abstract
Context: Baicalein is a major compound in extracts derived from Scutellaria baicalensis Georgi (Lamiaceae) which are used in the Traditional Chinese Medicine for the treatment of inflammatory and gastrointestinal diseases. This flavonoid is an activator of the Nrf2 signalling pathway but the molecular mechanism is not clearly established.
Objective: We investigated the molecular mode of baicalein-mediated Nrf2-activation in Hct116 cells by the analysis of proteasomal activity, radical-scavenging activity and the comparison with baicalein derivatives.
Materials and methods: The radical-scavenging activity (TEAC, DCF) up to 25 μM, cytotoxicity (MTT assay, 48 h) up to 100 μM, proteasomal activity and the Nrf2-activation (luciferase assay, ubiquitinylation, western blot, Ser40-phosphorylation; incubation for 1 or 4 h) by concentrations up to 40 or 50 μM of the compounds were analysed in Hct116 human colon carcinoma cells.
Results: No change in the ubiquitinylation of Nrf2, proteasomal activity and transcription of the NRF2 gene were detectable. Baicalein decreased the phosphorylation of Nrf2 (IC50-value approximately 20 μM) suggesting an inhibitory effect of the flavonoid on protein kinases. Since the activation of the Nrf2 pathway by baicalein might be also due to redox-activity of the compound, we investigated the effects of methylated baicalein derivatives oroxylin A, negeletein and baicaleintrimethylether. Oroxylin A and negletein showed a comparable redox-active potential, but only negletein (50 μM, 4 h) was able to activate Nrf2.
Conclusion: This result confirms the hypothesis that baicalein, a component of extracts derived from Baical Skullcap, causes an activation of Nrf2 independent of a modulation of the cellular redox potential.
Introduction
Extracts derived from Scutellaria baicalensis Georgi (Lamiaceae) are used in the Traditional Chinese Medicine (TCM) for the treatment of inflammatory, cardiovascular and gastrointestinal diseases. Preparations are listed in the Chinese Pharmacopeia as ‘huangqin’ (Li et al. Citation2009) and are used in Korea as ‘hwang-keum’ (Kang et al. Citation2012). A major compound of Baical Skullap extracts is baicalein, a flavonoid with three hydroxyl groups (position 5, 6 and 7; ). We have previously shown that baicalein is an activator of the redox sensitive Nrf2/ARE pathway in Hct116 human colon carcinoma cells and activates SKN-1 (the Nrf2 homologue) in the nematode Caenorhabditis elegans (Rhabditida, Rhabditidae) (Havermann et al. Citation2013). The transcription factor Nrf2 mediates the cellular response to oxidative and electrophilic stress by binding to antioxidant responsive elements (AREs) (Rushmore et al. Citation1991) and thereby regulates the expression of several antioxidant and phase II drug metabolising enzymes (Alam et al. Citation1999; Hayes et al. Citation2000; Sekhar et al. Citation2000; Bloom & Jaiswal Citation2003). Under physiological conditions, the transcription factor is sequestered in the cytosol by binding to its inhibitor protein Keap1 (Itoh et al. Citation1999) which steadily mediates ubiquitinylation, and therefore proteasomal breakdown of Nrf2 (Kobayashi et al. Citation2004). To maintain a steady state, the protein Nrf2 is continuously synthesised (Nguyen et al. Citation2003). Oxidative stress or electrophilic xenobiotics activate this signalling pathway resulting in the accumulation of Nrf2 (Motohashi & Yamamoto Citation2004). Several molecular mechanisms might explain the activation of Nrf2 by secondary plant compounds: The activation can be attained by an increased expression of the transcription factor which is inducible due to binding-sites, like AREs, in its promotor region (Kwak et al. Citation2002) or by a higher translation of the mRNA (Purdom-Dickinson et al. Citation2007). Further, stabilisation of Nrf2 can be achieved by the inhibition of the proteasome (Itoh et al. Citation2003), or by a decreased ubiquitinylation of Nrf2, and therefore a reduced degradation (Kobayashi & Yamamoto Citation2006). The latter is influenced, e.g., by an increased breakdown of the inhibitor protein, a change in conformation or cleavage of the Nrf2–Keap1 complex by oxidative stress or a direct reaction with compounds (Dinkova-Kostova et al. Citation2001) or a change in Nrf2 phosphorylation by modulation of kinases or phosphatases (Nguyen et al. Citation2003; Jaiswal Citation2004). A molecular interaction of Nrf2 with different kinases, e.g., MAPK, ERK (Zipper and Mulcahy Citation2000), PKC (Bloom and Jaiswal Citation2003) and PI3K (Kang et al. Citation2002), has been reported.
The activation of Nrf2 by baicalein might also be explained with redox-activity of the compound. It could directly interact with Nrf2/Keap-1 via a quinoid-like structure (= oxidised baicalein). To determine the role of redox-activity for the mechanism of Nrf2 activation by baicalein, we have used three methylated baicalein derivatives: Baicalein-7-O-methylether (negletein) with a catechol structure (OH groups in ortho position), baicalein-6-O-metylether (oroxylin A) with two free OH–groups in meta position and baicalein-5,6,7-O-trimethylether (5,6,7-trimethoxyflavone) without free OH groups in the molecule. The determination of their redox-activity as well as their impact on Nrf2 activation will give evidence which molecular structure of baicalein is necessary for the compound’s effect on the Nrf2 signalling pathway. This is necessary for a better understanding of the molecular mechanisms of baicalein in extracts derived from Baical Skullcap.
Materials and methods
Materials
Baicalein (≥98%) and oroxylin A (≥98%) were obtained from Sigma (Steinheim, Germany), negletein (≥90%) and baicaleintrimethylether (≥90%) were bought from Extrasynthese (Genay Cedex, France). The ARE GST-Ya luciferase reporter construct containing a 41-bp GST-Ya ARE inserted into the pTI-luc plasmid was kindly provided by Dr. Ming Zhu (UC Davis Cancer Center, Sacramento, CA) and was constructed as described elsewhere (Wasserman & Fahl Citation1997).
Trolox equivalent antioxidative capacity (TEAC) assay
Equal volumes of 14 mM 2,2′-azinobis-(3-ethylbenzthiazolin)-6-sulphonic acid (ABTS; Sigma, Steinheim, Germany) and 4.9 mM ammoniumperoxodisulphate (Merck, Darmstadt, Germany) were mixed and were allowed to form stable ABTS radicals overnight. This solution was diluted with 70% ethanol to an absorption of 1.4 at a wavelength of 734 nm. Five hundred microlitre of this dilution were mixed with equal volumes of compound solutions and the absorption of the mixture was measured after 2 min. The synthetic vitamin E derivative Trolox (Calbiochem, Darmstadt, Germany) was used as a positive control.
Cell culture
Hct116 colon carcinoma cells (Brattain et al. Citation1981) were obtained from the DSMZ (Braunschweig, Germany) and were cultured in DMEM (Gibco, Karlsruhe, Germany) supplemented with 10% heat-inactivated foetal calf serum (PAA Laboratories, Pasching, Austria), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Karlsruhe, Germany) at 37 °C in a humidified atmosphere with 5% CO2. Cells were passaged twice a week before reaching confluence. FHC foetal normal colon cells (Siddiqui & Chopra Citation1984) were obtained from LGC Standards (Wesel, Germany) and were cultured in DMEM:F12 (Gibco, Karlsruhe, Germany) supplemented with additional 10 mM HEPES (Sigma, Steinheim, Germany), 10 ng/mL choleratoxin (Enzo LifeSciences, Lörrach, Germany), 5 μg/mL human insulin (Sigma, Steinheim, Germany), 5 μg/mL human transferrin (Sigma, Steinheim, Germany), 100 ng/mL hydrocortisol (Sigma, Steinheim, Germany), 10% heat-inactivated FCS, 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were passaged before reaching confluence; the medium was changed twice a week.
Cytotoxicity testing (MTT assay)
Hct116 cells/well (n = 10 000) were seeded into the wells of a 96-well plate and were allowed to attach for 24 h. The cells were then incubated with various concentrations of the compounds for 48 h and were then treated with MTT for 30 min. The medium was removed and the produced formazane was extracted from the cells with DMSO. Absorption was measured at 560 nm in a plate reader (Sunrise, Tecan, Grödig, Austria).
Intracellular accumulation of ROS (DCF assay)
Hct116 cells/well (n= 500 000) were seeded into a 6-well plate and were allowed to attach for 24 h. The cells were incubated with the compounds (25 μM) for 4 h followed by washing with serum-free DMEM and loading with 10 μM 2,7-dichlorodihydrofluoresceine acetate (H2DCF-DA; Sigma, Steinheim, Germany) for 15 min. After washing with serum-free medium, the cells were stressed with 500 μM H2O2 for 1 h followed by washing with PBS. The cells were trypsinised (0.025% trypsin-EDTA), harvested in PBS and analysed in an Accuri C6 flow cytometer (Accuri Cytometers, St. Ives. Cambs, UK) for DCF fluorescence; excitation: 488 nm, emission: 530 ± 15 nm (FL1).
Luciferase assay
Hct116 cells were transiently transfected using JetPei™ transfection reagent (Polyplus Transfection) in a batch protocol. The complex forming took place in 2 μL reagent per μg plasmid DNA. These complexes were mixed with 5 × 105 cells and were seeded into 6-well plates. The 24 h transfection period was followed by 24 h incubation with the compounds. Afterwards cells were washed with ice-cold PBS and were then lysed by shaking in reporter lysis buffer (Promega, Madison, WI) for 15 min. The lysate was centrifuged for 1 min at 13 200 rpm (4 °C) und the luciferase activity in the supernatant was analysed using a luciferase assay kit (BioThema AB, Sweden, Sweden) according to the protocol of the manufacturer in a Victor2 1420 multilabel counter equipped with a dispenser. The measured chemiluminescence was normalised to protein content (Bio-Rad Protein Assay, Bio-Rad, Hercules, CA).
Total protein isolation and western blot analysis
Hct116 cells (n = 750 000) were seeded into 35 mm cell culture dishes and were then allowed to attach for 24 h before incubating the cells with the compounds for 4 h. For FHC cells, 150 000 cells were seeded into 60 mm cell culture dishes and were allowed to attach for 3 d. Afterwards the cells were rinsed with ice-cold PBS and collected in RIPA buffer (50 mM Tris-HCl, 150 mM NaCL, 5 mM EDTA, 1% Nonidet P40, 0.1% SDS, 0.5% deoxycholate; pH 8.0) supplemented with 1 mM DTT, 1 mM PMSF, proteinase inhibitor cocktail and 0.01% okadaic acid. The cells were subjected to two freeze-thaw-cycles and the fraction containing the total protein was retrieved by centrifugation at 13 200 rpm for 10 min (4 °C). Protein content was measured using the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA) according to the protocol of the manufacturer. Forty microgram of protein were separated by SDS-PAGE and were then transferred to a PVDF membrane (Roche, Mannheim, Germany). After blocking for 1 h at room temperature with 5% BSA in TBS supplemented with 0.1% Tween20 (TBST), the membranes were incubated with an anti-Nrf2 antibody (1:5000; #2178-1, Epitomics, Burlingame, CA) or an anti-phospho-Nrf2 antibody (1:5000; #2073-1, Epitomics, Burlingame, CA) for 1 h at room temperature. After washing with TBST, the membranes were incubated with a 1:3000 dilution of secondary antibody (goat anti-rabbit, SouthernBiotech, Birmingham, UK) over night at 4 °C. The signals were visualised with the BM Chemiluminescence Western Blotting Kit (Roche, Mannheim, Germany) and detected using a Fusion FX7 imaging system (Vilbert Lourmat, Eberhardzell, Germany) followed by densitometric analysis (Bio1D software, Vilbert Lourmat, Eberhardzell, Germany).
Immunoprecipitation
Hct116 cells (n= 2 × 106) were seeded into 60 mm cell culture and dishes were allowed to attach for 24 h. After 4 h incubation with the compound and washing with ice-cold PBS, the cells were collected in RIPA buffer supplemented with an additional 5 mg/mL deoxycholate, 15 U/mL DNase I, proteaseinhibitor cocktail and 1 mM PMSF. After shearing with a 21-Gauge needle and additional incubation on ice, the samples were centrifuged. The protein concentration was measured using the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). Proteins of 250 μg were mixed with anti-Nrf2 antibody (1:20) and was rotated for 1.5 h and 15 rpm at 4 °C. Twenty microlitres of protein A/G plus-agarose immunoprecipitation reagent (sc-2003, Santa Cruz Biotechnology, Santa Cruz, CA) were added followed by overnight rotation at 4 °C at 15 rpm. After centrifugation, the pellet was washed with PBS and was then denatured at 95 °C in a SDS loading dye. The agarose was removed by centrifugation and the samples were subjected to SDS-PAGE with following Western Blot analysis as described earlier. Next to probing with an anti-Nrf2 antibody, an anti-ubiquitin antibody (1:250; sc-6085, Santa Cruz Biotechnology, Santa Cruz, CA) was used.
Proteasomal activity assay
Proteasomal activity was analysed using the Proteasome-Glo chymotrypsin-like, trypsin-like and caspase-like cell-based assay kit (Promega, Madison, WI) according to the protocol of the manufacturer. About 10 000 Hct116 cells/well were seeded into the wells of a white 96-well plate with a clear base and were allowed to attach for 24 h. After incubating the cells with the compound for 1 or 4 h, the medium was replaced by a 1:1 mixture of medium and substrates. Chemoluminescence was measured in a Victor2 1420 multilabel counter equipped with a dispenser (Promega, Madison, WI).
RNA isolation and reverse transcription-polymerase chain reaction
Hct116 cells/well (n = 750 000) were seeded into 6-well plates and were allowed to attach for 24 h before treating the cells with 40 μM baicalein or 0.5% DMSO, for different time points. Total RNA was isolated by a Trizol-based method (Tri®Reagent, Sigma, Steinheim, Germany). One microgram of total RNA was transcribed into complementary DNA using the M-MLV reverse transcriptase kit from Promega (Mannheim, Germany; 2 μM oligo-d(T) primers, 0.5 mM of each dNTP, 25 U ribonuclease inhibitor and 200 U reverse transcriptase in a final volume of 25 μl). Subsequent polymerase chain reaction was performed using the GoTaq® Flexi DNA-polymerase kit from Promega (Mannheim, Germany; 0.2 mM of each dNTP, 3 mM MgCl2, 0.05 U Taq DNA polymerase and 0.2 μM of each PCR primer). Thirty-two PCR cycles were carried out as follows: 1 min 95 °C, 1 min 62 °C and 50 s 72 °C. Primers for NRF2 were sense, 5′-CTG CTT TCA TAG CTG AGC CC -3′; anti-sense, 5'-CCT GAG ATG GTG ACA AGG GT-3′; primers for GAPDH were sense, 5′-ACC ACA GTC CAT GCC ATC AC-3′; anti-sense: 5′-TCC ACC ACC CTG TTG GCT GTA-3′. The amplified products were analysed by agarose gel electrophoresis and densitometric analysis was performed using the Bio1D software from Vilbert Lourmat (Eberhardzell, Germany).
Statistical analysis
Results are expressed as means ± standard deviation. Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Inc., La Jolla, CA). The minimum level of significance was p < 0.05. Statistical significance was assessed by unpaired Student's t-test with two-sided testing. When appropriate, one-way ANOVA with Dunnet's post-test was used.
Results and discussion
Effect of baicalein on the proteasomal degradation of Nrf2
We have previously shown that baicalein leads to increased Nrf2 levels in Hct116 cells, induces nuclear translocation of the transcription factor, activates the ARE and induces HO-1 (Havermann et al. Citation2013). Other studies have shown that baicalein also is an activator of Nrf2 in other cell lines: the flavonoid was able to increase the Nrf2 level and showed a transcriptional activity in a luciferase assay in human hepatoma cells (Qin et al. Citation2012) and rat pheochromocytoma cells (Zhang et al. Citation2012). Our previously published results showed that the cytosolic amount of Nrf2 remains constant (Havermann et al. Citation2013), while the total and nuclear Nrf2 protein amounts are increased by baicalein, indicating that rather than translocation of the transcription factor, its stability, expression or degradation might be of importance. Other authors have also discussed that the stabilisation of Nrf2 is of more importance than its sequestration (Nguyen et al. Citation2003).
To investigate the molecular mechanism by which this secondary plant compound increases the Nrf2 protein level, we first analysed its effect on the proteasomal activity: the 20S units of the proteasome have three major proteolytic activities: a chymotrypsin-like, a trypsin-like and a caspase-like activity. All three were analysed with specific substrates showing that treatment with baicalein (40 μM) did not modulate their activity after 1 or 4 h of incubation. The proteasomal inhibitor MG132 (1 μM) resulted in a nearly complete inhibition (97.75%) of the chymotrypsin-like activity () as well as an inhibition of 63.8% and 86.4% in case of trypsin-like and caspase-like proteasomal activity (). Incubation of Hct116 cells with MG132 strongly increased the amount of Nrf2 protein (data not shown).
Figure 2. Modulation of the cellular proteasomal activity and ubiquitinylation of Nrf2 by baicalein. Cells were incubated with baicalein (40 μM) for 1 h or 4 h before adding protease-specific substrate probes for (A) chymotrypsin-like, (B) trypsin-like or (C) caspase-like proteasomal activity according to the protocol of the manufacturer. Data are mean ± SD, n = 3, *p <0.05, **p < 0.01 versus DMSO-treated control cells, MG132 (1 μM, 4 h) was used as positive control. (D) Total protein derived from Hct116 cells treated with baicalein (40 μM) for 4 h was subjected to immunoprecipitation (anti-Nrf2 antibody) and immunostaining (anti-Nrf2 antibody and anti-ubiquitin antibody). One representative blot of three is shown with essentially the same results.
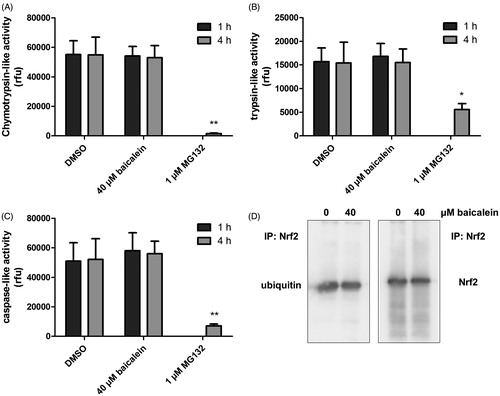
Since baicalein did not influence the general proteasomal activity, we further investigated if the compound might specifically target Nrf2 by a modulation of the ubiquitinylation status: under physiological conditions, Nrf2 is constantly marked for the proteasomal breakdown by ubiquitinylation. Interference of baicalein with this process might, therefore, be responsible for the increase in Nrf2 protein in Hct116 cells. We have applied immunoprecipitation followed by western blot to analyse the effect of baicalein on the ubiquitinylation of Nrf2. An incubation with baicalein (40 μM, 4 h) did not alter the quotient of detected Nrf2 and ubiquitin in the precipitate showing that the flavonoid does not modulate the ubiquitinylation of Nrf2 (). Similar results have been published for the flavonoid quercetin (Tanigawa et al. Citation2007). Tanigawa et al. (Citation2007) did not show a defined Nrf2–ubiquitin band but a smear what might indicate that there are differences in the ubiquitin chain length. Similar results could not be found in our study.
Another major protagonist in the process of Nrf2 degradation is the inhibitor protein Keap1 which plays the role of an intracellular sensor for ROS (Itoh et al. Citation2004). Keap1 mediates the steady ubiquitinylation of Nrf2, thereby marking the transcription factor for proteasomal degradation (Eggler et al. Citation2005). A possible scenario is a decrease in cellular Keap1 protein leading to a lower ability of the cell to ubiquitinylate Nrf2 for degradation. Zhang et al. (Citation2012) published a concentration-dependent decrease in Keap1 protein levels in rat pheochromocytoma cells starting at 100 μM baicalein. In contrast, baicalein did not alter Keap1 mRNA expression or protein in HepG2 cells, although ubiquitinylation of Keap1 was affected (Qin et al. Citation2014).
Effect of baicalein on the transcription of the NRF2 gene and the phosphorylation status of Nrf2
Cellular Nrf2 levels are regulated not only via protein turnover by the proteasome but can also be regulated via expression of the NRF2 gene. It is known that AREs are localised in the promotor region of NRF2, meaning that the activation of the pathway can be enhanced by Nrf2 itself (Kwak et al. Citation2002), what could explain the higher cellular Nrf2 protein level. This mechanism has been reported for the flavonoid quercetin in human hepatoma cells (Tanigawa et al. Citation2007). A study by Nguyen et al. (Citation2003) using the same cell line and the established Nrf2 activator tBHQ showed no increase in NRF2 transcription, thereby indicating that the activation at the transcriptional level is no general mechanism of Nrf2 activation. We have applied RT-PCR to analyse the kinetics of NRF2 transcription over 4 h of incubation with 40 μM baicalein. No increase or difference compared with the control cells was detected ().
Figure 3. Phosphorylation of Nrf2 and NRF2 expression. (A) Impact of baicalein on NRF2 expression: RNA isolated from Hct116 cells incubated with 40 μM baicalein for various timespans was subjected to RT-PCR using nrf2 and gapdh primers. One representative agarose gel of three is shown. (B) Modulation of Nrf2 phosphorylation (Ser40): western blot analysis (anti-phospho-Nrf2 antibody) of Hct116 cells incubated with baicalein for 4 h. One representative blot of three and the corresponding anti-Nrf2 stained blot are shown. (C) Data are given as ratio phospho-Nrf2:Nrf2 after densitometric analysis. Data are mean ± SD, n = 3, *p < 0.05, **p < 0.01 versus DMSO-treated control cells.
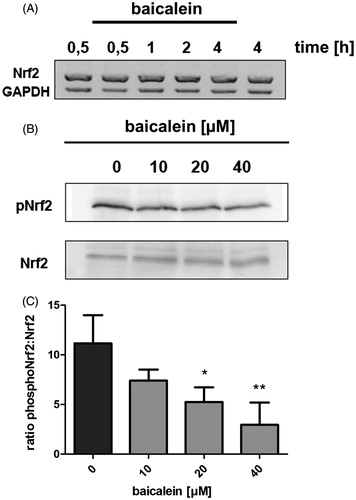
Activation of the Nrf2 pathway is often connected with phosphorylation of the transcription factor at specific residues, e.g., Ser40, by various kinases. Using a phospho-specific antibody, we have measured the quotient of phosphorylated Nrf2 (P-Nrf2) to Nrf2 in lysates from Hct116 cells treated with different concentrations of baicalein for 4 h. The detected amount of P-Nrf2 decreased in a concentration-dependent manner, also resulting in a concentration-dependent decrease of the quotient. Incubation with 40 μM baicalein results in a ratio of P-Nrf2:Nrf2 of only 26.3% compared with the DMSO-treated control value ().
It has previously been shown that activation of Nrf2 in human hepatoma cells by tBHQ is accompanied by phosphorylation of the transcription factor at Ser40 (Niture et al. Citation2009). It has been reported that phosphorylation at Ser40 leads to disruption of the Nrf2–Keap1 complex but is not necessary for stabilisation, nuclear accumulation or transcriptional activation (Bloom & Jaiswal Citation2003). However, Li et al. (Citation2012) reported that application of sulphoraphane to bladder cancer cells lead to an inhibition of Ser40 phosphorylation but stabilisation of Nrf2. We were able to show a concentration-dependent decrease of the ratio of phosphorylated (Ser40) to total Nrf2 indicating that baicalein modulates at least one kinase connected to Nrf2 activation. An interaction of baicalein with PKC and AKT but not PI3K in PC12 cells (Zhang et al. Citation2012) or MEK1/2, PI3K and JNK1/2 but not PKC and p38 in HepG2 cells (Qin et al. Citation2014) has been reported.
It might be possible that tumour cells are more sensitive to Nrf2-activating chemicals compared with non-transformed cells. To analyse if the Nrf2-modulating effect of baicalein may be specific for carcinoma cells (Hct116), which may possess a deregulated metabolism and signal transduction, we analysed the effect of baicalein on Nrf2-level in FHC immortalised normal faetal colon cells. In this cell line, incubation with baicalein (50 μM) does not increase the Nrf2 protein level (Figure S1) which leads to the speculation that baicalein selectively acts on tumour cells as discussed in other publications, e.g., Huang et al. (Citation2012). Since an activation of Nrf2 is known to increase the chemo-resistance of tumour cells (Wang et al. Citation2008), Nrf2 activators as nutritional supplements should be reviewed critically, especially during chemotherapy. However, we believe that this aspect has to be judged carefully due to the foetal origin of the cell line and a possible role of the transcription factor in development.
Antioxidative properties of the methylated baicalein derivatives
We further aimed to elucidate the structural feature of importance for the biological activity of baicalein and have, therefore, compared the effects of the three methylated baicalein derivatives baicalein-6-O-metylether (oroxylin A), baicalein-7-O-methylether (negletein) and baicalein-5,6,7-O-trimethylether (5,6,7-trimethoxyflavone) on Hct116 cells to those of our lead compound.
Oroxylin A is a compound found in Scutellaria baicalensis. An antioxidant capacity has been reported in the cell-free DPPH assay as well as the ability to suppress the LPS-induced formation of superoxide in whole blood as well as lipid peroxidation (Huang et al. Citation2006). Hu et al. (Citation2012) published that oroxylin A increased the level of reactive oxygen species, and activates the Nrf2 pathway in Hct116 colon carcinoma cells after incubation with oroxylin A (100 μM). Further, antithrombotic activities of oroxylin A in vitro and in vivo were reported (Ku et al. Citation2014) as well as inhibitory effects on colitis-associated carcinogenesis (Yang et al. Citation2013). The baicalein-7-O-methylether negletein has been identified as a degradation product after application of baicalin in blood (Chen et al. Citation2011), and is a constituent of, e.g., Desmon chinensis (Kiem et al. Citation2005) and Actinocarya tibetica Benth (Singh et al. Citation2013). Antioxidant effects in a cell-free assay have been reported (Lombardo et al. Citation2013) but to our knowledge, no studies concerning interaction with the Nrf2 pathway have been published. Baicaleintrimethylether is a flavonoid from natural origin that has been isolated from various sources (Bastos et al. Citation2009; Lan et al. Citation2011) with no published data concerning antioxidant potential as far as we know. It is known that baicalein-5,6,7-trimethylether activates peroxisomal fatty acid beta-oxidation (Morita et al. Citation2008) and suppresses pro-inflammatory mediators in lipopolysaccharide-induced RAW 264.7 macrophages (Rim et al. Citation2013).
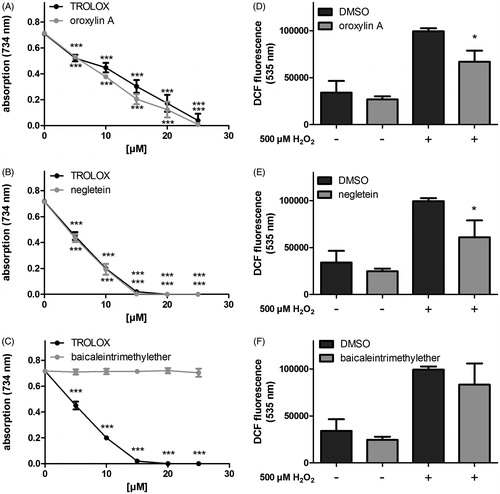
First, the toxicity of the compounds was analysed by a MTT assay showing no significant toxicity of the compounds at 50 μM after 48 h (Figure S2). We have previously been able to show that baicalein possesses a remarkable antioxidant potential (Havermann et al. Citation2013). Baicalein has been shown to be a potent antioxidant in various assays and species (Hanneken et al. Citation2006; Kang et al. Citation2012). For this feature, the three vicinal hydroxyl functions are held responsible (Kang et al. Citation2012). We have now analysed that two vicinal OH groups (or two separated OH groups) are adequate to maintain an antioxidative capacity comparable with baicalein. In the cell-free TEAC assay, both mono-methylated compounds reduced the green ABTS radical at the lowest concentration tested (5 μM), negletein being slightly more effective. The potential of oroxylin A is comparable with that of baicalein itself. Methylation of all three hydroxyl functions (baicaleintrimethylether) suppressed the antioxidant capacity in all concentrations analysed. Similar effects were observed in the DCF assay using a cellular system (Hct116 cells). While basal intracellular ROS levels were not significantly changed, both negletein and oroxylin A were able to reduce the accumulation of ROS after the application of H2O2. In coherence with the results obtained in the TEAC assay, baicaleintrimethylether was not effective (). Negletein exhibits significant antioxidative effects, which is in accordance with the finding that vicinal diols usually have the ability to scavenge radicals. Oroxylin A also possesses significant antioxidative property in both systems which is unexpected as the compound does not feature a typically antioxidant structure. The hydroxyl group at C6, which is blocked in oroxylin A, was thought to be of major importance as Marković et al. (Citation2011) calculated that dehydrogenation at this position forms the most stable radical. Nevertheless, Huang et al. (Citation2006) were also able to determine an antioxidative character in various test systems. Methylation of all three hydroxyl groups blocked the antioxidative activity as expected.
Figure 4. Antioxidative properties of the methylated baicalein derivatives. (A–C) The radical scavenging capacities were analysed in a cell free system (TEAC assay: detection of ABTS radical decolorisation, Trolox was used as positive control) and (D–F) in Hct116 cells: The intracellular ROS accumulation was measured using the fluorescent probe H2DCF-DA in cells treated with the compounds (4 h) followed by incubation with 500 μM H2O2 (1 h). DCF fluorescence as extent of oxidative stress was measured by flow cytometry (FL1-channel 530 ± 15 nm). Data are given as mean ± SD, n = 3, *p < 0.05, ***p < 0.001 versus the corresponding DMSO values.
Effects of the baicalein derivatives on Nrf2 amount and ARE activation in Hct116 cells
Epidemiological studies have shown that a diet rich in fruit and vegetables can lower the incidence of various ageing- and stress-associated diseases, due to the content of secondary plant compounds (Crozier et al. Citation2009), especially their antioxidant potential seems to be of importance. Next to radical scavenging, compounds can operate as indirect antioxidants by modulating redox-sensitive pathways that regulate the expression of several antioxidative and detoxifying enzymes as, e.g., the Nrf2 pathway (Hur et al. Citation2010; Sykiotis et al. Citation2011), and therefore strengthen natural defence mechanisms resulting in an increased resistance towards assaults. The activation of the Nrf2/ARE signalling pathway may be an important molecular parameter for the anti-inflammatory effects of baicalein; Tsai et al. (Citation2014) demonstrated that baicalein (20 mg/kg, i.p.) protects against LPS-induced acute lung injury in rats. Treatment with the flavonoid markedly attenuated LPS-induced lung oedema as well as the elevation of IL-1β, TNF-α, and IL-6. Since these effects were accompanied by an upregulation of Nrf2/HO-1 pathway, they suggested that this pathway may be involved in the inhibition of the NF-κB-mediated inflammatory responses. We have previously been able to show that baicalein increases the cellular Nrf2 level and further leads to transcriptional activity as shown using a reporter gene assay (Havermann et al. Citation2013). To identify the active substructure of baicalein needed for this biological activity, we have applied O-methylated derivatives.
Masking of the hydroxyl groups leads to a reduced reactivity which is thought to be pivotal for the ability of compounds to modulate the Nrf2 pathway. Incubation of Hct116 cells with negletein for 4 h slightly increased the protein amount at 5 μM and showed a significant increase at 50 μM in total cell lysates indicating an effect of this compound on the Nrf2 pathway, while the other derivatives did not have any effects (). O-Methylation of baicalein at position 7 (negletein) further leads to a more than three-fold higher ARE-driven luciferase activity comparable with that of baicalein itself, while oroxylin A and baicaleintrimethylether are inactive (). The weak and not statistical significant activity caused by oroxylin A was surprising as the compounds reactivity towards thiol groups should be similar to that of the flavonoid chrysin, which we found to be inactive in the reporter gene assay in Hct116 cells (data not shown). These results indicate that the critical substructure for Nrf2 activation might be the catechol structure at C5 and C6, potentially in combination with the carbonyl function at C4. This further points towards a radical mechanism as Marković et al. (Citation2011) calculated that the major radical is formed in the hydroxyl group at C6, while the most stable anion would be produced by abstraction of a proton from the hydroxyl group at C7. This study aimed to point out the flavonoid baicalein’s substructure of importance for the activation of Nrf2. Our results indicate that the catechol moiety at positions 5 and 6 is sufficient but that an additional hydroxyl function increases the compounds potency.
Figure 5. Effects of the baicalein derivatives on the cellular Nrf2 amount and ARE activation in Hct116 cells. Western blot analysis of Hct116 cells incubated with different concentrations of (A) oroxylin A, (B) negletein or (C) baicaleintrimethylether for 4 h. One representative blot of three is shown. Data are given as fold-increase of Nrf2 protein amount compared with the vehicle control (D). Mean ± SD, n = 3, *p <0.05 versus DMSO-treated control cells. (E) Modulation of ARE activation caused by the compounds: Hct116 cells were transfected with an ARE-luciferase construct followed by an incubation with the compounds (24 h). Luciferase activity as measure of ARE activation is shown, data are mean ± SD, n = 3, **p < 0.01 versus DMSO-treated control cells.
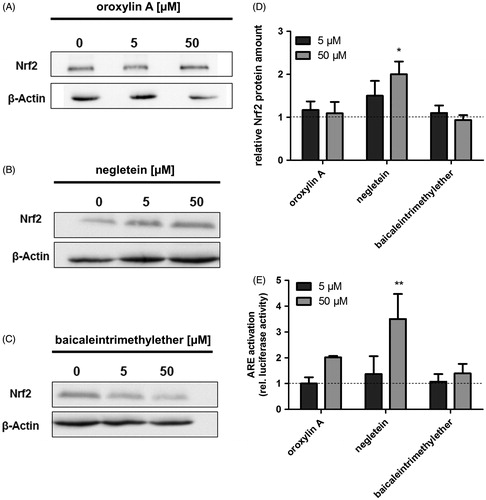
Our results obtained with the methylated derivatives show that oroxylin A and negletein possess similar antioxidative effects, which are comparable with the effects of the non-methylated compound baicalein (Havermann et al. Citation2013). Nrf2 can be activated by the redox-activity of secondary plant compounds, which may be a possible mechanism also in case of baicalein. However, we can exclude this mechanism, since the two derivatives oroxylin A and negletein showed comparable redox-active potentials (TEAC assay and DCF assay), but only negletein was able to activate the Nrf2 pathway (western blot and ARE activation) comparable with baicalein. These results therefore strengthen our hypothesis that baicalein activates Nrf2 independent of a modulation of the cellular redox potential.
Conclusion
Baicalein is an active compound in extracts derived from Scutellaria baicalensis which modulates different signalling pathways. The activation of the important antioxidative Nrf2 signalling pathway by baicalein shown in Hct116 colon carcinoma cells is not mediated via (i) inhibition of proteasomal activity, (ii) change in Nrf2 ubiquitinylation or (iii) change in the transcription of the NRF2 gene. Activation of Nrf2 by baicalein seems to be mediated via a distinct modulation of protein kinases as suggested by the decrease in Ser40 phosphorylation observed after incubation with baicalein. Experiments with methylated baicalein derivatives suggest that the redox-active potential has a minor impact on the Nrf2 activation and that concerning baicalein the catechol moiety at positions 5 and 6 seems to be sufficient. These results are useful to understand molecular mechanisms of baicalein and methylated derivatives in extracts used, e.g., in TCM.
Acknowledgements
The authors thank Ricarda Rohrig for design of the primers and Ingrid Köhler for excellent technical assistance.
Disclosure statement
The authors report that they have no conflict of interests. This work was supported by the Deutsche Forschungsgemeinschaft (DFG research training group GRK1427: “Food constituents as triggers of nuclear receptor-mediated intestinal signalling”).
References
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. 1999. Nrf2, aCap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 274:26071–26078.
- Bastos ML, Lima MR, Conserva LM, Andrade VS, Rocha EM, Lemos RP. 2009. Studies on the antimicrobial activity and brine shrimp toxicity of Zeyheria tuberculosa (Vell.) Bur. (Bignoniaceae) extracts and their main constituents. Ann Clin Microbiol Antimicrob. 8:16.
- Bloom DA, Jaiswal AK. 2003. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H: quinone oxidoreductase-1 gene expression. J Biol Chem. 278:44675–44682.
- Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. 1981. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 41:1751–1756.
- Chen L, Wang Q, Tang Y. 2011. Study of degradation behaviour of baicalin during simulative blood sample processing with solid phase extraction technology and identification of the degradation product. Drug Metab Lett. 5:276–279.
- Crozier A, Jaganath IB, Clifford MN. 2009. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 26:1001–1043.
- Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. 2001. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. 98:3404–3409.
- Eggler AL, Liu G, Pezzuto JM, van Breemen RB, Mesecar AD. 2005. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci USA. 102:10070–10075.
- Hanneken A, Lin FF, Johnson J, Maher P. 2006. Flavonoids protect human retinal pigment epithelial cells from oxidative-stress-induced death. Invest Ophthalmol Vis Sci. 47:3164–3177.
- Havermann S, Rohrig R, Chovolou Y, Humpf HU, Wätjen W. 2013. Molecular effects of baicalein in Hct116 cells and Caenorhabditis elegans: activation of the Nrf2 signaling pathway and prolongation of lifespan. J Agric Food Chem. 61:2158–2164.
- Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. 2000. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans. 28:33–41.
- Hu R, Chen N, Yao J, Zhao Q, Zhang F, Li ZY, You QD, Guo QL. 2012. The role of Nrf2 and apoptotic signaling pathways in oroxylin A-mediated responses in HCT-116 colorectal adenocarcinoma cells and xenograft tumors. Anticancer Drugs 23:651–658.
- Huang W, Lee A, Yang C. 2006. Antioxidative and anti-inflammatory activities of polyhydroxyflavonoids of Scutellaria baicalensis GEORGI. Biosci Biotechnol Biochem. 70:2371–2380.
- Huang WS, Kuo YH, Chin CC, Wang JY, Yu HR, Sheen JM, Tung SY, Shen CH, Chen TC, Sung ML, et al. 2012. Proteomic analysis of the effects of baicalein on colorectal cancer cells. Proteomics 12:810–819.
- Hur W, Sun Z, Jiang T, Mason DE, Peters EC, Zhang DD, Luesch H, Schultz PG, Gray NS. 2010. A small-molecule inducer of the antioxidant response element. Chem Biol. 17:537–547.
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76–86.
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379–391.
- Itoh K, Tong KI, Yamamoto M. 2004. Molecular mechanism activating Nrf2–Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 36:1208–1213.
- Jaiswal AK. 2004. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 36:1199–1207.
- Kang KW, Choi SH, Kim SG. 2002. Peroxynitrite activates NF-E2-related factor 2/antioxidant response element through the pathway of phosphatidylinositol 3-kinase: the role of nitric oxide synthase in rat glutathione S-transferase A2 induction. Nitric Oxide 7:244–253.
- Kang KA, Zhang R, Piao MJ, Chae S, Kim HS, Park JH, Jung KS, Hyun JW. 2012. Baicalein inhibits oxidative stress-induced cellular damage via antioxidant effects. Toxicol Ind Health. 28:412–421.
- Kiem PV, Minh CV, Huong HT, Lee JJ, Lee IS, Kim YH. 2005. Phenolic constituents with inhibitory activity against NFAT transcription from Desmos chinensis. Arch Pharm Res. 28:1345–1349.
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 24:7130–7139.
- Kobayashi M, Yamamoto M. 2006. Nrf2–Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 46:113–140.
- Ku SK, Lee IC, Bae JS. 2014. Antithrombotic activities of oroxylin A in vitro and in vivo. Arch Pharm Res. 37:679–686.
- Kwak MK, Itoh K, Yamamoto M, Kensler TW. 2002. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2promoter. Mol Cell Biol. 22:2883–2892.
- Lan YH, Leu YL, Peng YT, Thang TD, Lin CC, Bao BY. 2011. The first bis-retrochalcone from Fissistigma latifolium. Planta Med. 77:2019–2022.
- Li C, Zhou L, Lin G, Zuo Z. 2009. Contents of major bioactive flavones in proprietary traditional Chinese medicine products and reference herb of Radix Scutellariae. J Pharm Biomed Anal. 50:298–306.
- Li Y, Paonessa JD, Zhang Y. 2012. Mechanism of chemical activation of Nrf2. PLoS One 7:e35122.
- Lombardo E, Sabellico C, Hájek J, Staňková V, Filipský T, Balducci V, De Vito P, Leone S, Bavavea EI, Silvestri IP, et al. 2013. Protection of cells against oxidative stress by nanomolar levels of hydroxyflavones indicates a new type of intracellular antioxidant mechanism. PLoS One 8:e60796.
- Marković ZS, Dimitrić Marković JM, Milenković D, Filipović N. 2011. Mechanistic study of the structure-activity relationship for the free radical scavenging activity of baicalein. J Mol Model 17:2575–2584.
- Morita M, Kanai M, Mizuno S, Iwashima M, Hayashi T, Shimozawa N, Suzuki Y, Imanaka T. 2008. Baicalein 5,6,7-trimethyl ether activates peroxisomal but not mitochondrial fatty acid beta-oxidation. J Inherit Metab Dis. 31:442–449.
- Motohashi H, Yamamoto M. 2004. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 10:549–557.
- Niture SK, Jain AK, Jaiswal AK. 2009. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J Cell Sci. 122:4452–4464.
- Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. J Biol Chem. 278:4536–4541.
- Purdom-Dickinson SE, Sheveleva EV, Sun H, Chen QM. 2007. Translational control of nrf2 protein in activation of antioxidant response by oxidants. Mol Pharmacol. 72:1074–1081.
- Qin S, Chen J, Tanigawa S, Hou DX. 2012. Gene expression profiling and pathway network analysis of hepatic metabolic enzymes targeted by baicalein. J Ethnopharmacol. 140:131–140.
- Qin S, Deng F, Wu W, Jiang L, Yamashiro T, Yano S, Hou DX. 2014. Baicalein modulates Nrf2/Keap1 system in both Keap1-dependent and Keap1-independent mechanisms. Arch Biochem Biophys. 559:53–61.
- Rim HK, Yun CH, Shin JS, Cho YW, Jang DS, Ryu JH, Park H, Lee KT. 2013. 5,6,7-Trimethoxyflavone suppresses pro-inflammatory mediators in lipopolysaccharide-induced RAW 264.7 macrophages and protects mice from lethal endotoxin shock. Food Chem Toxicol. 62:847–855.
- Rushmore TH, Morton MR, Pickett CB. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 266:11632–11639.
- Sekhar KR, Soltaninassab SR, Borrelli MJ, Xu ZQ, Meredith MJ, Domann FE, Freeman ML. 2000. Inhibition of the 26S proteasome induces expression of GLCLC, the catalytic subunit for gamma-glutamylcysteine synthetase. Biochem Biophys Res Commun. 270:311–317.
- Siddiqui KM, Chopra DP. 1984. Primary and long term epithelial cell cultures from human fetal normal colonic mucosa. In Vitro 20:859–868.
- Sykiotis GP, Habeos IG, Samuelson AV, Bohmann D. 2011. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr Opin Clin Nutr Metab Care. 14:41–48.
- Singh B, Sidiq T, Joshi P, Jain SK, Lawaniya Y, Kichlu S, Khajuria A, Vishwakarma RA, Bharate SB. 2013. Anti-inflammatory and immunomodulatory flavones from Actinocarya tibetica Benth. Nat Prod Res. 27:2227–2230.
- Tanigawa S, Fujii M, Hou DX. 2007. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. 42:1690–1703.
- Tsai CL, Lin YC, Wang HM, Chou TC. 2014. Baicalein, an active component of Scutellaria baicalensis, protects against lipopolysaccharide-induced acute lung injury in rats. J Ethnopharmacol. 153:197–206.
- Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. 2008. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29:1235–1243.
- Wasserman WW, Fahl WE. 1997. Functional antioxidant responsive elements. Proc Natl Acad Sci USA. 94:5361–5366.
- Yang X, Zhang F, Wang Y, Cai M, Wang Q, Guo Q, Li Z, Hu R. 2013. Oroxylin A inhibits colitis-associated carcinogenesis through modulating the IL-6/STAT3 signaling pathway. Inflamm Bowel Dis. 19:1990–2000.
- Zhang Z, Cui W, Li G, Yuan S, Xu D, Hoi MP, Lin Z, Dou J, Han Y, Lee SM. 2012. Baicalein protects against 6-OHDA-induced neurotoxicity through activation of Keap1/Nrf2/HO-1 and involving PKCα and PI3K/AKT signaling pathways. J Agric Food Chem. 60:8171–8182.
- Zipper LM, Mulcahy RT. 2000. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 278:484–492.