Abstract
Retinoic acid is regarded as the retinol metabolite that controls proliferation and differentiation of epithelial cells. In the present study, we investigated the potential role of xanthine dehydrogenase (XDH) in retinoic acid biosynthesis in human thyroid glandular cells (HTGC). In particular, we observed that cellular retinoids binding proteins (CRBPs) are also implicated in the biosynthetic pathway leading to retinoic acid formation in primary cultures of HTGC, as we have already reported for human mammary epithelial cells (HMEC). After partial protein purification, the enzyme responsible for retinoic acid biosynthesis was identified and quantified as XDH by immunoassay, by its ability to oxidize xanthine to uric acid and its sensitivity to the inhibitory effect of oxypurinol. The evidence of XDH-driven formation of retinoic acid in HTGC cultures further corroborates the potential role of XDH in retinoic acid biosynthesis in the epithelia.
Introduction
Retinoids fulfil a myriad of functional roles in many important events including embryonic development, reproduction, differentiation, apoptosis, as well as photo-isomerisation in the vision processCitation1–3 and are vital for the maintenance of various epithelia and the immune system. In particular, all-trans- (atRA) and 9-cis-retinoic acid (9-cisRA) are considered the active metabolites of retinol, being implicated in a variety of biological functionsCitation4 and having preventive or curative potential in some pathological conditionsCitation5–8. Retinoic acid homeostasis is controlled by a complex metabolic pathway, consisting of multiple steps and different enzymesCitation9,Citation10. As previously reported by many authors, retinol (ROL) dehydrogenation into retinaldehyde (RAL) is catalyzed by retinol dehydrogenases (RDH) that belongs to the short-chain dehydrogenase/reductase gene family and is responsible for initiation of RA biosynthesis. The dehydrogenation of all-trans-retinaldehyde (atRAL) is catalyzed by retinal dehydrogenases (RALDH), which belongs to the Aldh gene family and produces atRA. Both the reactions are catalyzed by multiple RDH isozymes, including RDH2, RDH10, and DHRS9. Knock-out or inadequate expression of individual RDH produces a specific phenotype associated with impaired retinoid function, including enhanced adiposity (mRdh1), defects in head and body development (RDH10), or increased tumorigenesis (DHRS9)Citation11–13. Three retinaldehyde dehydrogenases (RALDH(s)) have also been associated with generation of atRA, by modifying adiposity (RALDH1)Citation14 or supporting embryonic development (Raldh -2 and -3)Citation15,Citation16. The same role of RDH(s) and RALDH(s) isozymes could be ascribed to both xanthine oxidase (XO) (E.C. 1.17.3.2.)Citation17,Citation18, that oxidizes all-trans and 9-cis-isomers of ROL and RAL, and to xanthine dehydrogenase (XDH) (E.C. 1.17.1.4.), that can directly oxidize ROL to RACitation19.
It is now recognized that retinoid binding proteins participate in RA biosynthesis and there is compelling evidence that retinoids bound to cellular retinoid binding proteins (CRBPs) are better substrates for the enzyme catalysis with respect to free, unbound retinoidsCitation19,Citation20. In particular, Ottonello et al.Citation20 revealed the presence of a cytosolic enzyme system in calf liver that uses CRBP-bound retinol for its direct oxidation to retinoic acid.
The role of XDH in the retinoic acid biosynthesis and the implication of CRBP(s) in this process have been described by us in epithelial tissues characterized by a functional dynamism as the breast tissueCitation19. The under expression of CRBP(s)Citation21–23 and/or XDHCitation23 in human breast cancer has been considered to be responsible for RA-deficient biosynthesis. Based on the evidence that XDH, CRBP, and CRABP are expressed in the human thyroid glandular cells and that these cells lack the expression of RDH1, RDHE2, RDH10 or DHRS9Citation24, in the present study we evaluated whether XDH, cooperated by CRBP(s), is able to catalyze the direct oxidation of ROL to RA in human thyrocytes.
Experimental procedures
Chemicals
All chemicals were from Sigma-Aldrich (Milan, Italy) except NaCl, sucrose, hydroxylamine-HCl, ammonium and sodium acetate and potassium phosphate that were from Merck Group (Milan, Italy). HPLC-grade acetonitrile, ethanol, 1,4-dioxane, hexane and dichloromethane were from J.T. Baker. Centriprep, Centricon (YM-50 and YM-10 membranes) and Millex-FG13 filter (0.22 mm pore size), were provided by Millipore. [11,12-3H]-trans-retinol ([3H]-atROL) and [11,12-3H]-trans-retinoic acid ([3H]-atRA) were from NEN Life Science Research Products, Perkin Elmer. Scintillation cocktails were obtained from Canberra Packard.
Chromatography and immunoassay equipment
For chromatographic analysis, we used a Gilson analytical liquid chromatograph that consists of two Model 306 pumps, one 811B dynamic mixer, a Model 234 auto injector equipped with a 100 or 20 µl injection loop, a 122 fluorometric detector, a Waters 2487 dual λ absorbance detector, a radiomatic FLO-ONE beta 500 TR Series Flow Scintillation Analyzer (Packard Canberra Company) and Unipoint LC System Software (v. 3.3) (Gilson Italia, Milan, Italy) and a Flo-One/beta F1B IC program (Radiomatic, Tampa, FL) for acquisition and elaboration of data. The Automated Micro plate Reader Lambda E for immunoassay was obtained from Bio-Tek Instruments – Coordination Center Europe (France).
Primary culture of human thyroid glandular cells
Normal thyroid tissue was obtained from the contra lateral lobe of thyroid glands surgically removed for a euthyroid nodule (n = 14; age 49 ± 8) in accordance with the ethical standards of the institutional committee responsible for human experimentation.
Normal thyroid tissues were digested for 2 h with collagenase (1.5 mg/mL; Life Technologies) and hyaluronidase (20 μg/mL; Sigma Chemical Co.) in DMEM as described previouslyCitation25. To obtain normal thyroid cells, digest was cultured in DMEM containing 10% heat-inactivated foetal bovine serum (FBS), antibiotics, and l-glutamine in adherent conditions at 37 °C in a humidified atmosphere of 5% CO2 for 12 h. Fibroblasts were depleted exposing cell cultures to trypsin/EDTA for 2 min. Then, thyroid cells were cultured in standard DMEM as previously reported and after 24 h they were detached with trypsin + EDTA and collected.
Purification of xanthine dehydrogenase and CRBP(s) from thyroid glandular cells
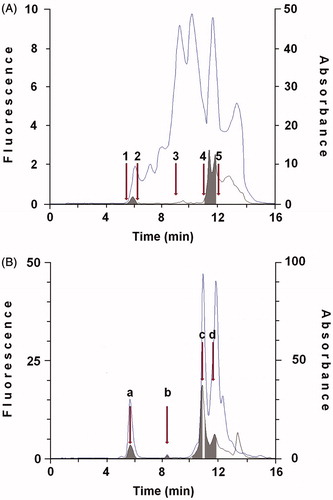
Homogenate was obtained through cavitation of a cell suspension (12 × 106 cells in 1 mL of 0.25 M sucrose, 1 mM glutathione (GSH), 20 mM Tris-HCl pH 7.0, 0.4 mM EDTA) in a pre-cooled cell disruption bomb (Parr Instrument Company) at a nitrogen pressure of 600 psi for 10 min. Homogenate was then centrifuged at 110 000 × g for 60 min in a L8-M Beckman ultracentrifuge and 1 mL of supernatant corresponding to about 10 × 106 cells were gel filtered on Shodex KW 804 column (Waters) injecting cytosol aliquots of 100 µl to gain separation of XDH and CRBP(s). Protein content of cytosol was determined using the Sigma 690-A kit. Proteins eluted with 50 mM Tris-HCl pH 7.4, at a flow rate of 1 mL/min, were monitored by measuring their fluorescence (e.g. 350 nm/em. 480 nm) and absorbance at 280 nm and at 350 nm to identify XDH and CRBP(s). A kit of protein markers with mass in the range of 12.4 and 200 kDa and dextran blue (2000 kDa) were used for column calibration. Fractions were collected, pooled and concentrated to a third of their initial volume (about 300 µL) with Centriprep YM-50 membrane (Fraction I) and Centripep YM-10 membrane (Fraction II). Concentrated Fraction I aliquots were gel filtered again on Shodex KW 804 column and eluates corresponding to XDH fraction (see peak a in ) were collected, pooled and concentrated to a fifth of their volume (about 600 µL) with Centripep YM-50 membrane. After addition of 1 mM GSH, Fraction I was flushed with N2 and gel filtered on Shodex KW 804 to perform a further purification of the enzyme preparation. The column eluates corresponding to Fraction I were pooled, re-flushed with N2 and saved at −80 °C. Further purification and subsequent analysis of fraction containing CRBP(s) (Fraction II) were performed as reported elsewhereCitation19.
Figure 1. Purification of protein showing retinol dehydrogenase activity and cellular retinoid binding proteins from cytosol of human thyroid glandular cells. A: 100 μL cytosol (1.0 × 106 cells) were gel filtered on a KW 804 column and eluted at 1 mL min−1 with 50 mM Tris HCl pH 7.4 containing 1 mM glutathione. Elution peaks were monitored by absorbance at 280 nm (top trace) and fluorescence (Ex 350 nm/Em 470 nm − bottom trace). Iterative analyses were carried out and fraction peaks were collected from the area containing XDH (Fraction I; RT 5.94 ± 0.3) or cellular retinoid binding proteins (Fraction II; RT 11.7 ± 0.3) (grey peaks). Positions of molecular weight markers were: (1) blue dextran (2.000 kDa); (2) β-amylase (200 kDa); (3) bovine serum albumin (66 kDa); (4) bovine erythrocytes carbonic anhydrase (29.3 kDa); (5) horse heart cytochrome C (12.4 kDa). B: 100 μL of concentrated Fraction I were gel filtered again on KW 804 column and several peaks were obtained. Iterative analyses were carried out and peaks containing XDH were collected and concentrated (Peak a; RT 5.94 ± 0.3). The elution profile shows a first peak (a), relative to the purified XDH, and peaks identifiable as lytic products of XDH (b, c, d) and CRBP(s) (see Purification and identification of xanthine dehydrogenase and CRBP(s) in Results section).
Identification of xanthine dehydrogenase by Western blot analysis
HTGC cytosol (input, 1% of the cytosol) was analyzed on an 8% SDS-polyacrylamide gel with a broad range pre-stained standard proteins (Bio-Rad, Segrate, Milano, Italia). Gel was stained with Ponceau S and blotted onto nitrocellulose membrane. In the same run, buttermilk XO (Sigma X-4500, grade III), pre-stained standard proteins and purified XDH (IB, Western blotting; input, 1.7% of the Fraction I) were directly blotted onto nitrocellulose membrane, blocked for 1 h with non-fat dry milk in TBS containing 0.05% Tween 20 and subsequently incubated for 3 h with a primary antibody raised against XO (mouse IgM; United States Biological, DBA Italia). After washing, the blot was incubated for 1 h with horseradish peroxidase-conjugated anti-mouse Ab (Amersham) and visualized using an enhanced chemiluminescence detection system (SuperSignal West Dura Extended duration Substrate; Pierce Chemical Co., Rockford, IL).
Quantification of xanthine dehydrogenase by immunoassay
The XDH content in cytosol and purified preparations from HTGC cells was determined using mono- and poly-clonal antibodies by a sandwich ELISA immunoassay, as previously reportedCitation19. Briefly, a polystyrene flat bottom well plate (IWAKI brand SciTech Division, Asahi Techno Glass, Japan) coated with IgM mouse anti-human XO (Mab) (United States Biological; Catalog No X0980-05X, DBA Italia) was incubated overnight at 4 °C and then rinsed with 50 mM phosphate buffered saline (pH 8.0) containing 0.05% Tween 20 (PBS/T). The remaining protein binding sites were blocked with 1% BSA in PBS/T incubating for 1 h at 4 °C. Antigen aliquots (50–100 µL) were then added and incubated for 2 h at 4 °C. Unbound antigen by was removed washing 4 times in PBS/T. After washing, a rabbit polyclonal anti-bovine XO/AO (United States Biological; Catalog No X0980-20, DBA Italia), conjugated with horseradish peroxidase, was added at dilution 1:20 000 and the plate incubated for 2 h at 4 °C. Unbound anti XO/AO antibody was removed by washing four times in PBS/T and the peroxidase substrate was added. After 30-min incubation at room temperature, the micro plate was read at 450 nm. A standard curve of XO was constructed as previously reportedCitation19, in the concentration range of 80–640 ng.
Homogenate and cytosol assays with free ROL or RAL
Cell homogenate or cytosol (100 µL) was incubated with 25 µL of 2 mM NAD+, 25 µL of 2 µM atROL, or atRAL in 0.12 mM Tween 80 and 100 µL of 50 mM Tris-HCl, pH 7.4, in a final volume of 250 µL. After 10-min incubation at 37 °C, the mixture was acidified by the addition of 20 µL of 0.5 M acetic acid and retinoids extracted by the addition of 3 mL of hexane, with 1 min shacking in the dark. The hexane phase was dried under nitrogen and stored at −20 °C. One third of the sample was analyzed as previously reportedCitation26.
atROL-CRBP(s) mixture preparation
50 µL of 2 µM atROL solution in 0.12 mM Tween 80–50 mM Tris-HCl (pH 7.4) were combined with 200 µL of cellular retinoid binding proteins fraction and kept at 22 °C a few minutes before the assay.
XDH assay with free and bound retinol
XDH preparation (200 µL) was incubated with 50 µL of 2 mM NAD+ and 50 µL of 2 µM atROL in 0.12 mM Tween 80, or 250 µL of atROL-CRBP(s) mixture, in a final volume of 500 µL. After 10-min incubation at 37 °C, the sample was treated as reported above.
XDH assay with purines
Partially purified XDH (200 µL) was incubated in a medium containing 50 mM Tris-HCl pH 7.4, 400 µM NAD+ and 20 µM xanthine in 0.005 N NaOH, and solubilised with an ultrasonic bath in a final volume of 500 µL. After 10-min incubation at 37 °C, the mixture was diluted 1:1 (v/v) with cold 47 mM potassium phosphate buffer (pH 4.65) and filtered through a Millex-FG13 membrane. 100 µL aliquots of samples were analyzed as described elsewhereCitation27. Chromatographic peaks of xanthine and uric acid were detected at 264 nm and 295 nm, respectively. For the inhibition study with purine analogs, 0.5–10 µM oxypurinol in 10 µL of 0.005 N NaOH were added to the incubation mixture prior to the addition of the enzyme protein.
Retinol dehydrogenase activity assay after XDH immune-precipitation
60 µL (3 µg) of a solution (1:2000) of rabbit anti-bovine XO/AO polyclonal antibodies were added to 800 µL of XDH preparation, incubated for 2 h at 4 °C, centrifuged at 20 000 rpm for 10 min, and finally filtered through Millex-FG13 membranes. 200 µL aliquots of the XDH preparation were then incubated with an atROL-CRBP(s) mixture as reported above. The same preparation was assayed with xanthine.
RAL oxime determination
XDH (Fraction I) was assayed with 500 nM free atROL in 50 mM Tris-HCL pH 7.4, 50 µL of 2 mM NAD+ in a medium volume of 500 µL. After 10-min incubation at 37 °C, the reaction was stopped using a 500 µM hydroxylamine-HCl solution buffered at pH 6.7 with sodium bicarbonateCitation28. After dilution of samples to 1 mL with water, retinoids were extracted through the addition of two volumes of a methanol/dichloromethane mixture (1:1 v/v), vortexed for 1 min, allowed to stand for 5 min, and mixed for one further min. Then, 0.8 mL of the lower dichloromethane phase was withdrawn and dried under N2. The dry residue was dissolved in 4% dioxane in hexane and analyzed on a µPorasil column (Waters) by eluting with the same 4% dioxane in hexaneCitation28.
Statistical analysis
Statistical significance of the data was evaluated using Student’s t-testCitation29 and probability values below 0.05 (p < 0.05) were considered significant. Results are expressed as mean ± SD.
Results
Purification and identification of xanthine dehydrogenase and CRBP(s) from HTGC cells
In order to determine whether human thyroid glandular cells express the catalytic system responsible for retinoic acid biosynthesis in human mammary epithelial cells, we purified from cell cytosol the enzyme protein catalyzing the ROL oxidation by gel permeation chromatography on Shodex KW 804 column. From the same source, we also purified a fraction containing specific cellular retinoid binding proteins to ascertain their potential contribution in the conversion of ROL to RA. The elution profile in shows two peaks positive for fluorometry, the first containing XDH, where proteins in the range of 200–400 kDa are present (Fraction I), the second containing the cellular retinoids binding proteins, where proteins with a mass between 14 and 20 kDa are present (Fraction II). Fraction I was gel filtered on Shodex KW 804 column to further purify XDH by removing the molecules modified by specific proteolytic cleavage and/or other proteins bound to the enzyme. The elution profile in shows a first peak, corresponding to the purified XDH (a) and three other peaks that may be identified as the MoCo-containing region (85 kDa) (b) the flavin-containing domain (40 kDa) (c) and a third peak with retention time of 11.47 where CRBP(s) and the N-terminal domain containing the two 2Fe/2S redox centres (20 kDa) are present (d)Citation30.
Since the CRBP(s) are characterized by similar mass and given that different isoforms exist, Fraction II was further purified and characterized through the use of Protein Pack I 60, as previously describedCitation19. Although HPLC with radiometric detection analysis allowed to reveal the presence of both CRBPs and CRABPs, we failed to achieve the separation of their different isoforms expressed in HTGC cells (CRBP1 or RBP1, CRBP4 or RBP7, CRABP1 or RBP5)Citation31 (). The amounts of CRBPs and CRABPs in the samples were determined by measuring the respective binding activity assayed versus the two [3H]-retinoids (). Overall, about 11.49 ng of CRBPs (7.6 pmoles) and 21.7 ng of CRABPs (14.8 pmoles) were detected in 50 µl aliquots of samples. This CRBP(s) preparation was also used to determine the oxidative activity of XDH against ROL.
Figure 2. Gel permeation chromatography of CRBP(s) bound to 3[H]-all-trans-retinol or 3[H]-all-trans -retinoic acid. In the Protein Pak I60 chromatograms, grey areas are radioactive peaks, and light areas are absorbance at 280 nm. In the chromatograms of CRBP-3[H]-all-trans-retinol (A) and CRABP-3[H]-all-trans-retinoic acid (B), radioactivity relative to the bound atROL or atRA was distributed in one big peak. Both the chromatographic profiles also show two small peaks, probably due to nonspecific binding. Results from one representative of three separate experiments are shown.
![Figure 2. Gel permeation chromatography of CRBP(s) bound to 3[H]-all-trans-retinol or 3[H]-all-trans -retinoic acid. In the Protein Pak I60 chromatograms, grey areas are radioactive peaks, and light areas are absorbance at 280 nm. In the chromatograms of CRBP-3[H]-all-trans-retinol (A) and CRABP-3[H]-all-trans-retinoic acid (B), radioactivity relative to the bound atROL or atRA was distributed in one big peak. Both the chromatographic profiles also show two small peaks, probably due to nonspecific binding. Results from one representative of three separate experiments are shown.](/cms/asset/81eab19c-0b7c-4e92-afd6-59c19bf61595/ienz_a_855928_f0002_b.jpg)
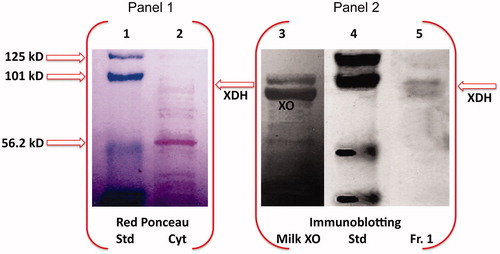
Considering that, like HTGC homogenate and cytosol, Fraction I expresses oxidative activity on hypoxanthine and xanthine, we inspected the presence of XDH/XO, through SDS-PAGE electrophoresis of HTGC cytosol and the purified Fraction I, followed by western blot analysis of XO. The immunoblot assay of Fraction I clearly displayed the immunofluorescent bands of XDH/XO (, line 5).
Figure 3. Western blot analysis of HTGC cytosol and XDH purified fraction. SDS-PAGE electropherogram relative to pre-stained standard proteins (Std), HTGC cytosol (Cyt) (input, 1% of the cytosol) stained with red Ponceau (Panel 1), is compared to Western blot of buttermilk XO (XO), Std, and the XDH purified fraction (IB, Western blotting; input, 1.7% of the Fraction I), immunostained with XO antibody, and detected by chemiluminescence (Panel 2). One representative of at least three independent experiments are shown.
Xanthine dehydrogenase quantification in cytosol of HTGC cells and purified Fraction I
XDH content was estimated in cytosol and purified preparations from HTGC cells using a mouse anti-human XO monoclonal antibody and rabbit anti-bovine XO/AO polyclonal antibodies. A significant amount of the enzyme protein was detected in cytosol aliquots obtained from 250 × 103 HTGC. As shown in , the XDH amount recovered in the purified fraction was about 80% as compared with the crude preparation.
Table 1. Content of xanthine dehydrogenase in human thyroid glandular cells.
Rates of HTGC retinoic acid biosynthesis
Crude HTGC cell preparation and Fraction I, when assayed with 200 nM atROL or atRAL, showed higher rates of activity for RAL with respect to ROL. The activity of the purified enzyme assayed with atROL-CRBP(s) was significantly greater than that observed with free atROL (). These results are in agreement with those obtained by assaying the purified XDH from HMECCitation19, for which we determined a higher Kcat value toward holo-CRBP(s) with respect to free ROL.
Table 2. Rates of retinoic acid biosynthesis rates in human thyroid glandular cells.
Xanthine dehydrogenase performs a direct catalytic conversion of all-trans-retinol to all-trans-retinoic acid
In order to clarify whether the catalytic behaviour of XDH in the oxidation of atROL to atRA in HTGC is comparable to that observed in HMEC cells, hydroxylamine was added to the purified XDH fraction (Fraction I) to evaluate the presence of atRAL oxime derivatives (see Experimental Procedures). The lack of atRAL formation is strongly indicative of the XDH ability to oxidize directly ROL to RA also in HTGC. The same analysis performed on the cytosol of HTGC showed no detectable formation of atRAL, suggesting that the conversion of atROL to atRA can be catalyzed in these cells by the combined presence of other proteins having RDH and RALDH activity.
Retinol dehydrogenase activity of xanthine dehydrogenase was inhibited by oxypurinol
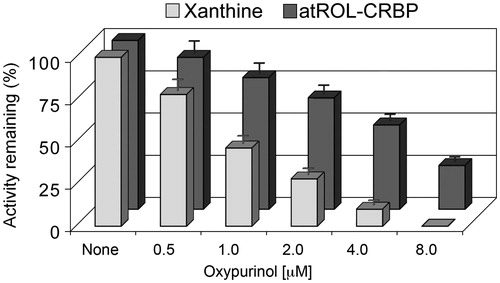
XDH was found to be expressed in confluent HTGC cells and when assayed with xanthine its activity was completely abrogated by the addition of 4 µM oxypurinol, while the inhibitory effect of oxypurinol tested on the oxidative reaction of atROL-CRBP was about 50%; the maximum inhibitory effect was observed at concentrations greater than 8 µM (). The same inhibitory effect was evidenced on the enzyme activity when assayed with ethanol, as previously reportedCitation17.
Figure 4. Comparison between the inhibitory effects of oxypurinol on the oxidation of xanthine or on all-trans-retinol in human thyroid glandular cells. Xanthine dehydrogenase purified from HTGC cytosol was assayed with 8 μM xanthine or 200 nM atROL bound to CRBP(s) at oxypurinol concentration of 0.5–8 μM. The inhibitory effect of oxypurinol against xanthine was already distinguished at the lowest concentration (0.5 μM) (p < 0.05 versus control) and was fully expressed at 4 μM concentration (p < 0.01 versus control). The inhibitory effect observed against atROL bound to CRBP(s) was instead accomplished at a higher oxypurinol concentration (8 μM) (p < 0.01 versus control). Results from a representative of three separate experiments are shown, reported as the mean ± SEM of triplicate determinations. Statistical significance, calculated with Student’s paired t-test, refers to a comparison of sample incubated with or without oxypurinol.
Discussion
It is well known that xanthine oxidoreductase (XOR), the complex enzyme system that catalyzes the two terminal steps in purine catabolism, exists in two interconvertible forms: XDH and XO. A substantial distinction between XDH and XO is that XDH, the physiological form of the enzyme in cells, reduces NAD+ without superoxide generation, whereas XO, the modified form, uses molecular oxygen as electron acceptor and generates superoxide anion as a product. Conversion of XDH into XO can occur through oxidation of sulfhydryl residues of cysteine that decreases NAD+ dependence or as a consequence of limited proteolysisCitation30. Over the past two decades, there has been growing interest in the physiological functions of XOR system and its potential role in adipogenesis and peroxisome proliferator-activated receptor-γ activityCitation32, cyclooxygenase-2 gene expressionCitation33, catalytic conversion of nitrate and nitrite to nitric oxideCitation34, catalytic hydroxylation of a wide range of N-heterocyclic alcoholCitation17 and aldehyde substratesCitation33, and as endogenous source of ROS formation for cell signallingCitation35,Citation36. XDH activity is subject to both transcriptional and translational control by mechanisms that are modulated by hormones (including oestrogen and/or angiotensin II), cytokines and oxygen tension, eventually leading to its transition to the oxidase form, which moves outside the cellCitation37,Citation38. The study by Cheung et al.Citation32 on the characterization of XOR as a novel regulator of adipogenesis in 3T3-L1 cells is extremely interesting. XOR lies in fact downstream of C/EBPβ and upstream of PPARγ, in the cascade of factors that control adipogenesis. The authors demonstrate that XOR expression and activity are closely regulated, with a robust increase during the first 24 h after initiation of differentiation and a return to basal levels within day 3. The authors also suggest that XOR produces an extractable component that has the ability to activate PPAR-γ. In addition, other reports indicate that the adipogenic activity of the xor gene may be mediated by the XDH activity of XORCitation32. These observations have prompted us to hypothesize that the extractable componentCitation33 produced by XDH is retinoic acid, this latter being responsible for the effects on adipogenesis and activity of PPAR-γCitation39–40. There is some evidence that atRA can be proadipogenic or antiadipogenic in vivo, depending on the dose. atRA is in fact a powerful inhibitor when used at relatively high doses (0.1–10 µM) in early stages of adipogenesisCitation41, whereas it promotes adipogenesis at lower doses (1 pM to 10 nM range)Citation42. Similar results have been obtained in developing myeloid cells, where retinoids induce a 3-fold increase of C/EBPβ levels, with a resulting rise of transcription of PPARγ target genesCitation43.
The role of CRBP1, which is specifically expressed in 3T3-L1 preadipocytes, in the regulation of adipocyte differentiation is also intriguingCitation44: in this cell line CRBP1 expression is highest during the first 2 days of differentiation and decreases thereafter, indicating the importance of CRBP1 during early differentiation. In a previous work we demonstrated that XDH and CRBP1, purified from human mammary epithelial cells, create a system capable of oxidizing directly retinol to retinoic acidCitation19. We have also indicated that malignant human mammary cell lines, MCF7 and MDA-MB231, which do not express XDH and CRBP1, are unable to oxidize retinol to retinoic acid but maintain their oxidative activity toward retinaldehyde thanks to the presence of XOCitation23. Furthermore, retinoic acid induces growth arrest of MCF7 cells, which express CRABP2, through a selective regulation of the IRS-1/PI3-kinase/AKT pathwayCitation45.
Multiple evidences also support a link between thyroid gland and retinoids. Several studies have reported that both a deficiency or an excess of vitamin A affect thyroid gland volume acting on the thyroid stimulating hormone synthesis and that high vitamin A doses result in a decline of serum levels of T3 and T4 in the ratCitation46,Citation47.
XDH role as enzyme involved in retinoic acid production and, consequently, in developmental events is noteworthily supported by the fact that 5′-flanking region of the human XDH gene contains several consensus sequences in its proximal region, indicating possible binding of specific regulatory factors involved in developmental eventsCitation48. These include the putative AP-1, AP-2, ETS-1, WGATAR and ATTA/TAAT (homeobox) promoter sites. Furthermore, it has been reported that AP2 factors regulate CRABP2 expression in HMECsCitation49.
The suggestion by Xu et al.Citation48 that XDH plays a role in human development is consistent with the present evidence that this enzyme also presides over retinoic acid biosynthesis in thyroid glandular cells. Our results demonstrate that in these cells, which do not express any retinol dehydrogenase, XDH, supported by CRBPs, directly oxidizes ROL to RA without release of RAL in the medium. Another finding in support of the equivalence between retinol dehydrogenase and XDH is the complete loss of catalytic activity, toward atROL and xanthine, of the purified enzyme fraction that we observe after treatment with anti-bovine XO/AO antibodies.
Until now, in the more described pathway retinol → retinoic acid, free retinol has been considered the preferred substrate of retinol dehydrogenases (RDH1, RDH10, DRSH9), operating in the endoplasmic reticulum, producing retinaldehyde. The second irreversible step to retinoic acid is catalyzed, in the cytoplasm, by retinaldehyde dehydrogenases (RALDH1, RALDH2, RALDH3)Citation10.
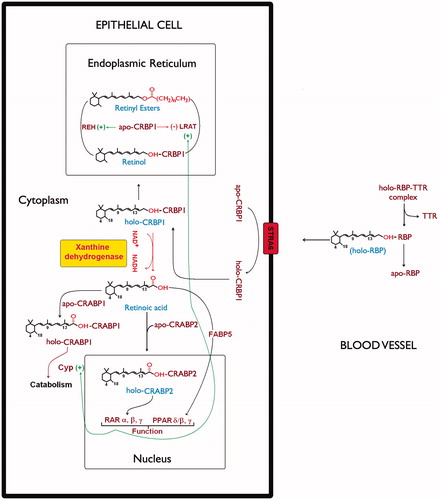
We have previously demonstrated that NAD+-dependent XDH, cooperated by CRBP and CRABP, is responsible for RA biosynthesis in HMEC, catalyzing the direct oxidation of ROL to RACitation19. Moreover, the non-release of RAL in the cytoplasm as intermediate substrate during retinol oxidation, preventing the competition with ROL for the binding to CRBP1, avoids the slow-down or the block of the biosynthetic pathway. Thus, we propose this enzyme as eligible for the direct oxidative catalysis of retinol bound to CRBP1 (holo-CRBP1) also in human thyroid glandular cells (see scheme in ).
Figure 5. Xanthine dehydrogenase involvement in retinol/retinoic acid homeostasis in epithelial cells. The scheme describes retinoid metabolism in the different cellular compartments. In the cell plasma membrane retinol exchange between apo-CRBP1 and holo-RBP is mediated by STRA6 receptor (Stimulated by Retinoic Acid 6)Citation50. In the endoplasmic reticulum, the relative ratio of holo-CRBP1 to apo-CRBP1 is influenced by retinyl ester hydrolase (REH) or by lecithin retinol acyltransferase (LRAT). On the other hand, the apo-CRBP1, through its modulating action reduces their activity by regulating the cellular levels and fluxes of retinol. In cells that do not express any RDH activity, xanthine dehydrogenase interacting with holo-CRBP1 catalyzes retinol oxidation to retinoic acid, which is then released by interaction with CRABP1 and/or CRABP2. Such a hypothetical scheme of reaction mechanism devised by xanthine dehydrogenase to oxidize retinol to retinoic acid has been described in our previous articleCitation19. Delivery of atRA to distinct nuclear receptors or for catabolism involves atRA binding proteins, CRABP1, CRABP2, and FABP5.
In conclusion, this study confirms the role of XDH in the biosynthesis of retinoic acid, in human thyroid glandular cells, and strengthens the importance of cellular retinoid binding proteins in supporting the retinol dehydrogenase activity of this system, in analogy to what we have previously observed in human mammary epithelial cells.
Declaration of interest
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of this article.
Acknowledgements
We thank Prof. Giorgio Stassi and Dr. Matilde Todaro (Cellular and Molecular Phatophysiology Laboratory, University of Palermo) for the generous gift of human thyrocytes and Dr. Monica Zerilli for the Western blot analysis.
References
- Gudas LJ. Retinoids and vertebrate development. J Biol Chem 1994;269:15399–402
- Dolle P. Developmental expression of retinoic acid receptors (RARs). Nucl Recept Signal 2009;7:e006
- Mark M, Ghyselinck NB, Chambon P. Function of retinoic acid receptors during embryonic development. Nucl Recept Signal 2009;7:e002
- Gudas LJ, Sporn MB, Roberts AB. Cellular biology and biochemistry of the retinoids. In: Sporn MB, Roberts AB, Goodman DS, eds. The retinoids: biology, chemistry, and medicine. New York (NY): Raven Press, Ltd; 1994:443–520
- Lotan R. Effect of vitamin A and its analogues (retinoids) on normal and neoplastic cells. Biochim Biophys Acta 1980;605:33–91
- Orphanos CE. Oral retinoids-present status. Br J Dermatol 1980;103:473–81
- Chytil F. Retinoic acid, biochemistry, pharmacology, toxicology and therapeutic use. Pharmacol Rev 1984;36:93S–100S
- Kraemer KH, Di Giovanna JJ, Moshell AN, et al. Prevention of skin cancer in xeroderma pigmentosum with the use of oral isotretinoin. N Engl J Med 1988;318:1633–7
- Napoli JL. Retinoic acid biosynthesis and metabolism. FASEB J 1996;10:993–1001
- Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta 2012;1821:152–67
- Jette C, Peterson PW, Sandoval IT, et al. The tumor suppressor adenomatous polyposis coli and caudal related homeodomain protein regulate expression of retinol dehydrogenase L. J Biol Chem 2004;279:34397–405
- Zhang M, Hu P, Krois CR, et al. Altered vitamin A homeostasis and increased size and adiposity in the rdh1-null mouse. FASEB J 2007;21:2886–96
- Siegenthaler JA, Ashique AM, Zarbalis K, et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell 2009;139:597–609
- Ziouzenkova O, Orasanu G, Sharlach M, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med 2007;13:695–702
- Dupé V, Matt N, Garnier JM, et al. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA 2003;100:14036–41
- Halilagic A, Ribes V, Ghyselinck NB, et al. Retinoids control anterior and dorsal properties in the developing forebrain. Dev Biol 2007;303:362–75
- Taibi G, Nicotra CMA. Xanthine oxidase catalyzes the oxidation of retinol. J Enzyme Inhib Med Chem 2007;22:471–6
- Taibi G, Paganini A, Gueli MC, et al. Xanthine oxidase catalyzes the synthesis of retinoic acid. J Enzyme Inhib 2001;16:275–85
- Taibi G, Di Gaudio F, Nicotra CMA. Xanthine dehydrogenase processes retinol to retinoic acid in human mammary epithelial cells. J Enzyme Inhib Med Chem 2008;23:317–27
- Ottonello S, Scita G, Mantovani G, et al. Retinol bound to cellular retinol-binding protein is a substrate for cytosplasmic retinoic acid synthesis. J Biol Chem 1993;268:27133–42
- Kuppumbatti YS, Bleiweiss IJ, Mandeli JP, et al. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst 2000;92:475–80
- Hayden LJ, Satre MA. Alterations in cellular retinol metabolism contribute to differential retinoid responsiveness in normal human mammary epithelial cells versus breast cancer cells. Breast Cancer Res Treat 2002;72:95–105
- Taibi G, Carruba G, Cocciadiferro L, et al. Low Levels of both xanthine dehydrogenase and cellular retinol binding protein are responsible for retinoic acid deficiency in malignant human mammary epithelial cells. Steroid Enzym Cancer: Ann NY Acad Sci 2009;1155:268–72
- Human Protein Atlas, by the Knut & Alice Wallenberg foundation, Uppsala University. Version: 10.0. Available from: http://www.proteinatlas.org/ [last accessed 12 Sept 2012]
- Todaro M, Iovino F, Eterno V, et al. Tumorigenic and metastatic activity of human thyroid cancer stem cells. Cancer Res 2010;70:8874–85
- Frolik CA, Tavela TE, Peck GL, Sporn MB. High-pressure liquid chromatographic determination of 13-cis-retinoic acid and all-trans-retinoic acid in human plasma. Anal Biochem 1978;86:743–50
- Kock R, Delvoux B, Greiting H. A comparative study of the concentrations of hypoxanthine, xanthine, uric acid and allantoin in the peripheral blood of normals and patients with acute myocardial infarction and other ischaemic diseases. Eur J Clin Chem Clin Biochem 1993;31:303–10
- Groenendijk GWT, Jensen PAA, Bonting SL, Daemen FJM. Analysis of geometrically isomeric vitamin A compounds. Methods Enzymol 1980;67:203–20
- Snedecor G, Cochran W. Statistical methods. Ames (IA): Iowa State University Press; 1967:120–34
- Nishino T, Okamoto K, Egere BT, et al. Mammalian xanthine oxidoreductase – mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J 2008;275:3278–89
- Chen Q, Park HC, Goligorsky MS, et al. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One 2012;7:e37149,1–14
- Cheung KJ, Tzameli I, Pissios P, et al. Xanthine oxidoreductase is a regulator of adipogenesis and PPAR-γ activity. Cell Metab 2007;5:115–28
- Ohtsubo T, Rovira, II, Starost MF, et al. Xanthine oxidoreductase is an endogenous regulator of cyclooxygenase-2. Circ Res 2004;95:1118–24
- Webb AJ, Milsom AB, Rathod KS, et al. Mechanisms underlying erythrocyte and endothelial nitrite reduction to NO in hypoxia: role for xanthine oxidoreductase and eNOS. Circ Res 2008;103:957–64
- Zhang Z, Blake DR, Stevens CR, et al. A reappraisal of xanthine dehydrogenase and oxidase in hypoxic reperfusion injury: the role of NADH as an electron donor. Free Radic Res 1998;28:151–64
- Zhang R, Pinson A, Samuni A. Both hydroxylamine and nitroxide protect cardiomyocytes from oxidative stress. Free Radic Biol Med 1998;24:66–75
- Taibi G, Carruba G, Miceli V, et al. Estradiol decreases xanthine dehydrogenase enzyme activity and protein expression in non-tumorigenic and malignant human mammary epithelial cells. J Cell Biochem 2009;108:688–92
- Landmesser U, Spiekermann S, Preuss C, et al. Angiotensin II induces endothelial xanthine oxidase activation-role for endothelial dysfunction in patients with coronary disease. Artherioscler Thromb Vasc Biol 2007;27:943–8
- Yasmeen R, Jeyakumar SM, Reichert B, et al. The contribution of vitamin A to autocrine regulation of fat depots. Biochim Biophys Acta 2012;1821:190–7
- Bonet ML, Ribot J, Palou A. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim Biophys Acta 2012;1821:177–89
- Xue JC, Schwarz EJ, Chawla A, Lazar MA. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARγ. Mol Cell Biol 1996;16:1567–75
- Safonova I, Darimont C, Amri EZ, et al. Retinoids are positive effectors of adipose cell differentiation. Mol Cell Endocrinol 1994;104:201–11
- Szanto A, Nagy L. Retinoids potentiate peroxisome proliferator-activated receptor γ action in differentiation, gene expression, and lipid metabolic processes in developing myeloid cells. Mol Pharmacol 2005;67:1935–43
- Zizola CF, Frey SK, Jitngarmkusol S, et al. Cellular retinol-binding protein type I (CRBP-I) regulates adipogenesis. Mol Cell Biol 2010;30:3412–20
- del Rincòn SV, Rousseau C, Samanta R, Miller WH Jr. Retinoic acid-induced growth arrest of MCF-7 cells involves the selective regulation of the IRS-1/PI 3-kinase/AKT pathway. Oncogene 2003;22:3353–60
- Drill VA. Interrelations between thyroid function and vitamin metabolism. Physiol Rev 1943;23:355–79
- Morley JE, Melmed S, Reed A, et al. Effect of vitamin A on the hypothalamo-pituitary-thyroid axis. Am J Physiol-Cell Ph 1980;238:E174–9
- Xu P, Huecksteadt TP, Hoidal JR. Molecular cloning and characterization of the human xanthine dehydrogenase gene (XDH). Genomics 1996;34:173–80
- McPherson LA, Woodfield GW, Weigel RJ. AP2 Transcription factors regulate expression of CRABPII in hormone hesponsive breast carcinoma. J Surgical Res 2007;138:71–8
- Kawaguchi R, Zhong M, Kassai M, et al. STRA6-catalyzed vitamin A influx, efflux and, exchange. J Memb Biol 2012;245:731–45
