Abstract
When located in the DNA minor groove, dimeric bisbenzimidazoles DB(n) effectively inhibited in vitro the Dnmt3a catalytic domain (IC50 5–77 μM). The lowest IC50 value was observed for compound DB(11) with an 11-unit methylene linker joining the bisbenzimidazole fragments. Increased time of incubation of DNA with DB(n) as well as the presence of AT-clusters in DNA enhances the inhibitory effect.
Keywords::
Introduction
The DNA methyltransferase (MTase) Dnmt3a (EC 2.1.1.37) belongs to the family of mammalian Dnmt3Citation1,Citation2. MTase Dnmt3a can recognize a DNA СрG site and methylate the carbon 5 of cytosine residue (DNA-(cytosine C5)-methyltransferase (C5-MTase)) thus forming the de novo methylation profileCitation3. Also, Dnmt3a together with MTase Dnmt1 is involved in maintenance DNA methylationCitation4–6. X-ray studies revealed that complex C-terminal domain of Dnmt3a with its regulatory factor Dnmt3L exists as a 2:2 heterotetramerCitation7. The importance of tetramer formation for S-adenosyl-l-methionine (AdoMet) binding, DNA binding and methylation has been shownCitation7. The Dnmt3a-Dnmt3L complexes polymerize on the DNA forming nucleoprotein filamentCitation8.
The correct distribution of methylated СрG sites in the genome is crucial for cell development. One of the mechanisms of tumour cell formation involves inactivation of tumour suppressor genes due to de novo hypermethylation of promoter СрG islands of these genes in the process of carcinogenesisCitation9–11. As changes in the DNA methylation status are affected by MTase functioning, much attention was given to inhibition of these enzymesCitation12,Citation13.
The DNA methylation process includes the formation of a covalent bond between a cysteine residue of the enzyme active site and the C6 position of the target cytosine in DNACitation14. At present the so called mechanism-based inhibitors (5-aza-2′-deoxycytidine, 5-fluo-2′-deoxycytidine, and 2-pyrimidinone-1-β-d-2′-deoxyribofuranoside) are the most effective inhibitors of C5-MTasesCitation12,Citation13. In this case inhibition is irreversible and the formed covalent adducts either cannot be degraded or their degradation proceeds extremely slowly. However, because of incorporation of these compounds into DNA, they are very toxic and mutagenic in vivo. Therefore the search of new effective MTase non-nucleoside inhibitors is an actual task.
Several low molecular weight non-nucleoside MTase inhibitors are known to date: epigallocatechin 3-gallate and genistein (4′,5,7-trihydroxyisoflavone), whose IC50 towards Dnmt1 are 20 and 67 μM, respectivelyCitation15, disulfide derivatives of l-bromtyrosine (psammaplins; IC50 3,4-18,8 nM for Dnmt1)Citation16, and l-tryptophan derivative RG108 [2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-3-(1H-indol-3-yl)propionic acid] (IC50 115 nM for DNA methyltransferase SssI (M.SssI))Citation17.
Based on the structure of the complex of prokaryotic MTase HhaI with DNA and S-adenosyl-l-homocysteine, most of the DNA-MTase contacts providing specific MTase binding to DNA occurs in the major groove of the double helix, and transfer of the methyl group to the target cytosine is accompanied by motion of the MTase catalytic loop towards the minor groove side of the DNA substrateCitation18. Hence, DNA ligands capable of disturbing this motion may be effective MTase inhibitors. One of such ligands is Hoechst 33258, a bisbenzimidazole dye, which can bind to DNA AT sites in the minor groove with a binding constant 108–109 M−1 Citation19,Citation20.
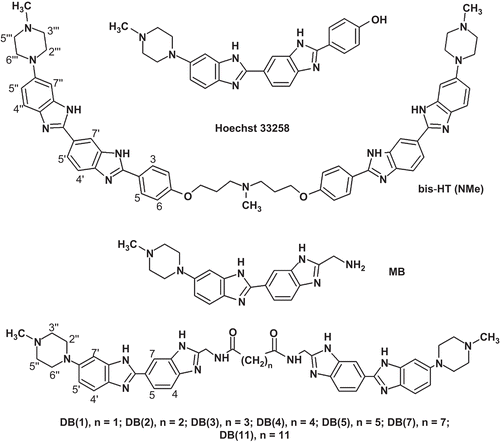
In this work we tested, as murine catalytic domain of Dnmt3a (Dnmt3a-CD) inhibitors, a series of dimeric benzimidazoles DB(n), namely, DB(1–5,7,11), differing in the length of methylene linkers joining bisbenzimidazole fragments and bis-HT(NMe), which contains two Hoechst 33258 fragments joined by means of a linker containing six methylene groups and one N(Me) group ().
Materials and methods
General experimental
Reagents and solvents for syntheses were reagent-grade and were used without further purification. AdoMet, (32 mM stock solution) was from New England Biolabs. Buffer A (10 mM Tris-HCl (pH 7.9), 50 mM NaCl), B (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.0), 100 mM KCl, 1 mM ethylenediaminetetraacetic acid, and 0.2 mM 2-mercaptoethanol), C (10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 0.2 mM 2-mercaptoethanol), and Tango (Fermentas, Vilnius, Lithuania) were used.
Compound DB(5) was synthesized according to reported proceduresCitation21. Oligonucleotides 5′-FAM-CTGAATACTACTTGCGCTCTCTAACCTGAT, 5′ ATCAGGTTAGAGAGCGCAAGTAGTATTCAG, 5′-FAM-TGGACACCACCTGCGCTCTCTGACCTGAC, and 5′-GTCAGGTCAGAGAGCGCAGGTGGTGTCCAG were from Syntol (Moscow, Russia). The fluorescent label 6(5)-carboxyfluorescein (FAM) is joined with the oligodeoxyribonucleotides 5′-end with a (CH2)6-linker. DNA duplexes were prepared by mixing of FAM-labelled strand with 1.1-fold excess of complementary strand up to 5 µM duplex concentration in buffer A, heating to 85°C and allowing the sample to cool slowly over several hours to room temperature.
The murine C-terminal Dnmt3a-CD, which retained its catalytic activity in the absence of the N-terminal regulatory domainCitation22, was used. The plasmid pET28a containing the sequence encoding murine Dnmt3a-CD was provided by A. Jeltsch. Recombinant Dnmt3a-CD which contains amino acid residues 608-908 and the (His)6 cluster at the N-terminus was expressed in the Escherichia coli BL21(DE3) strain (Novagen, Merck KGaA, Darmstadt, Germany). Dnmt3a-CD was purified to ~95% purity by affinity chromatography using Co2+ containing TALON affinity resin (Clontech, CA, USA) as described inCitation23. Restriction endonuclease HhaI (R.HhaI) was purchased from Fermentas (Vilnius, Lithuania).
Hydrogen-nuclear magnetic resonance (1H-NMR) Spectra; BrukerAMX-400 spectrometer (Rheinstetten, Germany), in (D6)dimethyl sulfoxide; δ in ppm rel. to the residual solvent signal, J in Hz. Matrix assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI TOF-MS): time-of-flow Vision-2000 mass spectrometer (ThermoBioanalysis, Hemel Hempstead, UK) with registration of positive ions; the matrix was 2,5-dihydroxybenzoicacid; N2 laser, 337 nm; m/z.
Synthesis of MB, DB(1–5, 7, 11), and bis-HT(NMe)
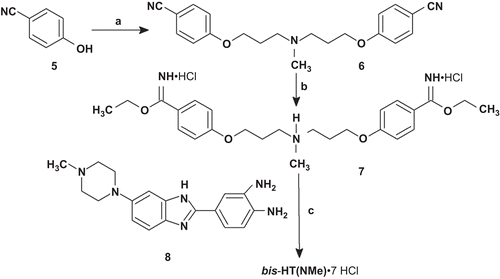
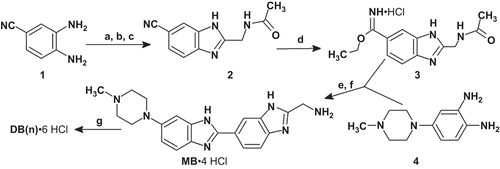
Compound DB(5) was synthesized as described in ref. Citation21. Monomeric bisbenzimidazole (MB) was prepared by coupling of 4-(4-methylpiperazin-1-yl)benzene-1,2-diamine 4Citation24 and imidate 3 followed by removal of an N-acetyl protective group (scheme 1). Compound 3 was obtained by the Pinner reaction from nitrile 2, which was synthesized from 3,4-diaminobenzonitrile 1Citation25 by coupling/cyclization with tert-butyloxycarbonyl-glycine (Boc-Gly-OH). Dimerization of MB molecules by coupling with the corresponding reagents led to the formation of compounds DB(1–4,7,11; ). Compound bis-HТ(NMe) was synthesized according to starting from the previously described diamine 8Citation21 and diimidate 7. Compound 7 was obtained by the Pinner reaction from dinitril 6. Compound 6 was prepared from 4-hydroxybenzonitrile 5 by coupling with 3-chloro-N-(3-chloropropyl)-N-methylpropan-1-amine. The structures of the synthesized compounds were confirmed by NMR spectroscopy and mass-spectrometry data.
Scheme 1. Synthesis of DB(1–5,7,11). Reagents and conditions: (a) iBuOCOCl, NMM, Boc-Gly-OH; (b) AcOH, 65–70°C, 1 h; (c) AcOH, 120°C, 4 h, 58% (for a–c); (d) HClgas/EtOH, 0–4°C, 1 h, 23°C, 3 days; (e) AcOH/EtOH, 95°C, 1 h, N2, 78% (for d–e); (f) HClconc, 105°C, 20 min, 89%; (g) for DB(1,2): DMF, DIPEA, BOP, HOOC-(CH2)n-COOH (n = 1,2), 0°C, 1 h, 23°C, 1 day, HCl/MeOH, DB(1), 22%, DB(2), 24%; for DB(3–5,7,11): DMF, Et3N, XOOC-(CH2)n-COOX (X = Np; Pfp; Su; n = 3,4,5,7,11), HCl/MeOH, DB(3), 73%, DB(4), 68%, DB(5), 51%, DB(7), 78%, DB(11), 61%. All DB(n) were purified by refluxing in methanol followed by cooling and filtration of the target precipitate. AcOH, acetic acid; Boc-Gly-OH, tert-butyloxycarbonyl-glycine; BOP, (benzotriazole-1-yloxy)tris(dimethylamino)phosphonium hexafluorophosphate; DIPEA, N,N-diisopropylethylamine; DMF, dimethylformamide; Et3N, triethylamine; EtOH, ethanol; iBuOCOCl, isobutyl chloroformate; MeOH, methanol; NMM, N-methylmorpholine; Np, p-nitrophenyl; Pfp, 2,3,4,5,6-pentafluorophenуl; Su, N-oxysuccinimide.
NMR and mass spectra characteristics of MB, DB(1–5, 7, 11), and bis-HT(NMe)
MB·4 HCl: 1H-NMR (400 MHz, 97°C): δ 2.83 (3H, s, CH3); 3.36 (4H, m, H(3′″, 5″)); 3.58 (4H, m, H(2″,6″)); 4.40 (2H, s, СН2NH2); 7.19 (1H, d, J = 8.7, Н5′); 7.24 (1H, s, H7′); 7.65 (1H, d, J = 9.3, Н4′); 7.80 (1H, d, J = 8.1, Н4); 8.23 (1H, d, J = 8.7, Н5); 8.67 (1H, s, H7). MALDI TOF-MS (monoisotop.): calcd for C20H23N7: 361.40; found: m/z (M)+ 361.3.
DB(1)·6 HCl: 1H-NMR (400 MHz, 32°C): δ 2.84 (6H, brs, CH3); 3.23 (8H, m, H(3″, 5″)); 3.48 (2H, s, COCH2); 3.54 (4H, m, H(2″, 6″)); 3.88 (4H, m, H(2″, 6″)); 4.79 (4H, d, J = 5.0, CH2NH); 7.19 (2H, brs, H7′); 7.34 (2H, d, J = 9.3, H5′); 7.69 (2H, d, J = 8.7, H4′); 7.92 (2H, d, J = 8.7, H4); 8.34 (2H, d, J = 8.7, H5); 8.75 (2H, s, H7); 9.19 (2H, t, J = 5.3, CONH); 11.23 (2H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C43H16N14O2: 790.92; found: m/z (M + H)+ 793.4, (M + Na + K) + 855.7.
DB(2)·6 HCl: 1H-NMR (400 MHz, 32°C): δ 2.63 (4H, s, CH2CO); 2.84 (6H, brs, CH3); 3.23 (8H, m, H(3″, 5″)); 3.54 (4H, m, H(2″, 6″)); 3.87 (4H, м, (2″, 6″)); 4.74 (4H, d, J = 5.6, CH2NH); 7.17 (2H, brs, H7′); 7.32 (2H, d, J = 8.7, H5′); 7.68 (2H, d, J = 9.3, H4′); 7.87 (2H, d, J = 8.7, H4); 8.32 (2H, d, J = 8.7, H5); 8.72 (2H, s, H7); 8.99 (2H, t, J = 5.6, CONH); 11.24 (1H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C44H48N14O2: 804.94; found: m/z (M + H)+ 806.3.
DB(3)·6 HCl: 1H-NMR (400 MHz, 23°C): δ 1.86 (2Н, quintet, J = 7.5, СН2СН2СН2), 2.33 (4Н, t, J = 7.5, СН2СО), 2.84 (6Н, d, J = 3.7, CH3), [3.38 (8H, m, H(3″,5″)), 3.58 (8H, m, H(2″,6″)—at 97°С)], 4.72 (4Н, d, J = 5.0, СН2NH), 7.18 (2H, brs, H7′), 7.34 (2Н, dd, J = 1.9, J = 9.3, H5′), 7.70 (2Н, d, J = 9.3, H4′), 7.94 (2Н, d, J = 8.7, H4), 8.34 (2Н, d, J = 8.7, H5), 8.73 (2Н, s, H7), 8.97 (2Н, t, J = 5.3, CONН), 11.13 (2H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C45H50N14O2: 818.88; found: m/z (M + H)+ 820.2.
DB(4) 6 HCl: 1H-NMR (400 MHz, 32°C): δ 1.60 (4H, м, CН2CH2CO), 2.32 (4H, м, CH2CO), 2.83 (6H, brs, CH3), 3.24 (8H, m, H(3″,5″)), 3.53 (4H, m, H(2″,6″)), 3.86 (4H, m, H(2″,6″)); 4.77 (4H, d, J = 5.0, СН2NH), 7.18 (2H, brs, H7′), 7.33 (2H, d, J = 9.3, H5′), 7.69 (2Н, d, J = 8.7, H4′), 7.96 (2Н, d, J = 8.7, H4), 8.47 (2Н, d, J = 9.3, H5); 8.84 (2Н, s, H7), 8.95 (2Н, t, J = 5.3, CONН), 11.46 (1H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C46H52N14O2: 832.91; found: m/z (M + H)+ 833.7.
DB(5) 6 HCl: 1H-NMR (400 MHz, 32°C): δ 1.32 (2Н, quintet, J = 7.5, CO(CH2)2CН2), 1.58 (4Н, quintet, J = 7.5, COCH2CH2), 2.26 (4H, t, J = 7.5, COCH2,), 2.85 (6H, s, CH3), [3.39 (8H, m, H(3″,5″)); 3.78 (8H, m, H(2″,6″)—at 84°C], 4.63 (4H, d, J = 5.6, CH2NH,), 7.20 (2H, d, J = 1.9, H7′), 7.31 (2H, dd, J = 1.9, J = 8.7, H5′), 7.68 (2H, d, J = 8.7, H4′), 7.89 (2H, d, J = 8.7, H4), 8.26 (2H, d, J = 8.7, H5), 8.66 (2H, s, H7), 8.68 (2H, t, J = 5.3, CONH), 11.03 (1H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C47H54N14O2: 847.02; found: m/z (M + H)+ 848.3.
DB(7) 6 HCl: 1H-NMR (400 MHz, 32°C): δ 1.28 (6Н, m, CO(CH2)2(CН2)3), 1.55 (4Н, m, COCH2CH2), 2.23 (4H, t, J = 7.5, COCH2), 2.83 (6H, s, CH3), [3.38 (8H, m, H(3″,5″)); 3.55 (8H, m, H(2″,6″))—at 84°C], 4.58 (4H, d, J = 5.0, CH2NH), 7.19 (2H, s, H7′), 7.26 (2H, d, J = 8.7, H5′), 7.66 (2H, d, J = 8.7, H4′), 7.80 (2H, d, J = 8.1, H4), 8.20 (2H, d, J = 8.1, H5), 8.60 (4H, m, CONH, H7), 11.19 (1H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C49H58N14O2: 875.08; found: m/z (M + H)+ 876.5.
DB(11)·6 HCl: 1H-NMR (400 MHz, 23°C): δ 1.22 (14Н, m, СО(СН2)2(СН2)7), 1.52 (4Н, m, СОСН2СН2), 2.23 (4Н, t, J = 7.2, СОСН2), 2.83 (6Н, brs, СH3), 3.21 (8H, m, H(3″, 5″)), 3.53 (4H, m, H(2″,6″)), 3.88 (4H, m, H(2″,6″)), 4.66 (4Н, d, J = 5.0, СН2NH), 7.19 (2H, s, H7′), 7.33 (2Н, dd, J = 1.2, J = 8.7, H5′), 7.70 (2Н, d, J = 8.7, H4′), 7.91 (2Н, d, J = 8.7, H4), 8.34 (2Н, d, J = 8.7, H5), 8.73 (2Н, s, H7), 8.78 (2Н, t, J = 5.3, CONН), 11.19 (2H, brs, NH(Bim)). MALDI TOF-MS (monoisotop.): calcd for C53H66N14O2: 931.18; found: m/z (M + H)+ 931.8.
bis-HT(NMe)·7 HCl: 1H-NMR (400 MHz, 23°C): δ 2.25 (4H, quint, J = 6.5, OСН2СН2); 2.85 (9H, brs, CH3); 3.12 (8H, m, H(3′″, 5′″)); 3.35 (4H, m, СН2N); 3.36 (8H, m, H(2″′, 6″′)); 4.23 (4H, m, OСН2); 7.18 (6H, m, H(2, 6′, 7″)); 7.28 (2H, dd, J = 1.9, J 9.3, H5″); 7.67 (2H, d, J = 9.3, H4″); 7.86 (2H, d, J = 8.7, H4′); 8.24 (2H, d, J = 8.7, H5′); 8.30 (4H, d, J = 8.7, H(3, 5)); 8.62 (2H, s, H7′). MALDI TOF-MS (monoisotop.): calcd for C57H61N13O2: 960.09; found: m/z (M + H)+ 961.0.
Enzyme inhibition assay
Duplexes A or B (300 nM) were incubated in the presence of increasing concentrations of a DB(n) inhibitor (0.1–200 µM) in buffer B (for Dnmt3a-CD) or C (for M.SssI) for 3 days at 4°C or for 10 min at 25°C. Dnmt3a-CD (2 µM of monomers) or M.SssI (2 µM) and AdoMet (25 µM) were added to the reaction mixtures and the mixtures were incubated for 40 min at 37°C. Reaction mixtures lacking either the enzyme or the inhibitor were used as controls. The DNA was precipitated with ethanol (EtOH) in the presence of 0.4 M sodium acetate. The precipitate was washed with 80% EtOH and evaporated on a SpeedVac to remove remained EtOH. The methylation was studied by protection of the methylated DNA from cleavage by 2 U of R.HhaI in the 20 µL Tango buffer for an hour at 37°C. The cleavage products were suspended in 10 µL 80% formamide and separated in 20% polyacrylamide gel under denatured conditions (7 M urea) followed by gel imaging on a FUJIFILM FLA-3000 device.
The methylation % (M) was determined as the ratio of methylated to total DNA based on the intensities of the corresponding bands in gel using the Image Quant 5.0 program. To determine inhibitor concentration which reduced enzyme activity by 50% (IC50) the dependence of relative methylation percentage (R, %) versus inhibitor concentration were plotted. R values were calculated from the formula:
The curves were subjected to regression analysis in Origin 6.0 program using simple exponential function:
where input parameters were R and inhibitor concentration, [I] (µM); R0, A, and t were dependent parameters for fitting. All data were fitted with correlation coefficients more than 0.95. The IC50 values were then determined by interpolation. For DB(4,5) when IC50 values exceed the inhibitor concentration range, they were estimated by extrapolation of the above mentioned function to 50% inhibition. The values of standard errors were calculated using 3–6 independent experiments.
Results and discussion
The inhibitory activities of DB(n) and bis-HТ(NMe) were studied in the reaction of Dnmt3a-CD catalyzed methylation of a 30-mer DNA duplex: 5′-FAM-CTGAATACTACTTGCGCTCTCTAACCTGAT, 3′-GACTTATGATGAACGCGAGAGATTGGACTA (A) containing a CpG site (shown in bold; cytosine residues to be methylated are underlined, clusters of AT pairs are indicated by larger letters) and a fluorescent FAM label at the 5′-end of the upper strand. The pre-incubation time of DB(n) with the 30-mer DNA duplex A, varied from 10 min to 3 days. Compounds bis-HТ(NMe) and DB(1–5,7,11) demonstrated inhibitory activity at micromolar concentrations (). The increase of incubation time resulted in 3–9-times decrease of IC50 value.
Table 1. Inhibition of the methylation of duplex A by Dnmt3a-CD in the presence of DB(n).
Compound bis-HТ(NMe) could form with AT-rich DNA complexes of three types depending mainly on the incubation timeCitation26. Type I complexes were formed by bis-HТ(NMe) in the open linear form; type II complexes were formed by intramolecular sandwiches and type III, by dimers of intramolecular sandwiches. Within complexes I, II, and III, the bis-HТ(NMe) occupies on DNA 10–11, 5–6, and 2–3 bp, respectivelyCitation26. In the case of immediate addition to bis-HТ(NMe) of a large DNA excess the formation of type III complex was observedCitation26. This type of complex is metastable and it rearranges slowly to the mixture of type I and II complexes. At bis-HТ(NMe) concentrations used in the experiments type I and II complexes were prevalent in the solution in 3 days, after thermodynamic equilibrium had been set. The inhibitory effect of bis-HТ(NMe) () was higher in the case of types I and II complexes (3 days incubation) than it was in the case of type III complex (10 min incubation). One can suggest that compounds DB(1–5,7,11) could form types I and II complexes which displayed higher inhibitory effects under the conditions of long-term incubation.
It is noteworthy that MB () only negligibly inhibited Dnmt3a-CD even under prolonged incubation with DNA (the enzymatic activity was reduced by 7% in the presence of 200 μM MB; data not shown). This fact supported the hypothesis that it was dimerization of MB molecules that provided the formation of DB(n) molecules with high inhibitory activity.
The observed inhibitory activities of DB(n) were affected by the length of methylene linkers (). DB(n) with the shortest (DB(1–3)) and the longest (DB(11)) methylene linkers showed the lowest IC50 values.
It is known that a bisbenzimidazole molecule binds preferably to the DNA fragment containing two AT or TA pairsCitation19. For binding of DB(n) two such fragments are required. Several AT-clusters in duplex A located at different distances relative to each other can be involved in the DB(n) binding. One can suppose that the most effective binding occurs when the distance between AT-clusters corresponds to the length of the methylene linkers joining bisbenzimidazole fragments. For example, the 5′-TACTTGCG/3′-ATGAACGC…fragment of duplex A would be preferable for binding to DB(1–3) and the 5′-TACTACTTGCG/3′-ATGATGAACGC fragment of duplex A would be preferable for binding to DB(11). It is likely that a high IC50 value for binding of DB(4,5) may be explained by a non-optimal distance between AT-clusters in duplex A (). Within the tested DB(n) bisbenzimidazoles, DB(11) was the most effective inhibitor with the IC50 value close to that of bis-HТ(NMe) (). The distance between bisbenzimidazole fragments in these molecules is also similar.
Compound bis-HТ(NMe) occupies 10–11 bp in the DNA minor groove in open linear formCitation26. Therefore, high inhibitory properties of DB(11) and bis-HТ(NMe) seem to be due to the open linear conformation of DB(n) in the complex with DNA.
For evaluation if the inhibition effect depends on the presence of AT-clusters in DNA, we obtained duplex B lacking AT-clusters: 5′-FAM-TGGACACCACCTGCGCTCTCTGACCTGAC, 3′-GACCTGTGGTGGACGCGAGAGACTGGACTG (B). The IC50 value of the DB(11) complex with duplex B (49.0 ± 2.0 μM) was one order of magnitude higher than that of the complex of DB(11) with duplex A (5.0 ± 1.5 μM). Therefore, AT-clusters within DNA are necessary for the effective inhibition of the DNA methylation by Dnmt3a. However, weak inhibition might be possible for any nucleotide sequence as it is observed for duplex B whose IC50 value is still in micromolar range.
With the goal to explain the mechanism of Dnmt3a inhibition we took into account the minor groove localization of DB(n). This assumption is based on the fact that Hoechst 33258, a MB derivative, is known to be located in the minor groove of the d(CGCGAATTCGCG)2 double helixCitation19,Citation27. In addition, disposition of Hoechst 33258 in the DNA minor groove was demonstrated for type I and II complexes with poly(dG-dC)·poly(dG-dC)Citation28. Recently we found that the presence of a bulky benzo[a]pyrene residue in the DNA minor groove caused a dramatic decrease in prokaryotic MTases SssI and HhaI methylating capacity because of distortion of DNA contacts with the enzyme catalytic loopCitation29,Citation30. Superimposition of the DNA methyltransferase HhaI (M.HhaI–DNA) complex structure onto the Dnmt3a-CD structure showed high similarity of M.HhaI and Dnmt3a-CD catalytic loop conformationsCitation7. Thus, the observed DB(n) inhibitory effects might be explained by perturbation of minor groove-Dnmt3a loop interactions due to the presence of a bulky ligand. This type of inhibition may be considered as the substrate protection mechanism.
In the case of addition of DB(11) inhibitor to the pre-incubated DNA-Dnmt3a-CD complex we did not observe significant reduce of methylation activity (data not shown). This fact suggests competition between DB(n) molecules and enzyme for DNA binding to DNA minor groove.
Also, we studied if the observed inhibitory effect of DB(n) was only inherent for Dnmt3a-CD. The interaction of M.SssI with duplex A was studied in the presence of DB(11) (). We showed that DB(11) inhibited M.SssI (IC50 4.6 ± 0.6 μM) at the same concentrations as Dnmt3a-CD (IC50 5.0 ± 1.5 μM). It is noteworthy that MBs could inhibit human topoisomerase ICitation31, DNA repair enzyme O6-alkylguanine-DNA alkyltransferaseCitation32, and (A)BC excinuclease from E. coliCitation33 and DB(5) could inhibit HIV integraseCitation21. We suppose that inhibition of the above DNA operating enzymes is attained via the substrate protection mechanism.
To summarize, we first found that compounds DB (1–5,7,11) are in vitro inhibitors of Dnmt3a-CD at low micromolar concentrations which is consistent with concentrations reported by others for different inhibitors of C5-MTasesCitation15,Citation34. It is notable that the same range of concentrations (5–50 µM) of non-nucleoside MTase inhibitor was used in the case of cell culture assaysCitation15. It is noteworthy that the use of DB(n) as in vivo MTase inhibitors is promising due to their capacity to penetrate through cell and nuclear membranes of living cells and bind to chromatinCitation35.
Acknowledgement
The authors thank Dr. A. Jeltsch for kind gift of the Dnmt3a-CD plasmid, Dr. B. Jack from New England Biolabs for kind gift of the M.SssI plasmid, Dr. O. Kirsanova and E. Tamyar for the procedure of DNA methylation.
Declaration of interest
The work was supported by RFBR grants 10-04-00809a, 08-04-01096a, 08-03-12148-ofi; RFBR Grant 08-04-91109/CRDF RUB1-2919-MO-07 and the Program of Presidium of RAS on Molecular and Cell Biology.
References
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996;122:3195–3205.
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998;19:219–20.
- Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol 2004;60:55–89.
- Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. Embo J 2002;21:4183–95.
- Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem 2002;269:4981–84.
- Métivier R, Gallais R, Tiffoche C, Le Pé,ron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature 2008;452:45–50.
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 2007;449:248–51.
- Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res 2008;36:6656–63.
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–28.
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 2001;10:687–92.
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683–92.
- Gowher H, Jeltsch A. Mechanism of inhibition of DNA methyltransferases by cytidine analogs in cancer therapy. Cancer Biol Ther 2004;3:1062–8.
- Kirsanova OV, Cherepanova NA, Gromova ES. Inhibition of C5-cytosine-DNA-methyltransferases. Biochemistry (Mosc) 2009;74:1175–86.
- Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA 1984;81:6993–7.
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 2003;63:7563–70.
- Piña IC, Gautschi JT, Wang GY, Sanders ML, Schmitz FJ, France D, Cornell-Kennon S, Sambucetti LC, Remiszewski SW, Perez LB, Bair KW, Crews P. Psammaplins from the sponge Pseudoceratina purpurea: inhibition of both histone deacetylase and DNA methyltransferase. J Org Chem 2003;68:3866–73.
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res 2005;65:6305–11.
- Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 1994;76:357–69.
- Teng MK, Usman N, Frederick CA, Wang AH. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res 1988;16:2671–90.
- Loontiens FG, Regenfuss P, Zechel A, Dumortier L, Clegg RM. Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)], and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry 1990;29:9029–39.
- Ivanov AA, Strel’tsov SA, Prikazchikova TA, Gottikh MB, Zhuze AL. Synthesis and properties of a symmetric dimeric bisbenzimidazole, a DNA-specific ligand. Bioorg Khim 2008;34:285–8.
- Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem 2002;277:20409–14.
- Maltseva DV, Baykov AA, Jeltsch A, Gromova ES. Impact of 7,8-dihydro-8-oxoguanine on methylation of the CpG site by Dnmt3a. Biochemistry 2009;48:1361–68.
- Loewe H, Urbanietz J. Basic-substituted 2,6-bisbenzimidazole derivates, a novel class of substances with chemotherapeutic activity. Arzneimittelforschung 1974;24:1927–33.
- Fairley TA, Tidwell RR, Donkor I, Naiman NA, Ohemeng KA, Lombardy RJ, Bentley JA, Cory M. Structure, DNA minor groove binding, and base pair specificity of alkyl- and aryl-linked bis(amidinobenzimidazoles) and bis(amidinoindoles). J Med Chem 1993;36:1746–53.
- Streltsov SA, Gromyko AV, Oleinikov VA, Zhuze AL. The Hoechst 33258 covalent dimer covers a total turn of the double-stranded DNA. J Biomol Struct Dyn 2006;24:285–302.
- Pjura PE, Grzeskowiak K, Dickerson RE. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol 1987;197:257–71.
- Streltsov SA, Zhuze AL. Hoechst 33258–poly(dG-dC).poly(dG-dC) complexes of three types. J Biomol Struct Dyn 2008;26:99–114.
- Subach OM, Baskunov VB, Darii MV, Maltseva DV, Alexandrov DA, Kirsanova OV, Kolbanovskiy A, Kolbanovskiy M, Johnson F, Bonala R, Geacintov NE, Gromova ES. Impact of benzo[a]pyrene-2′-deoxyguanosine lesions on methylation of DNA by SssI and HhaI DNA methyltransferases. Biochemistry 2006;45:6142–59.
- Subach OM, Maltseva DV, Shastry A, Kolbanovskiy A, Klimasauskas S, Geacintov NE, Gromova ES. The stereochemistry of benzo[a]pyrene-2′-deoxyguanosine adducts affects DNA methylation by SssI and HhaI DNA methyltransferases. FEBS J 2007;274:2121–34.
- Chen AY, Yu C, Gatto B, Liu LF. DNA minor groove-binding ligands: a different class of mammalian DNA topoisomerase I inhibitors. Proc Natl Acad Sci USA 1993;90:8131–5.
- Link A, Tempel K. Inhibition of O6-alkylguanine-DNA alkyltransferase and DNase I activities in vitro by some alkylating substances and antineoplastic agents. J Cancer Res Clin Oncol 1991;117:549–55.
- Selby CP, Sancar A. Noncovalent drug-DNA binding interactions that inhibit and stimulate (A)BC excinuclease. Biochemistry 1991;30:3841–9.
- Suzuki T, Tanaka R, Hamada S, Nakagawa H, Miyata N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg Med Chem Lett 2010;20:1124–7.
- Popov KV, Egorova EI, Ivanov AA, Gromyko AV, Zhuze AL, Bolsheva NL, Yurkevitch OYu, Muravenko OV, Zelenin AV. Dimeric bisbenzimidazole Hoechst 33258-related dyes as novel AT-specific DNA-binding fluorochromes for human and plant cytogenetics. Biochemistry (Mosc) Ser A: Membr Cell Biol 2008;2:203–9.