Mol Teratol 2009;85:42–51